Abstract
Transcription factor IIIC (TFIIIC) (or τ) is a large multisubunit and multifunctional factor required for transcription of all class III genes in Saccharomyces cerevisiae. It is responsible for promoter recognition and TFIIIB assembly. We report here the cloning and characterization of TFC6, an essential gene encoding the 91-kDa polypeptide, τ91, present in affinity-purified TFIIIC. τ91 has a predicted molecular mass of 74 kDa. It harbors a central cluster of His and Cys residues and has basic and acidic amino acid regions, but it shows no specific similarity to known proteins or predicted open reading frames. The TFIIIC subunit status of τ91 was established by the following biochemical and genetic evidence. Antibodies to τ91 bound TFIIIC-DNA complexes in gel shift assays; in vivo, a B block-deficient U6 RNA gene (SNR6) harboring GAL4 binding sites was reactivated by fusing the GAL4 DNA binding domain to τ91; and a point mutation in TFC6 (τ91-E330K) was found to suppress the thermosensitive phenotype of a tfc3-G349E mutant affected in the B block binding subunit (τ138). The suppressor mutation alleviated the DNA binding and transcription defects of mutant TFIIIC in vitro. These results indicated that τ91 cooperates with τ138 for DNA binding. Recombinant τ91 by itself did not interact with a tRNA gene, although it showed a strong affinity for single-stranded DNA.
Transcription of class III genes involves multiple interactions between promoter elements, auxiliary transcription factors, and RNA polymerase III (Pol III). Prototypical class III genes, like tRNA and 5S RNA genes, have intragenic promoter elements or internal control regions, differing in this way from class I and class II genes. The internal control regions of tRNA genes are composed of two sequence elements, termed the A and B blocks, that are separated by variable distances (31 to 93 bp in the 273 tRNA genes of Saccharomyces cerevisiae). The process of gene activation has been much investigated in vitro with purified components, especially in the yeast system (19, 25, 61). Two multisubunit factors, transcription factor IIIB (TFIIIB) and TFIIIC, are required for transcription of yeast tRNA genes. Transcription complex assembly is initiated by the binding of TFIIIC to the A and B block elements. Through its interaction with the B block, which functions to some extent in a distance- and orientation-independent manner (11), TFIIIC behaves as an enhancer binding factor and relieves repression by chromatin (10). Via A block binding, TFIIIC recruits TFIIIB at an upstream position and hence indirectly influences start site selection (4, 24, 26, 27). TFIIIB-DNA complexes stripped of TFIIIC are competent and sufficient to direct correct initiation and multiple rounds of transcription by Pol III (26).
Yeast TFIIIC (τ factor) is a large, multisubunit protein of about 550 to 600 kDa. It is made of two large domains with distinct DNA binding specificities, Tau A (τA) and Tau B (τB), that can be visualized by electron microscopy in a free or DNA-bound form (11, 47). The chromatographic separation of τA and τB has never been observed, but limited proteolysis releases the τB domain which retains B block binding specificity (40). Affinity-purified TFIIIC consistently contains six polypeptides of 138, 131, 95, 91, 60, and 55 kDa (3, 17, 42). An additional polypeptide of 75 kDa has been noted occasionally in highly purified TFIIIC fractions. The TFIIIC subunit status of the three largest polypeptides is well established, based on gene cloning (33, 38, 41, 53), mutagenesis (34, 44, 48, 60), protein-DNA cross-linking (3, 9, 17) and coimmunoprecipitation (13, 41) experiments. The τ138 subunit resides in the τB domain and interacts with DNA at the level of the B block (3, 17). A mutation in the τ138 subunit, encoded by TFC3, decreased TFIIIC-tRNA gene (DNA) binding affinity and also affected 5S RNA synthesis in vitro (34). All three components of TFIIIB (TATA binding protein [TBP], TFIIIB70/BRF1, and TFIIIB90/B"), when overexpressed, were found to suppress this defect in vivo (34, 46). τ131 is the TFIIIC subunit that can be cross-linked to the most upstream position within the TFIIIB binding site, at the level of the start site and further downstream (3, 4). It interacts with TFIIIB70/BRF1 and with TFIIIB90/B" (12, 28, 46) and therefore presumably ensures TFIIIB assembly on the DNA. Indeed, mutations in τ131 influence the level of active TFIIIB or the rate of TFIIIB recruitment (44). τ95 is thought to be responsible for A block recognition (3, 17, 53). Little is known about the other TFIIIC-associated polypeptides. Site-specific DNA-protein cross-linking indicated that the 91-kDa component is located at the 3′ end of 5S RNA or tRNATyr genes (9) and that the 55-kDa protein lies in the vicinity of the A block, close to τ95 (3, 4).
