Abstract
ζ-Crystallin is a taxon-specific crystallin, an enzyme which has undergone direct gene recruitment as a structural component of the guinea pig lens through a Pax6-dependent mechanism. Tissue specificity arises through a combination of effects involving three sites in the lens promoter. The Pax6 site (ZPE) itself shows specificity for an isoform of Pax6 preferentially expressed in lens cells. High-level expression of the promoter requires a second site, identical to an αCE2 site or half Maf response element (MARE), adjacent to the Pax6 site. A promoter fragment containing Pax6 and MARE sites gives lens-preferred induction of a heterologous promoter. Complexes binding the MARE in lens nuclear extracts are antigenically related to Nrl, and cotransfection with Nrl elevates ζ-crystallin promoter activity in lens cells. A truncated ζ promoter containing Nrl-MARE and Pax6 sites has a high level of expression in lens cells in transgenic mice but is also active in the brain. Suppression of the promoter in the brain requires sequences between −498 and −385, and a site in this region forms specific complexes in brain extract. A three-level model for lens-specific Pax6-dependent expression and gene recruitment is suggested: (i) binding of a specific isoform of Pax6; (ii) augmentation of expression through binding of Nrl or a related factor; and (iii) suppression of promoter activity in the central nervous system by an upstream negative element in the brain but not in the lens.
Guinea pig (Cavia porcellus) ζ-crystallin (ζ) is a quinone reductase (49), which, like several other enzymes in different vertebrate lineages, has undergone gene recruitment (4, 63, 64) to serve an additional structural function in the lens (25, 37). In addition to being present in the guinea pig, ζ is present at crystallin levels in some related species including rock cavy (Kerodon rupestris) and degu (Octodon degus) but is not present as a crystallin in other hystricomorph rodents, such as the coypu (Myocastor coypu), or in other rodents (37). The relatively restricted distribution of this taxon-specific crystallin suggests that its gene recruitment occurred fairly recently in rodent evolution. An interesting example of what appears to be parallel, independent recruitment of ζ has also been observed in camelids (11, 15).
From an evolutionary point of view, the molecular mechanism of such a gene recruitment presents some interesting questions. The final product of recruitment, the high-level expression of a protein in the lens, may have real selective benefits, modifying the properties of the lens to enhance its function in a particular environment (64). However, if several steps are required to achieve this goal, each would need to provide some selectable advantage to the organism in order to be retained before full recruitment was achieved. To gain some insight into this process and into tissue-specific gene expression in the lens in general, we have been examining the expression of guinea pig ζ in the lens.
Comparisons of gene sequences from guinea pigs, humans, and mice and functional analyses of the guinea pig promoter show that the gene recruitment of ζ occurred through acquisition of a lens-specific promoter in what would otherwise have been the first intron of the enzyme gene, while nonlens expression remained under the control of an upstream housekeeping promoter (14, 38). There is no similarity between guinea pig and mouse ζ genes in the region of the guinea pig lens promoter.
This separation of functions between two promoters makes the ζ lens promoter an attractive target for examination of the mechanisms of recruitment and lens-specific expression. The ζ lens promoter is highly tissue specific, without a requirement for remote enhancers (38). It functions in transgenic mice and in mouse and rabbit lens-derived cells (38), which shows that the recruitment is a cis process of promoter modification making use of evolutionarily conserved common transcription factors rather than a species-specific modification of the transcription machinery of the guinea pig lens.
Previously, we defined the lens-specific promoter (38) and then showed that ζ is a target gene for the key eye development factor Pax6 (3, 19, 39, 52). Pax6 has also been implicated in the lens-specific expression of several other crystallin genes (3), although it may not be essential for all of these genes and some of the binding sites identified are in promoter regions previously thought not to be important for gene expression in the lens (53). In the case of ζ however, Pax6 is essential for function. The ζ promoter contains an element (ζ-protected element [ZPE]) which is identically protected by nuclear protein extracts from both mouse and rabbit lens-derived cells and is differently protected by extracts from fibroblasts (38). The ZPE has a maximum size of 50 bp (−202 to −152) on the upper DNA strand and 35 bp on the lower strand. In electrophoretic mobility shift assays (EMSA), the ZPE forms two distinct complexes (I and II). Extracts from nonlens sources unable to support ζ lens promoter expression produce only complex I, while mouse lens extract produces only complex II (52). Extracts of lens-derived cultured cells, N/N1003A (50) and αTN4-1 (66), and of brain produce both complexes, although complex II often predominates in lens-derived cells (52). Different antisera to Pax6 specifically abolish the formation of complex II without affecting complex I, and the recombinant Pax6 paired domain (PD) can bind the ZPE (52). Mutation within the ZPE abolishes both complex II formation and Pax6 binding and causes complete loss of promoter activity in both transient transfections and transgenic mice (52). These results show that Pax6 is essential for tissue-specific expression of the ζ lens promoter. Furthermore, in addition to its important role very early in eye embryogenesis in both vertebrates and invertebrates (19, 20, 22, 39, 48, 59, 62), we found that Pax6 has continuing expression in adult lens and in cultured cells able to support ζ expression (52).
However, Pax6 is not lens or eye specific; it is also expressed in various parts of the central nervous system and even in the pancreas (57, 60, 62), and Pax6 binding sites have been found in noncrystallin genes (2, 24). Clearly, tissue-specific gene expression dependent on Pax6 requires fine-tuning. Consequently, the process of crystallin gene recruitment must have been multistep. Here we describe a three-part mechanism for lens-specific gene expression in a taxon-specific crystallin. Although reconstruction of the actual evolutionary path taken in this gene may be impossible, these results show, at least in principle, how each stage could have occurred as discrete steps leading toward full gene recruitment of an enzyme as a major structural component of a mammalian eye lens.
MATERIALS AND METHODS
Brain and cultured lens cell nuclear extracts.
Brain tissue was obtained from 4-week-old mice, and nuclear extract was prepared by the procedure of Sierra (55). A 1-g portion of tissue was homogenized in 10 ml of homogenization buffer (10 mM HEPES [pH 7.6], 15 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, 2 M sucrose, 10% glycerol, 1% low-fat milk). The homogenate was then added to 10 ml of homogenization buffer in SW27 tubes and ultracentrifuged at 24,000 × g for 60 min at 2°C to pellet the nuclei. The nuclei were resuspended in lysis buffer (10 mM HEPES [pH 7.6], 100 mM KCl, 0.1 mM EDTA, 10% glycerol, 3 mM MgCl2, 200 nM phenylmethylsulfonyl fluoride [PMSF], 4 ng of aprotinin per ml, 100 ng of chymostatin per ml, 4 ng of pepstatin per ml, 40 ng of bestatin per ml, 4 ng of leupeptin per ml) and homogenized in a glass homogenizer. The sample was diluted to a DNA concentration of 0.5 mg/ml, and 1/10 volume of 4 M (NH4)2SO4 was added. After incubation at 30 min on ice with occasional mixing, the lysate was centrifuged for 60 min at 35,000 × g. Nuclear protein was precipitated from the supernatant by addition of 0.3 g of (NH4)2SO4 per ml of supernatant followed by centrifugation for 20 min at 85,000 × g. The pellet was dissolved in dialysis buffer (25 mM HEPES [pH 7.6], 40 mM KCl, 0.1 mM EDTA, 20% glycerol, 1 mM dithiothreitol [DTT]) and dialyzed twice against 100 volumes for 2 h. The nuclear extract was aliquoted and stored in liquid nitrogen.
Nuclear extract from rabbit lens N/N1003A cells (50) was prepared by the method of Shapiro et al. (54). Twenty 150-mm plates of confluent monolayer cultures were washed twice with cold phosphate-buffered saline (PBS). The cells were harvested by scraping in PBS and pelleted at 300 × g for 10 min. Subsequent steps were performed at 4°C. The cells were resuspended in 5 packed-cell volumes of hypotonic buffer (10 mM HEPES [pH 7.9], 0.75 mM spermidine, 0.15 mM spermine, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 10 mM KCl), incubated for 10 min on ice, pelleted, and resuspended in 2 packed-cell volumes of hypotonic buffer. The cells were ruptured by three strokes in a glass homogenizer with a tight pestle. Then 1/10 volume of sucrose restore buffer (9 volumes of 75% sucrose; 1 volume consisting of 500 mM HEPES [pH 7.9], 7.5 mM spermidine, 1.5 mM spermine, 2 mM EDTA, 0.1 mM EGTA, 10 mM DTT, 100 mM KCl, 200 nM PMSF, 4 ng of aprotinin per ml, 100 ng of chymostatin per ml, 4 ng of pepstatin per ml, 40 ng of bestatin per ml, and 4 ng of leupeptin per ml) was added, and the cells were subjected to two strokes with a loose-pestle homogenizer. The lysate was centrifuged for 30 s at 16,000 × g and the nuclear pellet was resuspended in 3 ml of nuclear resuspension buffer (9 volumes consisting of 20 mM HEPES [pH 7.9], 0.75 mM spermidine, 0.15 mM spermine, 0.2 mM EDTA, 2 mM EGTA, 2 mM DTT, and 25% glycerol; 1 volume of saturated ammonium sulfate solution) per 106 cells. The resulting suspension was rocked gently at 4°C for 30 min and then sedimented at 150,000 × g for 45 min. The supernatant was recovered, and nuclear protein was precipitated by addition of 0.3 g of (NH4)2SO4 per ml of supernatant and centrifugation for 20 min at 85,000 × g. The pellet was dissolved in dialysis buffer and dialyzed twice against 100 volumes for 2 h. The nuclear extract was aliquoted and stored in liquid nitrogen.