Human TFIIIC activity has been partially characterized but appears to be more complex than yeast TFIIIC activity. Human TFIIIC separates into two protein fractions, TFIIIC1 and TFIIIC2 (57, 62). TFIIIC2 is comprised of five subunits (α, β, γ, δ, and ɛ) of 230, 110, 100, 80, and 60 kDa, respectively (29, 57, 63); it binds to the B block element tightly (7, 15, 62) and thus appears to be similar to yeast TFIIIC. However, unlike yeast TFIIIC, this multiprotein complex is deficient in A block binding and transcription factor activities, both of which require the TFIIIC1 fraction. The polypeptide composition of TFIIIC1 and its role in promoter recognition and TFIIIB assembly are still unclear (14, 57). Intriguingly, the TFIIIC1 fraction extends the footprint of TFIIIC2 to and beyond the A block as well as to downstream sequences over the termination region (57, 62), to which it can bind by itself (57). The complexity of human TFIIIC thus makes it difficult to draw a simple correlation with yeast TFIIIC, inasmuch as the two largest subunits of TFIIIC2, TFIIICα and TFIIICβ (30, 35, 51), show no significant sequence similarity to any of the three cloned subunits of TFIIIC (τ138, τ131, and τ95).
In the present work, we have pursued the characterization of TFIIIC components by cloning a yeast gene, termed TFC6, which encodes the 91-kDa polypeptide (τ91). We show that τ91 is an essential subunit of TFIIIC that cooperates with τ138 for DNA binding.
MATERIALS AND METHODS
DNA constructions and yeast strains.
Two oligonucleotides (Ol 1 [5′TCTTCCTCTAGTTCGACCCG] and Ol 2 [5′ATTTAGGATCCCTTCTTCTCC]) were used to amplify the open reading frame (ORF) and surrounding sequences of the TFC6 gene by PCR. The resulting 3,400-bp S. cerevisiae genomic DNA fragment was cloned into a pGEM-T vector (Promega) to obtain pGEM128. Yeast centromeric and multicopy pYc91 and pYμ91 plasmids were obtained by cloning the pGEM128 KpnI-SpeI fragment harboring the TFC6 gene into Ycplac33 and YEplac112 vectors (20), respectively.
Oligonucleotides Ol 2 (see above) and Ol 3 (5′ATATATTAAGTTGTGCATATGTATCCTTACGACGTTCCTGATTATGCCATGGCAGTAATACCG) were used to add the epitope derived from the influenza virus hemagglutinin (HA) protein after the initiation codon of TFC6 by PCR-mediated mutagenesis. The NdeI-BglII fragment from the PCR-amplified DNA was cloned into pGEM128 to produce pGEM91-HA. The KpnI-SpeI fragment from pGEM91-HA was cloned into YCplac22 to obtain pYCt91-HA. The NdeI-BamHI or NcoI-BamHI fragments from pYCt91-HA were inserted into the corresponding sites of pET28a (Novagen) and pACTII and pAS1-CYH2 (kindly provided by S. Elledge) vectors, giving pET91, pACT91, and pAS91, respectively.
The yeast strains used in this study are listed in Table 1. They were constructed by genetic techniques based on transformation of lithium acetate-treated cells, sexual mating, and tetrad analysis with standard media and growth conditions (49).
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| YNN281 | MATa ade2-1 ura3-52 lys2-801 his3-Δ200 trp1-Δ1 | YGSCa |
| YNN282 | MATα ade2-1 ura3-52 lys2-801 his3-Δ200 trp1-Δ1 | YGSC |
| YNNRA1 | (YNN281 × YNN282) TFC6/tfc6-Δ::HIS3 | This work |
| YNRA2 | MATa ade2-1 ura3-52 lys2-801 his3-Δ200 trp1-Δ1 TFC6-Δ::HIS3 + pYc91 | This work |
| YNRA3 | MATa ade2-1 ura3-52 lys2-801 his3-Δ200 trp1-Δ1 TFC6-Δ::HIS3 + pYCt91-HA | This work |
| D135-2c | MATa ade1 ura3-52 leu2-3 his3-b trp1-Δ63 tfc3-G349E | This work |
| D135-3c | MATα ade1 ura3-52 lys2-801 his3-b trp1-Δ63 tfc3-G349E | This work |
| SRY15-5d | MATa ura3-52 trp4 leu2-3 ade2 tfc3-G349E | This work |
| D135-αg2 | MATα ade1 ura3-52 lys2-801 his3-b trp1-Δ63 tfc3-G349E TFC6-E330K | This work |
| YPH500 | MATα ade2-101 ura3-52 lys2-801 his3-Δ200 leu2-Δ1 trp1-Δ63 | 50 |
YGSC, Yeast Genetic Stock Center, Berkeley, Calif.
his3-Δ1 or his3-Δ200.
Disruption of TFC6.
The whole TFC6 ORF was disrupted by the direct-deletion method (6). Two 57-mer oligonucleotides were used to amplify by PCR a DNA fragment containing the HIS3 gene and stop modules flanked by TFC6 promoter and terminator sequences. The 1,078-bp PCR-amplified fragment was directly used to transform the strain YNN281 × YNN282 (22). The structure of the diploid His3+ disruptants (called YNNRA1) was verified by PCR analysis. YNNRA1 was transformed with pYc91, and after sporulation and dissection, a His+ spore containing pYc91 was chosen to give YNRA2. TFC6 disruption was performed in the same way in strain D135-αg2 × SRY15-5d.
Purification of TFIIIC.
TFIIIC was purified as previously described (23). Briefly, for preparative electrophoresis, HA-tagged-τ95-containing TFIIIC was purified from 1,300 g of cells. Three hundred to 400 pmol of tDNA affinity-purified factor was resolved on a sodium dodecyl sulfate (SDS)–8% polyacrylamide gel and stained with Coomassie blue. A gel slice containing the 91-kDa (τ91) polypeptide was incubated with proteinase K or trypsin, and four polypeptides, isolated by reversed-phase high-pressure liquid chromatography, were microsequenced (34). TFIIIC containing the HA-tagged τ91 subunit was purified from 20 g of YNRA3 cells by three chromatographic steps, i.e., Ultrogel-heparin, DEAE-Sephadex, and tDNA affinity chromatography, as previously described (23).
Expression and purification of recombinant His-HA-τ91.
Recombinant TFC6 protein (rTFC6p or rτ91) fused at its N terminus to six histidines and to the HA epitope was obtained from Escherichia coli BL21(DE3)(pLysS) transformed with the plasmid pET91. Cell culture, protein induction and crude extract preparation were performed essentially as described previously (46) except that buffer A10 (20 mM HEPES [pH 7.5], 500 mM NaCl, 10% glycerol, 10 mM imidazole) was used as the lysis buffer. Crude cell extract containing rτ91 was recovered after centrifugation at 145,000 × g for 45 min at 4°C and subjected to fast protein liquid chromatography in a 1-ml Ni2+-charged HiTrap chelating (Pharmacia) column. Proteins were eluted by a linear gradient of 53.5 to 300 mM imidazole. The peak of rτ91 was eluted at ∼100 mM imidazole. Fractions containing the recombinant protein were pooled and further purified with the Smart System. The Ni2+ eluate was diluted with buffer B0 (20 mM Tris-HCl [pH 8], 0.5 mM EDTA, 10 mM β-mercaptoethanol, 10% glycerol) to a salt concentration equivalent to 100 mM ammonium sulfate and applied on a 100-μl heparin HyperD (BioSepra) column. Proteins were eluted with buffer B600 (600 mM ammonium sulfate in B0 buffer) and loaded on a Superdex 75 column previously equilibrated in buffer B300 (300 mM ammonium sulfate in B0 buffer). The concentration and purity of rτ91 were estimated to be about 1 to 5 ng/μl and more than 95%, respectively, by visual analysis of a silver-stained SDS-polyacrylamide gel.