Lens protein extracts.
Eight-day postnatal lenses were dissected and cleaned of adhering pigmented tissue. For rat lenses, the lens capsule with the anterior lens epithelium attached was separated from the fiber mass by microdissection with sharpened jeweler’s forceps. Tissues from 80 animals were pooled for extraction of proteins. Cell extracts were prepared from both the rat lens epithelia and fiber mass. The cells were lysed in lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% [wt/vol] Nonidet P-40, 0.1 mM EGTA, 1 mM DTT, 200 nM PMSF, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 0.4 mM sodium fluoride, 0.4 mM orthovanadate). After brief microcentrifugation to remove cellular debris, the lysate was centrifuged at 100,000 × g, and the supernatant was made 20% in glycerol. The protein extract was aliquoted and stored in liquid nitrogen.
Recombinant Pax6 proteins.
DNA binding domains of Pax6 were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli. Sequences corresponding to amino acids 1 to 130 (PD) or 1 to 270 (PD plus homeodomain [HD]) of human Pax6 were prepared by PCR amplification of a cDNA clone. The products were cloned into pGEX2T (Amrad, Melbourne, Australia) and verified by sequencing. The bacteria were transformed with the expression vector, and fusion protein synthesis was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) as specified by the supplier. Fusion proteins were isolated with glutathione-agarose beads (Sigma, St. Louis, Mo.). The proteins were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide), and aliquots were stored at −80°C. The His-tagged Pax6 PD was produced as described previously (6).
Full-length Pax6 proteins were produced with a baculovirus system. The entire coding regions of human Pax6 and Pax6(5a+) cDNAs were amplified by PCR and cloned into pVL941 (Pharmingen, San Diego, Calif.) and pBlueBacHis (Invitrogen, San Diego, Calif.). Recombinant virus was isolated as specified by the supplier. Amplified baculovirus stock (1 ml) was used to infect 1.5 × 107 to 2.0 × 107 TN5B1-4 (High Five; Invitrogen) cells maintained in Ex-Cell 400 medium (JRH Biosciences, Woodland, Calif.) in 150-mm tissue culture plates. The cells were harvested after 48 to 72 h, resuspended in 500 μl of PBS containing the protease inhibitors antipain, leupeptin, E-64, and Pefabloc (Boehringer Mannheim, Indianapolis, Ind.), and disrupted by three freeze-thaw cycles of liquid nitrogen followed by 37°C for 5 min each. The lysate was cleared by centrifugation in a tabletop microcentrifuge at 4°C, and the supernatant was stored in aliquots at −80°C. Protein expression was verified by Western blotting with rabbit polyclonal antiserum to the C-terminal domain of human Pax6.
DNase I protection (footprinting).
Pax6 binding at the ZPE region used the −323 to −57 promoter fragment, as described previously (38). For the brain-protected element (BPE) region, the −535 to −373 fragment was prepared by digestion of the −756 to +70 promoter construct (38) with Alw441, 5′-end labelled with [γ-32P]ATP, and digested with StuI, generating the −535 to −373 fragment labelled on the lower strand. DNase I footprinting was performed as described previously (10, 38). DNA binding was carried out in a 20-μl volume with 32P-labeled DNA probe (2 × 104 to 3 × 104 cpm), 1 μg of poly(dI-dC), 5 mM MgCl, 1 mM DTT, 0.5 mM EDTA, 60 mM KCl, 10% glycerol, and ranges of concentrations of protein extracts or recombinant Pax6 proteins for 15 min on ice. Recombinant Pax6 proteins from each preparation were titrated over a range of approximately 0.1 to 2.5 μg, and tissue and cell extracts were titrated over a range of 1 to 60 μg. The DNase I concentration was also titrated for each reaction. Typically, 3 μl of DNase I mixture (5 to 50 U of DNase I in 25 mM CaCl2) was added for 5 min on ice, and then 80 μl of stop solution (25 mM Tris [pH 8.0], 20 mM EDTA, 250 mM NaCl, 0.5% sodium dodecyl sulfate) was added; this was followed by extraction with phenol-chloroform and precipitation with ethanol.
Transient transfections.
Deletions and other mutations of the ζ promoter fused to the bacterial chloramphenicol acetyltransferase (CAT) reporter gene were constructed from a −323/+70.CAT construct (38). This was digested with SalI, which cleaves at position −323, and with NsiI, which cleaves at position −187. Replacement DNA fragments for mutations and deletions were synthesized with the appropriate restriction sites and ligated into the truncated promoter construct.
To test the influence of the −245 to −152 fragment on the expression of a heterologous promoter, the fragment was constructed by PCR, sequenced, and cloned upstream of the herpes simplex virus thymidine kinase (tk) promoter fused to the CAT reporter gene in the pTKCAT plasmid (42).
By using calcium phosphate coprecipitation (16), 10 μg of promoter construct plasmids and of pCMVβ (Clontech, Palo Alto, Calif.) (used for normalization) was transfected into N/N1003A or NIH 3T3 cells seeded on 10-cm Falcon dishes at a density of 3 × 106. The cells were harvested 48 h after transfection and subjected to freeze-thawing (41). The β-galactosidase activity was measured (5), and volumes of extracts equalized for β-galactosidase activity were used for the CAT assays (17). Acetylated [14C]chloramphenicol (Amersham Life Science Inc., Arlington Heights, Ill.) was separated by thin-layer chromatography (TLC), and excised radioactive spots were measured by liquid scintillation. CAT activity was expressed as the percent conversion of [14C]chloramphenicol into 14C-acetylated chloramphenicol derivatives. All reported CAT activities were averages of three independent transfection experiments.
Cotransfections were performed in a similar way, with 15 μg of the −229/+70.CAT construct, 5 μg of the Nrl expression plasmid pMT-NRL (51), and 5 μg of the pCMVβ plasmid for normalization of transfection efficiency. As a control, the pMT-NRL plasmid was replaced with 5 μg of the parental pMT3 vector with no Nrl expression.
EMSA.
EMSA was performed with recombinant Pax6 proteins, mouse brain nuclear extract, N/N1003A nuclear extract, mouse lens cell extract, and rat lens epithelial and fiber cell extract. Double-stranded DNA oligodeoxynucleotides were 5′-end labeled with [γ-32P]ATP. The binding reaction was carried out, in a volume of 20 μl, with 8 to 10 μg of protein, 0.5 ng of DNA probe, 2 μg of poly(dI-dC), and 1 μg of 1-kbp DNA size ladder in a buffer containing 60 mM KCl, 15 mM HEPES (pH 7.9), 1 mM MgCl2, 0.1 mM EDTA, 5 mM spermidine, 0.66 mM DTT, 0.5 mM PMSF, and 4% Ficoll. The reaction mixture was incubated for 30 min at 4°C and run on a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) buffer at 4°C. In the case of the brain suppressor region (see below), 100 mM KCl was used to optimize the binding reaction. For competition experiments, double-stranded DNA competitor fragments were synthesized and used at 80-fold excess (40 ng).
The presence or absence of candidate DNA binding proteins in EMSA complexes was tested with specific antisera in the EMSA. Antisera to c-Fos, c-Jun, and NF-E2 were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). Antisera to Nrl were as described previously (51). Typically, protein extracts were incubated with 1 μl of antiserum for 10 min on ice before the addition of labelled probes as described previously (52) or as recommended by the supplier.
Transgenic mice.
Transgenic mice were produced in the National Eye Institute transgenic facility as a service. Promoter constructs were derived from the previously described −756/+70.CAT construct (38) by different strategies: exonuclease III digestion (21) from the 5′ end (−498 to +70), or removal of sequences to the PvuII site (−295) or the NsiI site (−187) followed by reconstruction with double-stranded synthetic DNA fragments (−323 to +70, −229 to +70, and −206 to +70) essentially as described previously (38). The enzymes and protocols for these procedures were from Life Technologies (Gaithersburg, Md.).
Genomic DNA from founder mice was assayed for the presence of the transgene by PCR with a 5′-sense-direction ζ primer (ATGCATCATTGCTAAACCAT) and a 3′ CAT antisense primer (CGGTCTGGTTATAGGTACATTGACC) with denaturation at 94°C for 90 s followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final extension step for 5 min. Transgenes were examined for integrity and number by Southern analysis, using NsiI digestion to cut inside the transgene and BamHI digestion to cut outside the transgene. Mice harboring intact transgenes were crossed with wild-type FVB/N mice to obtain heterozygous F1 progeny.
CAT gene expression in transgenic mice.
Brain, heart, lung, liver, kidney, pancreas, spleen, intestine, muscle, and lens tissues were isolated from adult transgenic mice as described previously (38). The tissues were homogenized in 100 to 800 μl of 0.25 M Tris-HCl (pH 7.8) and incubated for 15 min at 65°C, and cellular debris was removed by brief microcentrifugation. Extracts from each tissue were analyzed for CAT activity by TLC as described above. For analysis of the −229/+70.CAT lines, which were produced later than other lines, 1-deoxy[dichloroacetyl-1-14C]chloramphenicol (Amersham) was used. This newer reagent produces a single labelled product.
RESULTS
Pax6 site.
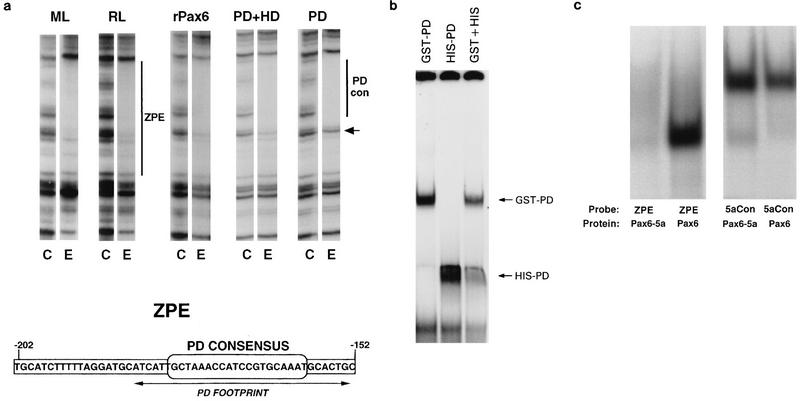
To confirm Pax6 binding and to determine how much of the ZPE protection, a maximum of 50 bp, −202 to −152 (38), is due to Pax6 alone, footprinting with recombinant Pax6 proteins was compared with that for lens-derived cell extracts. Three proteins were tested, full-length recombinant canonical human Pax6 (rPax6), a truncated Pax6 containing the DNA-binding PD and HD but lacking the PST-rich activating domain (13) (PD+HD), and a protein with a further truncation containing only the PD. For each recombinant protein, the optimal conditions for the protection assay were empirically determined for ranges of protein concentration and DNase I activity, essentially as described previously (38). For clarity, single representative lanes for each experiment are shown in Fig. 1a, in which results for rabbit and mouse lens cells are compared. All three proteins gave protection very similar to that for lens cell extracts (Fig. 1a). Footprinting on the lower strand was identical in all cases (data not shown). The longer protected region on the upper strand (52) was essentially identical for rPax6, PD+HD, and mouse and rabbit lens cell extracts. The PD alone gave a similar footprint, from −184 to −152, but did not completely protect bands at the 5′ end of the ZPE (Fig. 1a). This protected region corresponds closely to the consensus PD binding site derived in vitro (6) and to the core element essential for complex II formation, ZE1, identified previously (52).
FIG. 1.
Binding of canonical Pax6 to the ZPE. (a) Binding of lens cell extracts and recombinant Pax6 proteins to the ζ ZPE. DNase I footprints of the upper strand of the ZPE region are shown. Pairs of lanes show results for control with no added protein (C) and a representative lane for addition of lens cell nuclear extract or recombinant proteins (E). For each experiment, DNase I and protein concentrations were titrated, but for clarity only a single representative lane for each protein is shown. In each case, two- to fivefold increases in the DNase I concentration or twofold increases in the protein concentration gave no difference in protection. Extracts and proteins were as follows: ML, mouse lens-derived αTN4-1 cells; RL, rabbit lens N/N1003A cells; rPax6, full-length human Pax6; PD+HD, a truncated Pax6 containing PD and HD; PD, Pax6 PD. The small arrow shows a band which is efficiently protected by full-length rPax6 but not by PD alone. Bars show the maximal extent of the ZPE (−202 to −152) and the region corresponding to the Pax6 PD consensus binding site (−184 to −152). The sequence of the ZPE region of the ζ promoter is shown below. The position of the consensus PD binding sequence is indicated. Arrows show the extent of the experimental protection due to PD alone, which overlaps the PD consensus. (b) EMSA of the ZPE (−202 to −152) with two different-size Pax6 PD fusion proteins, GST-Pax6 and His-Pax6. The combination of the two forms produced no intermediate-size shifts. (c) The ZPE (−202 to −152) binds canonical Pax6 but not Pax6-5a. Results are shown for EMSA of the labelled ZPE and for 5aCon (a different sequence derived in vitro as a Pax6-5a binding sequence [7]), using recombinant Pax6 and Pax6-5a proteins as shown.
These results show that Pax6 alone is capable of fully protecting the ZPE without major involvement by other proteins. Most of the protection is due to the binding of a PD at a site matching the in vitro consensus (6). The PST domain does not appear to contribute significantly to the protection, but it is possible that sequences including the HD are required for full protection.
An experiment was also performed to look for any evidence that multiple Pax6 molecules bind at this site. Although some DNA binding motifs (such as leucine zippers) may form tight dimers incapable of exchange, the X-ray structure of the PD shows no indication of such tight-dimer formation (65). Therefore, two size forms of the recombinant PD, a GST fusion and a smaller His-tag fusion (6), were used separately and in combination in EMSA of the ZPE. Distinct complexes were formed, with no evidence in the mixing experiment for intermediate sizes resulting from heterodimers or other multimers (Fig. 1b).
Alternative forms of Pax6.
Pax6 is subject to alternative splicing (1, 7, 20, 27). One significant variant, Pax6-5a, contains an alternative exon (called 5a in mammals) which alters the sequence and binding specificity of the PD (7). The ZPE sequence closely matches the consensus for binding by the canonical form of Pax6, which lacks the alternative exon 5a (6, 52). As shown in Fig. 1a and b, Pax6 proteins and domains corresponding to the canonical form will bind to the ZPE. To see whether Pax6-5a can also bind the ZPE, full-length recombinant Pax6 proteins corresponding to both forms were tested in EMSA with the ZPE sequence. As a control a different binding sequence, 5aCon (7), representing an in vitro binding site for Pax6-5a, was used (Fig. 1c). While both proteins bound 5aCon efficiently, the ZPE sequence bound only canonical Pax6.
MARE site.
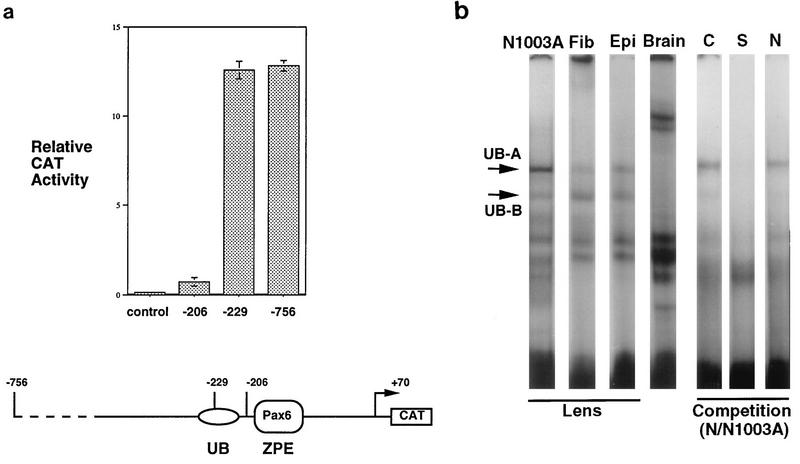
A promoter construct truncated immediately 5′ to the ZPE/Pax6 (−206 to +70) was tested for activity in lens-derived N/N1003A cells. The promoter was active but only at about fivefold the activity of the promoterless plasmid control (Fig. 2a). Earlier DNase I footprinting studies (38) showed no obvious protection upstream of the ZPE by lens cell extracts, although nonlens extract (fibroblast) protected a region from −245 to −210, which was designated the upstream box (UB). This suggested the presence of a nonlens, negative element at this site but did not immediately suggest the presence of a positive element in the lens. However, when sequences upstream of the Pax6 site (−229 to +70) were included in ζ promoter constructs, the reporter activity increased almost 20-fold over that of the −206 to +70 construct, reaching a level similar to that of the “full-length” −756 to +70 construct (Fig. 2a).