Anti-τ91 polyclonal antibodies.
Partially purified rτ91 was loaded on a preparative SDS–8% polyacrylamide gel, and the band containing the 91-kDa protein was excised and injected into mice for antibody production. A total of 40 μg of purified protein was injected in four injections at 3-week intervals. The mice were then inoculated with ascite cells, and three batches of about 5 to 10 ml each of ascitic fluid were collected. To purify anti-τ91 antibodies, immunoglobulins were adsorbed on a 300-μl protein A-Sepharose column and eluted with 0.1 M glycine, pH 3. Fractions (250 μl) were collected in tubes containing 25 μl of 1 M Tris-HCl, pH 8. The protein concentration (∼0.6 μg/μl) was estimated by Bradford analysis (8). Control ascitic fluid was treated similarly to give control antibodies.
DNA binding and in vitro transcription assays.
Unless otherwise indicated, TFIIIC-tDNA interaction was monitored by gel retardation analysis as described previously (34), using either Ultrogel-heparin or DNA-affinity purified TFIIIC fractions. τB-tDNA interaction was observed after limited proteolysis of TFIIIC-tDNA complexes. After 10 min of incubation at 25°C, 10 ng of α-chymotrypsin (Sigma) was added to the TFIIIC-tDNAGlu complex mixture and further incubated at 25°C for 10 min. Chymotrypsin digestion was stopped by addition of 1 ng of aprotinin (Sigma).
rτ91-nucleic acid interaction was investigated by gel retardation analysis. rτ91 (∼2 ng in 0.5 μl of the Superdex 75 fraction containing 300 mM ammonium sulfate) was incubated with the corresponding nucleic acid probe (∼10,000 cpm) in 15 μl of binding buffer containing 10 mM Tris-HCl (pH 8), 100 mM KCl, 10% glycerol, 10 μg of bovine serum albumin, and, when indicated, 2.5 mM MgCl2. The final ammonium sulfate concentration was 120 mM. Nucleic acid probes were as follows. The double-stranded DNA probe was a 200-bp PCR-amplified fragment from the pUC-Glu plasmid carrying the yeast tRNA3Glu gene (18). The same DNA probe was denatured by boiling for 3 min and quenching in ice. The tRNA probe was a 32P-labelled mammal liver tRNAMet (kindly provided by M. N. Thang). Complexes were formed at 25°C for 10 min, separated by nondenaturing gel electrophoresis in an 8% polyacrylamide gel at 16 V/cm for 90 min at 4°C, and autoradiographed.
Plasmid pGE2 harboring the yeast tRNA3Leu gene (1) was used for in vitro transcription. Transcription reactions were carried out as described previously (23) with 2 μl (∼0.7 μg) of Ultrogel-heparin TFIIIC fractions from wild-type, tfc3-G349E mutant, or suppressor strains.
In vivo UASG-U6 chimeric transcription system.
UASG-U6 RNA gene constructs were as described previously (39). The GAL4-(1-147)-τ91, GAL4-(AD)-τ91, GAL4-(1-147)-τ95, and GAL4-(AD)-τ95 fusions were expressed from the plasmids pAS91, pACT91, pAS95, and pACT95, respectively. pACT91 and pAS91 were constructed as described above. pACT95 and pAS95 were obtained by cloning a BglII fragment of a mutagenized form of TFC1 into BamHI-digested pACTII and pAS1-CYH2 vectors. Transformation of strain YPH500, RNA extraction, and Northern blot analysis were performed as described previously (39).
Isolation of tfc3-G349E suppressors and genetic methods.
An overnight liquid culture of the thermosensitive D135-3c strain (MATα tfc3-G349E [see Table 1 for the complete genotype]) was plated (106 cells/plate) on yeast extract-peptone-dextrose, UV irradiated with a Vilbert Lourmat lamp for 10 or 20 s at 254 nm (4 W/m2), and incubated in the dark at 37°C for 3 days (32). Survival was about 70 and 40%, respectively, as determined by cell plating efficiency. The efficiency of mutagenesis was estimated by measuring the frequency of CANr (canavanine-resistant) mutants, which was 1.2 × 10−4 cells. Viable colonies were reisolated at 37°C and backcrossed with the MATa tfc3-G349E strain D135-2c to determine their dominance and the monogenic character of suppression. Fifteen clones were retained for further characterization as described in Results.