FIG. 2.
Positive element for lens expression 5′ to the ZPE. (a) The −206 to +70 promoter is active in lens cells but is much less active than the −229 to +70 promoter. The CAT reporter activities of ζ lens promoter constructs transiently transfected in N/N1003A lens cells are shown. Constructs: control, pSV0ATCAT parent plasmid with no promoter; −206, −206/+70.CAT; −229, −229/+70.CAT; −756, −756/+70.CAT. The relative CAT activity, normalized for equal β-galactosidase activity to control for transfection efficiency, is shown. A schematic of the promoter is shown below. (b) EMSA of the −229 to −188 probe with nuclear extracts of rabbit N/N1003A cells (N1003A), extracts of rat lens fiber (Fib) and epithelial (Epi) cells, and nuclear extract of mouse brain (Brain). UB-A and UB-B complexes are indicated by arrows. C, S, and N show results of competition experiments: C, control with no additional competitor; S, self (−229 to −188) competition; N, nonself (−202 to −152) competition.
This result demonstrated that a binding site for a positive factor for the lens is present in the UB region, although it was not revealed by footprinting. As an alternative approach, EMSA of the −229 to −188 fragment was used. This gave similar complexes with nuclear extracts of N/N1003A rabbit lens-derived cells and protein extracts of rat lens fiber cells and rat lens epithelial cells (Fig. 2b). N/N1003A extract, in particular, gave a prominent complex, which was designated UB-A. Other minor complexes were also apparent, including one designated UB-B, which appeared to be more variable in abundance and apparent stability. In contrast, mouse brain extract gave different complexes and lacked UB-A and UB-B. Self competition eliminated the formation of the UB-A and UB-B complexes in N/N1003A nuclear extract, while the nonself competitor (the ZPE fragment) did not efficiently compete complex formation (Fig. 2b), suggesting the presence of a sequence-specific binding factor(s).
The −229 to −188 region contains a sequence, TCAGCA (−218 to −213), which is identical to that of a functional element in the chicken αA-crystallin gene, designated αCE2 (41). This sequence is identical to those of core half-sites of Maf response elements (MARE) of several genes (23, 33). The MARE has been defined in various ways, usually as a dyad, but recent results obtained with the interleukin-4 gene show that c-Maf binds to a TCAGCA MARE identical that of the ζ promoter (23) and the chicken αA-crystallin promoter (41).
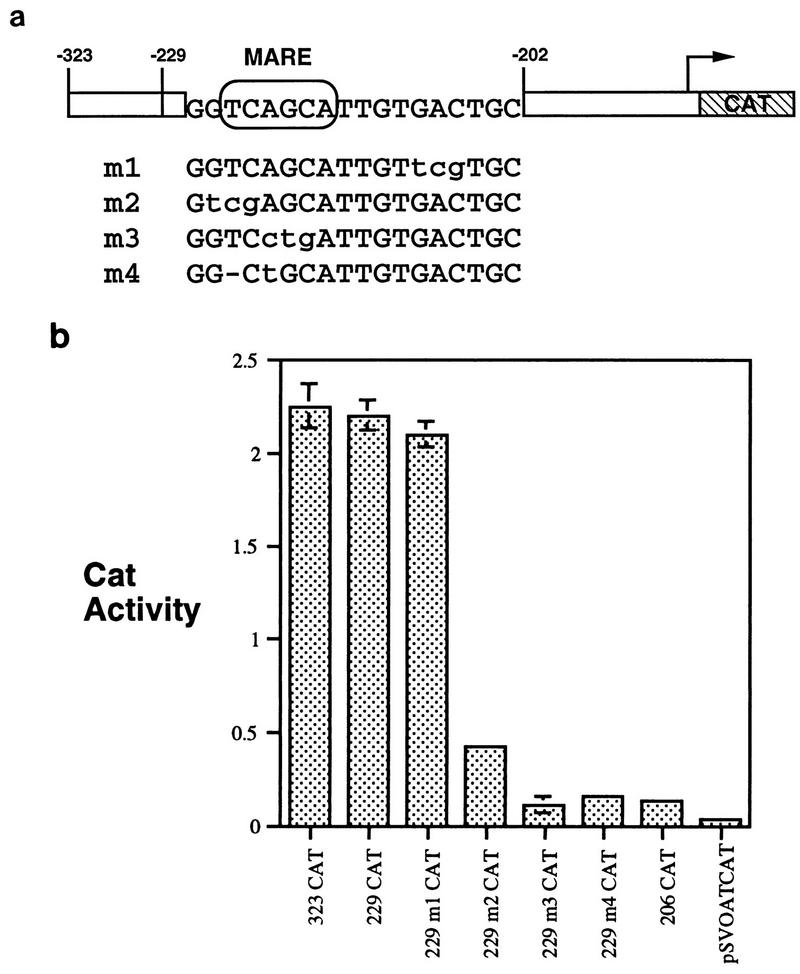
The significance of the MARE for activity of the ζ lens promoter was examined directly by incorporation of specific mutations into the −229/+70.CAT promoter construct. One mutation, m1, was outside the MARE, while others, m2, m3, and m4, were inside the MARE (Fig. 3a). (m4 incorporated a deletion arising accidentally in DNA synthesis.) The m1 mutation had no effect on promoter activity, but all three mutations in the MARE sequence (m2, m3, and m4) markedly reduced promoter activity (Fig. 3b). In particular, the m3 and m4 mutations reduced activity to the same level as that for the −206 to +70 promoter, which completely lacks the UB region.
FIG. 3.
The MARE is essential for high-level expression in lens cells. (a) Scheme for mutated sequences. Mutations are shown in the context of the −229 to +70 promoter. Mutations m2, m3, and m4 lie inside the MARE site, while m1 is outside. (b) Relative CAT activity of wild-type and mutated constructs in N/N1003A cells. Mutations m1 to m4 were incorporated in the −229 to +70 promoter as shown.
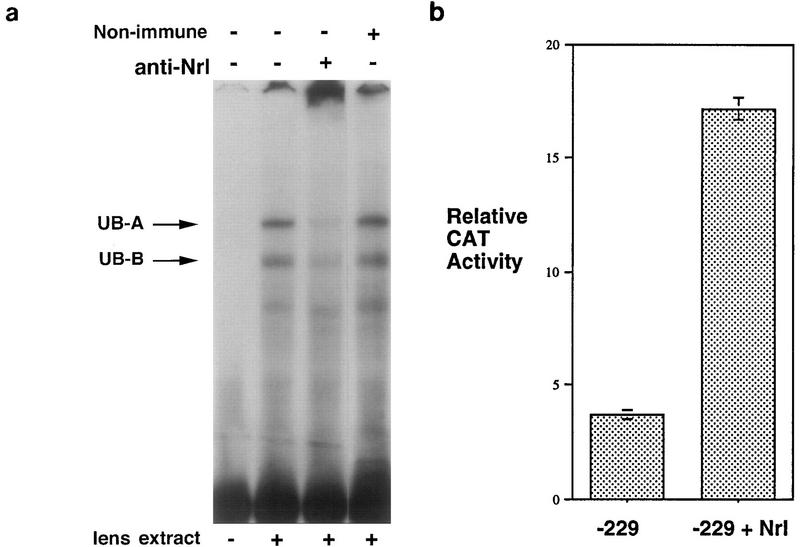
Thus, an intact MARE is essential for the function of a positive element in the ζ lens promoter. Candidate factors for participation in binding to the MARE were tested by using specific antisera in EMSA of the −229 to −188 probe with mouse lens nuclear extract (which gave a similar pattern to rabbit lens cells and rat lens extracts). The Maf family is regarded as a subset of the AP-1/CREB/ATF group, and Maf proteins may heterodimerize with c-Fos, c-Jun, and NF-E2 family proteins (26, 31–35). Antisera to the AP-1 components c-Fos and c-Jun did not affect complex formation, although they successfully abolished EMSA complexes when tested against a consensus AP-1 probe (data not shown). Similarly, antiserum to NF-E2 also failed to affect MARE complex formation (data not shown). In contrast, antiserum to Nrl (51) significantly reduced the formation of UB complexes, particularly UB-A (Fig. 4a). A possible “supershift” complex, with very low mobility in this gel system, was also apparent (Fig. 4a). This suggests that Nrl, or an antigenically related factor in lens and lens cell nuclear extracts, binds the ζ promoter upstream of the Pax6 site. Other factors may also bind this region.
FIG. 4.