RESULTS
Identification, cloning, and disruption of the TFC6 gene.
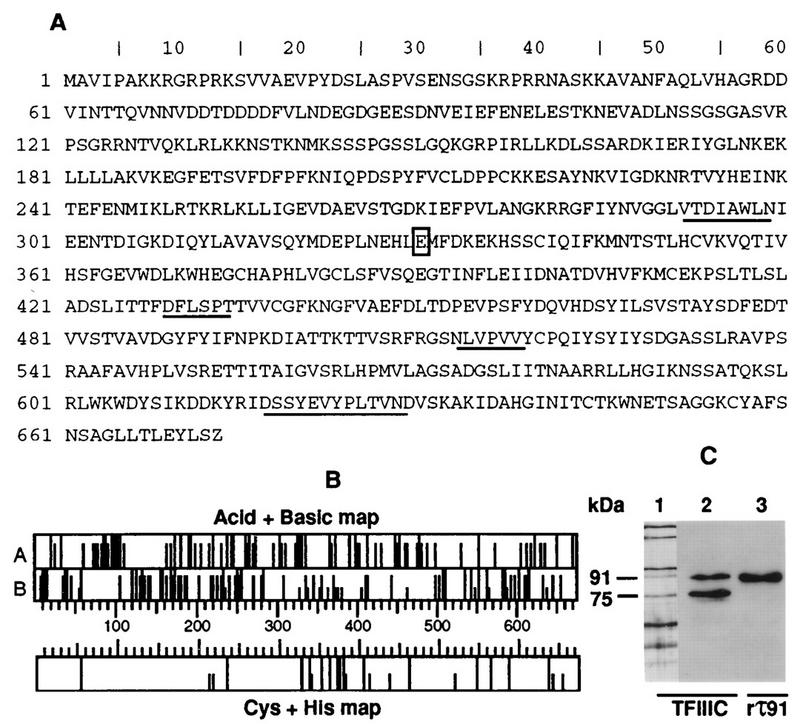
The 91-kDa polypeptide present in affinity-purified yeast TFIIIC (τ91) was isolated by preparative SDS-polyacrylamide electrophoresis (SDS-PAGE). The gel-purified protein was digested with proteinase K or with trypsin, and the amino acid sequences of four peptides (underlined in Fig. 1A) were determined. When compared to the National Center for Biotechnology Information nonredundant database, all of these peptide sequences were found to be contained in a protein encoded by a 2,019-bp ORF located on chromosome IV of S. cerevisiae (Fig. 1A). This gene, identified as YDR362C by the group of Mark Johnston, was renamed TFC6. Two remarkable regions were found in the predicted sequence of TFC6p (Fig. 1B): the N-terminal part (amino acids 7 to 180) contains a highly acidic region between two basic domains, and the central part contains a cluster of 13 cysteine and histidine residues (amino acids 328 to 411) that potentially binds zinc ions. However, TFC6p showed no similarity to any protein sequence in the EMBL/GenBank data bank or to current versions of the Schizosaccharomyces pombe and Caenorhabditis elegans genomic sequences. A direct comparison of TFC6p with the two largest subunits of human TFIIIC2, TFIIICα and -β, (30, 35, 51), by direct pairwise sequence alignment (43) also revealed no significant homology.
FIG. 1.