Nrl is a candidate for binding the ζ lens promoter MARE. (a) Antiserum to Nrl reduces the formation of the UB-A complex in mouse lens nuclear extract. Lanes show EMSA with the −229 to −188 probe with no added antiserum, with antiserum to Nrl, and with nonimmune serum. Other antisera were also tested and had no effect on complex formation. (b) Cotransfection with the Nrl expression vector pMTNRL increases ζ lens promoter activity in N/N1003A lens cells. The relative CAT reporter activity for the −229/+70.CAT construct cotransfected either with the empty pMT3 plasmid (−229) or with the Nrl expression plasmid pMTNRL (−229 + Nrl) is shown.
To determine whether Nrl itself can affect the activity of the ζ promoter, the −229/+70.CAT construct was cotransfected into N/N1003A cells with the Nrl expression plasmid pMT-NRL or, as control, with the parent pMT3 plasmid (51) (Fig. 4b). In the lens-derived cells, addition of the Nrl expression plasmid increased the CAT reporter activity fourfold over that produced by cotransfection with pMT3, while pMT-NRL and −229/+70.CAT cotransfection of NIH 3T3 fibroblast cells, which cannot support ζ promoter activity (38), gave no reporter gene expression (data not shown). Thus, Nrl exerts a positive effect on the ζ lens promoter in a permissive (lens) background, although Nrl alone cannot activate the promoter in fibroblasts.
Nrl, or a close relative, is a good candidate for involvement in high-level expression of the Pax6-dependent ζ lens promoter. Other, so far unidentified factors may also be involved, perhaps as heterodimeric partners in the complexes formed at this site. The presence of different complexes formed in brain extract (Fig. 2b) suggests that other MARE-binding proteins, not unexpectedly, may also be able to bind in other tissues.
Lens-preferred positive element.
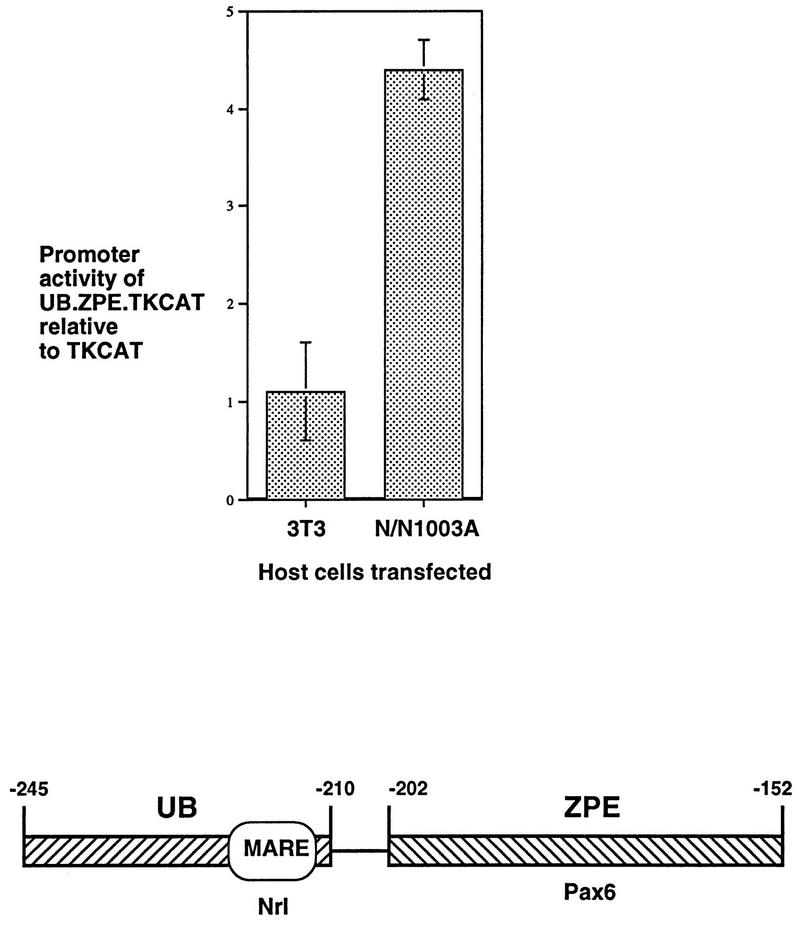
Having demonstrated the importance of the Pax6 and MARE sites acting together in the ζ promoter, a fragment of the promoter containing the complete UB and ZPE regions (covering the MARE and Pax6 sites, −245 to −152) was constructed and cloned into the pTKCAT plasmid (42) to test its effect on a heterologous promoter. The UB-ZPE-TKCAT construct was transiently transfected into N/N1003A lens cells and NIH 3T3 fibroblasts, and the results were compared with those obtained with the parent pTKCAT construct (Fig. 5). In the NIH 3T3 cells, which are not able to support expression of the ζ promoter (38), the UB-ZPE fragment produced no induction of reporter gene expression. However, in the lens-derived cells, this fragment caused a fourfold elevation in the activity of the heterologous promoter. Thus, the UB and ZPE regions together constitute a lens-preferred positive element. This result also suggests that sequences downstream of the Pax6 site (3′ to −152) in the ζ promoter are not essential for the lens-positive element.
FIG. 5.
A positive element for lens-preferred expression. Induction of a heterologous (TK) promoter by the composite UB.ZPE element, containing the MARE and Pax6 binding sites (−245 to −152). The results of transfection of pTKCAT and UB.ZPE.TKCAT into NIH 3T3 cells and N/N1003A cells are shown. The CAT activity of UB.ZPE.TKCAT relative to that of parent pTKCAT in the same experiment is shown for each cell type. The composite UB.ZPE positive element is illustrated below. Boxes show major footprinted regions: UB is footprinted in fibroblasts which do not support ζ expression, while ZPE is footprinted in lens-derived cells which do allow expression (38). The position of the MARE/Nrl binding site is shown. The ZPE is the binding site for canonical Pax6.
The −229 to +70 promoter is active in lens and brain tissue in transgenic mice.
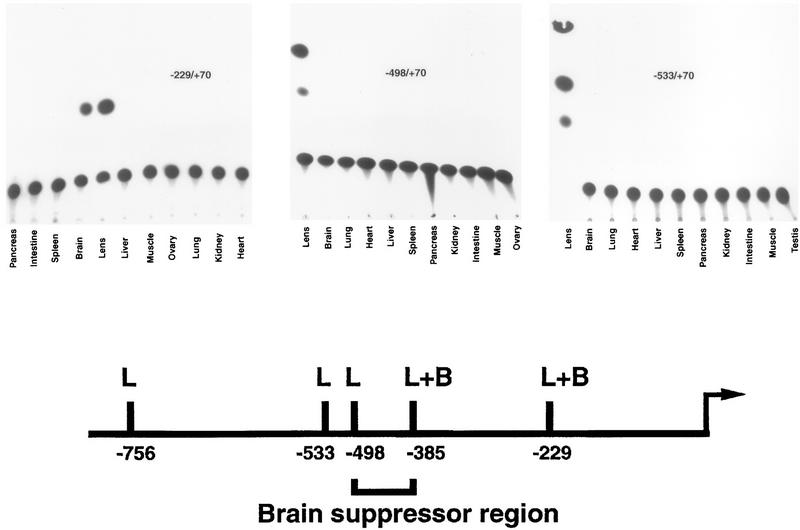
The −229/+70.CAT construct, which contains both the ZPE/Pax6 and MARE sites and which is highly active in transient transfections of lens-derived cells, was introduced into transgenic mice (Fig. 6). As with all transgenic constructs, three independent lines were examined. All three −229/+70.CAT lines gave identical results in TLC CAT analysis of tissues, with a high level of activity in lens tissue but also in brain tissue, another site of Pax6 expression (52). All the other tissues examined showed no activity. This result was reminiscent of the −385 to +70 promoter, which also showed expression in brain tissue in addition to that in lens tissue (38).
FIG. 6.
The −229 to +70 construct is expressed in lens and brain tissues in transgenic mice, while the −498 to +70 and −533 to +70 constructs are lens specific. TLC autoradiograms showing CAT activity in 50 μg of tissue extracts from representative examples of −229/+70.CAT, −498/+70.CAT, and −533/+70.CAT transgenic mice are shown. For all constructs, at least three independent lines were tested, and all gave identical results with strong expression in lens tissue and brain tissue (where appropriate), and no detectable expression in other nonlens tissues. The −229/+70.CAT lines were derived at a later time and were analyzed with 1-deoxy[dichloroacetyl-1-14C]chloramphenicol (Amersham), resulting in a single migrating spot. Note that the order of tissues is also different for −229/+70.CAT and the other constructs. Shown below is a summary of results for expression patterns in transgenic mice from previous (38) and present experiments. In a schematic of the ζ lens promoter, L indicates expression in lens tissue only while L+B indicates expression in both lens and brain tissues. Previous results showed that the −385/+70.CAT construct was expressed in lens and brain tissues in transgenic mice while the −765/+70.CAT construct was lens specific (38).
Suppression of promoter activity in brain tissue.
Previously, two other ζ promoter constructs had been tested in transgenic mice (38). These were the −756/+70.CAT construct, which was lens specific, and the −385/+70.CAT construct, which, in addition to high-level expression in lens tissue, showed some expression in brain tissue (38). To further define the basis for tissue specificity, two additional transgenic-mouse experiments were performed with −498/+70.CAT and −533/+70.CAT constructs (Fig. 6). Again, three independent lines were examined for each construct, and again, all three lines for each construct gave identical results in TLC CAT analysis of tissues. Like the −756 to +70 promoter, but in contrast to the −385 to +70 and −229 to +70 promoters, the −533 to +70 and −498 to +70 constructs were highly lens specific with no detectable expression in brain tissue. Thus, sequences between −385 and −498 are necessary to suppress the expression of the Pax6-dependent ζ lens promoter in brain tissue (Fig. 6).
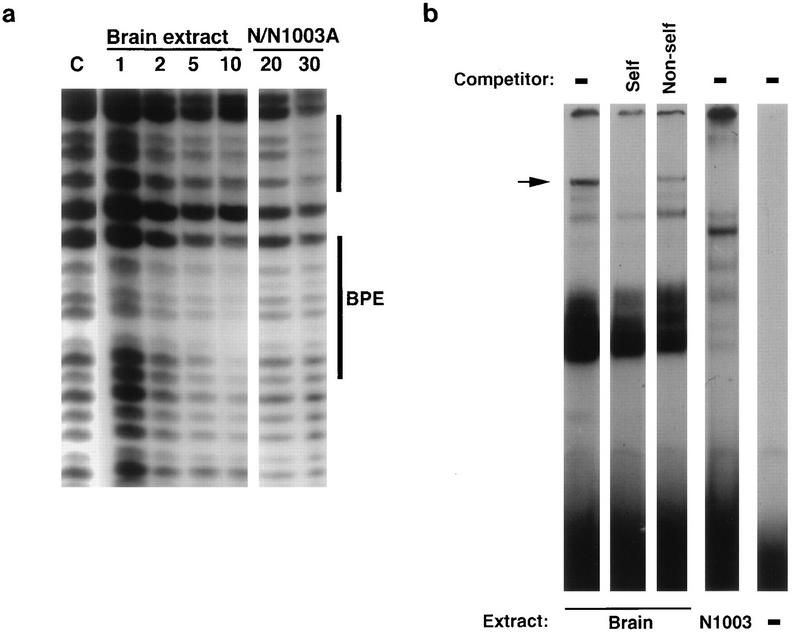
BPE.
DNase I footprinting was performed on the −535 to −373 promoter region with mouse brain and N/N1003A lens cell nuclear extracts to search for differential protection in the region required for suppression of the promoter in brain tissue (Fig. 7a). Several complex regions of protection were apparent, but only one possible region of difference could be identified between the protection produced by the two extracts; this region was an element between −411 and −401 which appeared to be protected by brain extract but not by N/N1003A extract. This was designated brain protected element (BPE). Just 5′ to the BPE is another region which appears to be similarly protected by both brain and lens extracts (Fig. 7a).
FIG. 7.
Brain-preferred complex formation in the brain suppressor region of the ζ lens promoter. (a) Mouse brain nuclear extract but not N/N1003A lens cell nuclear extract protects a site (lower bar, BPE) between −411 and −401 in a DNase I footprinting assay. The upper bar indicates a nearby region which appears to be similarly protected in both brain and lens tissue. The amounts of added protein extract (micrograms) are indicated. (b) In EMSA, the BPE region forms specific complexes with mouse brain extract. Labelled synthetic double-stranded DNA for the −418 to −394 fragment (TAAAAGCTCTGTGTTTTTTCCACCG) containing the BPE core sequence (italics) was incubated with nuclear extract derived from mouse brain and lens-derived N/N1003A cells (labelled below the panel). Competitions with self (S) and nonself (N) (−478 to −454) unlabelled fragments are shown (labelled above the panel). The dash indicates no addition of either competitor or extract. The arrow indicates a sequence-specific complex formed in brain but not lens extract.
The possibility of brain-preferred complex formation in this region was also examined by EMSA. A fragment containing the BPE (−418 to −394) produced a specific complex in brain extract (Fig. 7b) and gave different complexes with lens cell nuclear extract. The brain-preferred complex was eliminated by self-competition, while a different promoter fragment, −478 to −454, did not compete for complex formation. Taken together, these results suggest that this part of the brain suppressor region of the ζ promoter can bind a factor in brain tissue which is not present in lens tissue. As such, this constitutes a candidate region for future studies aimed at describing the mechanism of promoter suppression in brain tissue.
DISCUSSION
Our previous results (38, 52) have shown that Pax6 is essential for the lens-specific expression of the ζ lens promoter. pax6 has the characteristics of a master gene for eye development (3, 19, 39). The ζ promoter provides an opportunity to investigate the way in which a high-level factor such as Pax6 is able to influence tissue-specific expression of target genes downstream in a developmental cascade. It also provides a model for examining the multistep process of acquiring a new pattern of gene expression in molecular evolution.
A picture is now emerging of a mechanism for the Pax6-dependent tissue specificity of ζ. It seems that a single Pax6 is sufficient to occupy the ZPE (−202 to −152) and that binding depends principally on the PD, although other parts of the Pax6 protein, such as the HD, may also be involved in binding. Indeed, the 5′ end of the ZPE upstream of the minimal PD footprint contains the sequence TTTA (−194 to −191), which is similar to an HD binding consensus (12), and it is known that cooperation between PD and HD in Pax proteins can contribute to specificity in DNA recognition and target gene activation (9, 28, 43).
Like many other transcription factors, Pax6 exhibits alternative splicing, which increases its repertoire of recognition sequences (7). The ZPE of the ζ promoter shows specificity for the canonical form of Pax6 and does not bind the alternative Pax6-5a form. By itself, this provides a basis for some tissue specificity. Previously, using reverse transcription-PCR analysis of adult mouse lens, brain, and lens-derived cells, we noticed a strong preference for the canonical splice form of Pax6 mRNA in lens cells whereas in brain cells there was an approximately equal ratio of splice forms with and without the alternative exon 5a (52). Similar results have also been obtained for adult bovine eye tissue, in which the lens again shows a preferential abundance of canonical Pax6 while the iris, in contrast, shows a preference for Pax6-5a (27).
Canonical Pax6 is essential for ζ expression, but to achieve high-level expression, an adjacent element is also required. This TCAGCA sequence just upstream of the Pax6 site at −218 to −213 is identical to the MARE of the interleukin-4 gene (23) and to the αCE2 site of the chicken αA-crystallin gene (41). MARE, which are often found as palindromic dyads, are binding sites for members of the Maf family of proto-oncogene products, bZIP proteins which may heterodimerize with other leucine zipper proteins, including c-Jun, c-Fos, and NF-E2 (26, 31–35). Indeed, a lens-specific member of this family, designated L-maf, has recently been identified in chicken lens and is implicated in expression of the chicken αA-crystallin gene through the αCE2 site (45). No mammalian ortholog of L-maf has yet been reported, but another eye-preferred member of this family, Nrl (neural retina leucine zipper), has been identified in the adult human retina and in embryonic mouse lens and brain (8, 40, 58). Nrl has also been detected by reverse transcription-PCR in mature mouse lens (45a). Whether Nrl substitutes for L-maf in mammals or whether a direct mammalian ortholog exists remains to be determined.
Nrl has been implicated as a positive regulatory protein in rhodopsin gene expression (36) and is also a strong candidate for involvement in the ζ promoter. Specific antisera to Nrl (51) affect the formation of EMSA complexes in the −218 to −188 region, and coexpression of Nrl significantly increases the expression of the ζ promoter in a permissive lens cell background. In nonpermissive cells, which do not support expression of the ζ promoter and which lack Pax6, expression of Nrl has no effect, showing that it acts in concert with other factors and cannot activate the promoter alone.
Indeed, the ZPE and MARE sites combine to form the basis for a lens-preferred element. The two binding sites are so close that a direct protein-protein interaction between Pax6 and Nrl (or other factors binding at that position) is quite possible, and might constitute a lens-preferred core transcription complex. The −245 to −152 fragment containing these two binding sites is able to confer enhanced expression on a heterologous promoter in lens cells but not in fibroblasts. However, canonical Pax6 and MARE binding proteins (such as c-Maf) are also present in other tissues, particularly the brain, where similar complexes could also form. Indeed, transgenic-mouse experiments show that truncated ζ promoters, containing Pax6 and MARE sites but lacking upstream sequences, are expressed in the brain in addition to the lens.
Thus, while the ZPE-MARE region confers tissue-preferred activity on the lens promoter, fine-tuning of lens-specific expression requires another level of control. Transgenic-mouse experiments show that this is achieved through a brain suppressor region about 400 bp 5′ to the transcription start site. The identities of the factors binding the brain suppressor region are not yet known, and its characterization is still at an early stage. However, differential footprinting reveals a candidate BPE in protein extracts of brain but not lens tissue, and the same region produces different, tissue-specific EMSA complexes with brain and lens proteins.
The core of the BPE (TCTGTGTT) is similar to binding sites for HMG or Sox (SRY box) proteins (TCTTTGTT) (44, 46, 47, 56, 61, 67). Sox-2 is expressed in the developing lens of both chicken and mouse embryos and plays a positive role in the expression of some crystallin genes (29, 30). However, in contrast to the positive role of Sox proteins in the lens (30), the brain suppressor region presumably binds a repressor complex in the brain but not in the lens. Preliminary results of experiments with specific antisera (a gift from R. Lovell-Badge) suggest that Sox-2 does not bind the BPE region (data not shown). Functional analysis of the BPE and the rest of the brain suppressor region awaits development of a suitable neural cell culture system to mimic the behavior of ζ promoter constructs in brain tissue.
Taken together, these results illustrate how three levels of transcriptional regulation can combine to produce lens specificity. Furthermore, while it is extremely difficult to reconstruct evolutionary events such as those which led to the gene recruitment of ζ in guinea pigs, it is apparent, at least in principle, how this could have occurred in three discrete steps with some possible selective benefits along the way (Fig. 8).
FIG. 8.
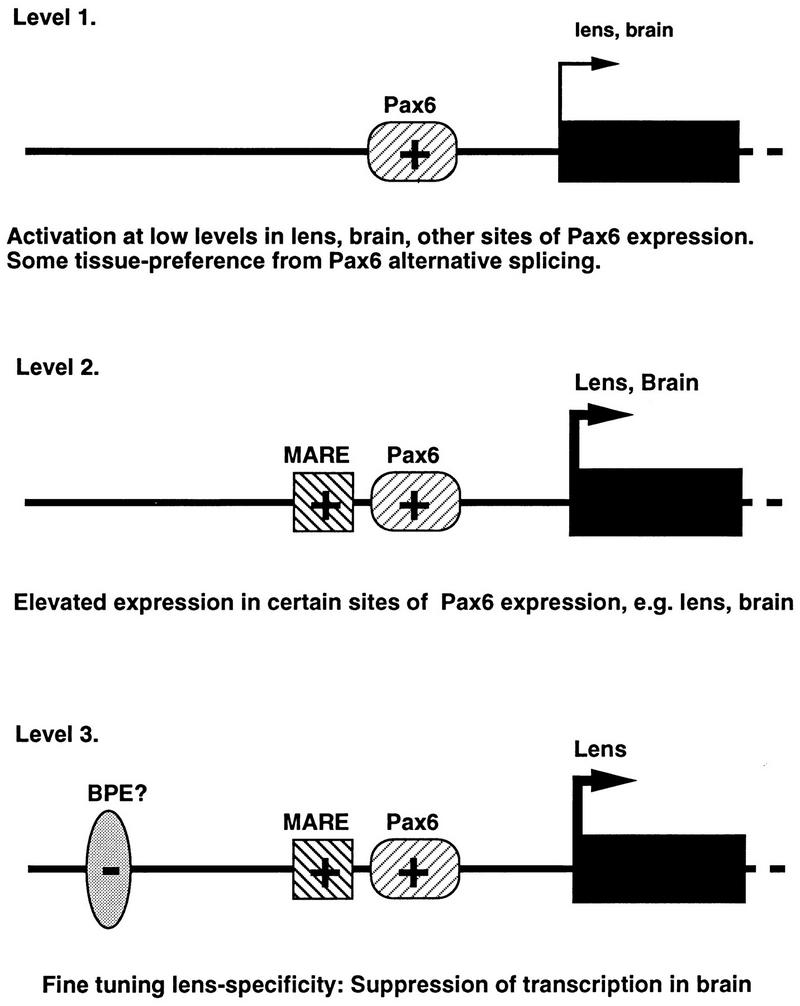
A possible pathway for multistep evolution of a new, tissue-specific gene promoter. Layering of positive (+) and negative (−) elements with some tissue preference can result in high levels of tissue-specific expression of a recruited gene in the lens.
First, in the context of suitable TATA or initiator sites, a new binding site for the canonical form of Pax6 could have conferred a small increase in the expression of an enzyme (in this case a quinone reductase) in Pax6-containing tissues, perhaps with some preference for lens tissue. Even moderately increased levels of a protective enzyme such as this could have been advantageous for the lens. A greater increase in the level of this enzyme in lens tissue may have had additional evolutionary benefits, reengineering the composition of the lens to fit changed behaviors or environmental conditions, as has been proposed (63, 64). Addition of a MARE binding site for Nrl or other Maf proteins adjacent to the Pax6 site would have facilitated such as increase. However, even though this might have led to an improvement in lens function, collateral expression (18) of high levels of ζ in the brain or other sites of Pax6 and Maf expression may actually have become disadvantageous. The “adaptive conflict” (63, 64) resulting from these opposing selective pressures could have been resolved by a third level of gene regulation: acquisition of a binding site for a negative factor in the central nervous system with a distribution overlapping that of Pax6 and the ability to suppress the activity of the promoter in brain tissue.
While the ζ lens promoter is a peculiar feature of guinea pigs and some related mammals, it illustrates some important general mechanisms in the development of tissue-specific gene expression in complex differentiated tissues. Even without tissue-specific transcription factors, alternative splicing and overlapping distributions of positive and negative factors in various tissues can produce fully specific expression in target genes. Crystallin gene recruitment can be looked on as a reenactment of processes which occurred at much earlier evolutionary stages for many other genes.
ACKNOWLEDGMENTS
We thank Peggy Zelenka, Ana Chepelinsky, and Cynthia Jaworski for critical reading of the manuscript. We acknowledge support from the Scientific Computing Resource Center of the NIH Division of Computer Research and Technology and from the National Eye Institute transgenic mouse facility.
A.S. is a recipient of an RPB Lew R. Wasserman Merit Award and is supported by NIH grant EY01115.
REFERENCES
- 1.Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalepakis G, Wijnholds J, Giese P, Schachner M, Gruss P. Characterization of Pax-6 and Hoxa-1 binding to the promoter region of the neural cell adhesion molecule L1. DNA Cell Biol. 1994;13:891–900. doi: 10.1089/dna.1994.13.891. [DOI] [PubMed] [Google Scholar]
- 3.Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 4.de Jong W W, Lubsen N H, Kraft H J. Molecular evolution of the eye lens. In: Chader G, Osbourne N, editors. Progress in retinal and eye research. Vol. 13. Oxford, United Kingdom: Elsevier Science Ltd.; 1994. pp. 391–442. [Google Scholar]
- 5.Edlund T, Walker M D, Barr P J, Rutter W J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science. 1985;230:912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- 6.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 7.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D S, Maas R L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2035. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 8.Farjo Q, Jackson A U, Xu J, Gryzenia M, Skolnick C, Agarwal N, Swaroop A. Molecular characterization of the murine neural retina leucine zipper gene, Nrl. Genomics. 1993;18:216–222. doi: 10.1006/geno.1993.1458. [DOI] [PubMed] [Google Scholar]
- 9.Fujioka M, Miskiewicz P, Raj L, Gulledge A A, Weir M, Goto T. Drosophila Paired regulates late even-skipped expression through a composite binding site for the paired domain and the homeodomain. Development. 1996;122:2697–2707. doi: 10.1242/dev.122.9.2697. [DOI] [PubMed] [Google Scholar]
- 10.Galas D J, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garland D, Rao P V, Del Corso A, Mura U, Zigler J S., Jr ζ-Crystallin is a major protein in the lens of Camelus dromedarius. Arch Biochem Biophys. 1991;285:134–136. doi: 10.1016/0003-9861(91)90339-k. [DOI] [PubMed] [Google Scholar]
- 12.Gehring W J, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 13.Glaser T, Jepeal L, Edwards J G, Young S R, Favor J, Maas R L. Pax6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez P, Hernandez-Calzadilla C, Rao P V, Rodriguez I R, Zigler J S, Jr, Borras T. Comparative analysis of the ζ-crystallin/quinone reductase gene in guinea pig and mouse. Mol Biol Evol. 1994;11:305–315. doi: 10.1093/oxfordjournals.molbev.a040111. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez P, Rao P V, Nunez S B, Zigler J S., Jr Evidence for independent recruitment of ζ-crystallin/quinone reductase (CRYZ) as a crystallin in camelids and hystricomorph rodents. Mol Biol Evol. 1995;12:773–781. doi: 10.1093/oxfordjournals.molbev.a040255. [DOI] [PubMed] [Google Scholar]
- 16.Gorman C, Padmanabhan R, Howard B H. High efficiency DNA-mediated transformation of primate cells. Science. 1983;221:551–553. doi: 10.1126/science.6306768. [DOI] [PubMed] [Google Scholar]
- 17.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham C, Hodin J, Wistow G. A retinaldehye dehydrogenase as a structural protein in a mammalian eye lens: gene recruitment of η-crystallin. J Biol Chem. 1996;271:15623–15628. doi: 10.1074/jbc.271.26.15623. [DOI] [PubMed] [Google Scholar]
- 19.Halder G, Callaerts P, Gehring W J. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 20.Hanson I, van Heyningen V. Pax6: more than meets the eye. Trends Genet. 1995;11:268–272. doi: 10.1016/s0168-9525(00)89073-3. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 22.Hill R E, Favor J, Hogan B L, Ton C C, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 23.Ho I C, Hodge M R, Rooney J W, Glimcher L H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 24.Holst B D, Wang Y, Jones F S, Edelman G M. A binding site for Pax proteins regulates expression of the gene for the neural cell adhesion molecule in the embryonic spinal cord. Proc Natl Acad Sci USA. 1997;94:1465–1470. doi: 10.1073/pnas.94.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Q L, Russell P, Stone S, Zigler J S. Zeta-crystallin, a novel lens protein from the guinea pig. Curr Eye Res. 1987;6:725–732. doi: 10.3109/02713688709034836. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 27.Jaworski C, Sperbeck S, Graham C, Wistow G. Alternative splicing of Pax6 in bovine eye and evolutionary conservation of intron sequences. Biochem Biophys Res Commun. 1997;240:196–202. doi: 10.1006/bbrc.1997.7623. [DOI] [PubMed] [Google Scholar]
- 28.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 29.Kamachi, Y. 1996. Involvement of SOX proteins in activation of crystallin genes and lens development. Tanpakushitsu Kakusan Koso 41(Suppl. 8):1113–1123. [PubMed]
- 30.Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–3519. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataoka K, Fujiwara K T, Noda M, Nishizawa M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol Cell Biol. 1994;14:7581–7591. doi: 10.1128/mcb.14.11.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataoka K, Igarashi K, Itoh K, Fujiwara K T, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kataoka K, Noda M, Nishizawa M. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos and small Maf proteins. Oncogene. 1996;12:53–62. [PubMed] [Google Scholar]
- 35.Kerppola T K, Curran T. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene. 1994;9:675–684. [PubMed] [Google Scholar]
- 36.Kumar R, Chen S, Scheurer D, Wang Q L, Duh E, Sung C H, Rehemtulla A, Swaroop A, Adler R, Zack D J. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J Biol Chem. 1996;271:29612–29618. doi: 10.1074/jbc.271.47.29612. [DOI] [PubMed] [Google Scholar]
- 37.Lee D C, Gonzalez P, Rao P V, Zigler Jr J S, Wistow G J. Carbonyl-metabolizing enzymes and their relatives recruited as structural proteins in the eye lens. Adv Exp Med Biol. 1993;284:159–168. doi: 10.1007/978-1-4615-2904-0_18. [DOI] [PubMed] [Google Scholar]
- 38.Lee D C, Gonzalez P, Wistow G. ζ-Crystallin: a lens-specific promoter and the gene recruitment of an enzyme as a crystallin. J Mol Biol. 1994;236:669–678. doi: 10.1006/jmbi.1994.1178. [DOI] [PubMed] [Google Scholar]
- 39.Li H S, Yang J M, Jacobson R D, Pasko D, Sundin O. Pax-6 is first expressed in a region of ectoderm anterior to the early neural plate: implications for stepwise determination of the lens. Dev Biol. 1994;162:181–194. doi: 10.1006/dbio.1994.1077. [DOI] [PubMed] [Google Scholar]
- 40.Liu Q, Ji X, Breitman M L, Hitchcock P F, Swaroop A. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene. 1996;12:207–211. [PubMed] [Google Scholar]
- 41.Matsuo I, Yasuda K. The cooperative interaction between two motifs of an enhancer element of the chicken αA-crystallin gene, αCE1 and αCE2, confers lens-specific expression. Nucleic Acids Res. 1992;20:3701–3712. doi: 10.1093/nar/20.14.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miksicek R, Heber A, Schmid W, Danesch U, Posseckert G, Beato M, Schutz G. Glucocorticoid responsiveness of the transcriptional enhancer of Moloney murine sarcoma virus. Cell. 1986;46:283–290. doi: 10.1016/0092-8674(86)90745-2. [DOI] [PubMed] [Google Scholar]
- 43.Miskiewicz P, Morrissey D, Lan Y, Raj L, Kessler S, Fujioka M, Goto T, Weir M. Both the paired domain and homeodomain are required for in vivo function of Drosophila Paired. Development. 1996;122:2709–2718. doi: 10.1242/dev.122.9.2709. [DOI] [PubMed] [Google Scholar]
- 44.Nasrin N, Buggs C, Kong X F, Carnazza J, Goebl M, Alexander-Bridges M. DNA-binding properties of the product of the testis-determining gene and a related protein. Nature. 1991;354:317–320. doi: 10.1038/354317a0. [DOI] [PubMed] [Google Scholar]
- 45.Ogino, H., and K. Yasuda. 1996. Involvement of maf gene family in crystallin gene regulation. Tanpakushitsu Kakusan Koso 41(Suppl. 8):1050–1057. [PubMed]
- 45a.Okataka-Maruyama, R., and A. Chepelinsky. Personal communication.
- 46.Pontiggia A, Whitfield S, Goodfellow P N, Lovell-Badge R, Bianchi M E. Evolutionary conservation in the DNA-binding and -bending properties of HMG-boxes from SRY proteins of primates. Gene. 1995;154:277–280. doi: 10.1016/0378-1119(94)00853-k. [DOI] [PubMed] [Google Scholar]
- 47.Prior H M, Walter M A. SOX genes: architects of development. Mol Med. 1996;2:405–412. [PMC free article] [PubMed] [Google Scholar]
- 48.Quiring R, Walldorf U, Kloter U, Gehring W J. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 49.Rao P V, Krishna C M, Zigler J S. Identification and characterization of the enzymatic activity of ζ-crystallin from guinea pig lens. A novel NADPH:quinone oxidoreductase. J Biol Chem. 1992;267:96–102. [PubMed] [Google Scholar]
- 50.Reddan J R, Chepelinsky A B, Dziedzic D C, Piatigorsky J, Goldenberg E M. Retention of lens specificity in long-term cultures of diploid rabbit lens epithelial cells. Differentiation. 1986;33:168–174. doi: 10.1111/j.1432-0436.1986.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 51.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack D J, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson J, Cvekl A, Wistow G. Pax-6 is essential for lens specific expression of ζ-crystallin. Proc Natl Acad Sci USA. 1995;92:4676–4680. doi: 10.1073/pnas.92.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sax C, Piatigorsky J. Expression of the α-crystallin/small heat shock protein/molecular chaperone genes in the lens and other tissues. Adv Enzymol Relat Areas Mol Biol. 1994;69:155–201. doi: 10.1002/9780470123157.ch5. [DOI] [PubMed] [Google Scholar]
- 54.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 55.Sierra F. BioMethods. 2: a laboratory guide to in vitro transcription. Boston, Mass: Birkhauser; 1990. [Google Scholar]
- 56.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 57.St. Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 58.Swaroop A, Xu J Z, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 60.Turque N, Plaza S, Radvanyi F, Carriere C, Saule S. Pax-QNR/Pax-6, a paired box- and homeobox-containing gene expressed in neurons, is also expressed in pancreatic endocrine cells. Mol Endocrinol. 1994;8:929–938. doi: 10.1210/mend.8.7.7984154. [DOI] [PubMed] [Google Scholar]
- 61.van de Wetering M, Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992;11:3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 63.Wistow G. Lens crystallins: gene recruitment and evolutionary dynamism. Trends Biochem Sci. 1993;18:301–306. doi: 10.1016/0968-0004(93)90041-k. [DOI] [PubMed] [Google Scholar]
- 64.Wistow G. Molecular biology and evolution of crystallins: gene recruitment and multifunctional proteins in the eye lens. Molecular Biology Intelligence Series. Austin, Tex: R.G. Landes Co.; 1995. [Google Scholar]
- 65.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 66.Yamada T, Nakamura T, Westphal H, Russell P. Synthesis of α-crystallin by a cell line derived from the lens of a transgenic animal. Curr Eye Res. 1990;9:31–37. doi: 10.3109/02713689009000052. [DOI] [PubMed] [Google Scholar]
- 67.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]