Sequence analysis of TFC6 and characterization of anti-τ91 antibodies. (A) Deduced amino acid sequence of TFC6. The four microsequenced peptides used for database searching are underlined (GenBank no. U28372). The TFC6 mutation, E330K, responsible for tfc3-G349E suppression is boxed. (B) Amino acid analysis of τ91. The acidic-basic map shows the position of acidic (A) (D, short bars; E, long bars), and basic (B) (R, long bars; K, short bars; H, smallest bars) amino acids. In the Cys-His map, histidine and cysteine residues are plotted as long and short bars, respectively. (C) Characterization of anti-τ91 antibodies by Western blotting. Lane 1, the polypeptide content of a tDNA affinity-purified TFIIIC fraction was analyzed in a silver-stained SDS–8% polyacrylamide gel. (Lanes 2 and 3, ∼100 ng of TFIIIC (lane 2) and ∼20 ng of purified rTFC6p (rτ91) (lane 3) were subjected to SDS-PAGE, electroblotted to a nitrocellulose membrane, and probed with anti-τ91 antibodies. Immune complexes were visualized with an ECL kit (Amersham). The positions of 91- and 75-kDa proteins are indicated. The amino acid sequence, acidic-basic map, and Cys-His map are printouts from DNA Strider 1.2 software (37).
The protein encoded by TFC6 has a theoretical pI of 6.8 and a predicted molecular mass of 74 kDa, which is significantly lower than the mass deduced from its migration rate on an SDS-polyacrylamide gel (91 kDa). rTFC6p was expressed in E. coli and in a wheat germ extract in vitro. rTFC6p was tagged at the N-terminal position with six His residues followed by 10 amino acids encompassing the thrombin cleavage sequence and by the 9 amino acids of the influenza virus HA epitope. The Mr of 75,000 estimated by gel filtration analysis of rTFC6p corresponded fairly well to its theoretical value (78,000). However, the tagged recombinant polypeptide migrated like unmodified yeast τ91 (Fig. 1C) and faster than the HA-tagged yeast τ91 polypeptide upon SDS-PAGE (results not shown). The slower migration of the yeast polypeptide may reflect a postranslational modification. We did not explore this possibility further.
Mouse polyclonal antibodies directed against rTFC6p were used to probe an affinity-purified fraction of TFIIIC. TFIIIC and the recombinant protein were subjected to SDS-PAGE; one half of the gel was silver stained, and the proteins from the other half were transferred to a membrane and incubated with protein A-Sepharose-purified antibodies. The recombinant protein and two polypeptides from TFIIIC were strongly bound by the antibodies (Fig. 1C, lanes 2 and 3). The slower protein band corresponded to the 91-kDa polypeptide, and the other corresponded to a 75-kDa protein band that has been observed in several purified TFIIIC preparations obtained in our laboratory and elsewhere (13, 17, 42, 53). In a TFIIIC fraction containing the HA-tagged TFC6p, both the 91- and 75-kDa polypeptides were bound by anti-TFC6p antibodies, but only the 91-kDa protein was bound by anti-HA antibodies. This result indicates that the 75-kDa polypeptide which is occasionally found in TFIIIC preparations is not another TFIIIC subunit but is related to the 91-kDa protein, probably as a breakdown product.
The three largest subunits of yeast TFIIIC were previously found to be essential for cell viability. We have deleted the TFC6 gene in the diploid S. cerevisiae strain YNN281 × YNN282 by a PCR method (6). The whole ORF of TFC6 (2,019 bp) was replaced by a DNA fragment of 1,078 bp containing the yeast HIS3 selectable marker surrounded by stop codon modules and inserted in the antisense direction with respect to TFC6. The resulting diploid cells, YNNRA1, had one chromosome with the deleted allele (tfc6-Δ::HIS3) and one chromosome harboring the wild-type TFC6+ gene. Tetrad analysis of the meiotic offspring generated two viable and two nonviable spores per meiosis. All viable segregants were His−, indicating that they bear the TFC6+ allele and that this allele encodes an essential gene product.
To confirm this conclusion, we cloned TFC6 by PCR amplification of genomic DNA with primers complementary to sequences located about 500 bp upstream and downstream of the coding sequence. This PCR-amplified fragment was inserted into a centromeric yeast vector, Ycplac33 (20), to produce the centromeric plasmid pYc91 (CEN URA3 TFC6). The heterozygous diploid tfc6-Δ::HIS3/TFC6 strain YNNRA1 was transformed with pYc91 and sporulated. Haploid segregants bearing the tfc6-Δ::HIS3 mutation but harboring the plasmid TFC6 gene (e.g., strain YNRA2 [Table 1]) were invariably viable.
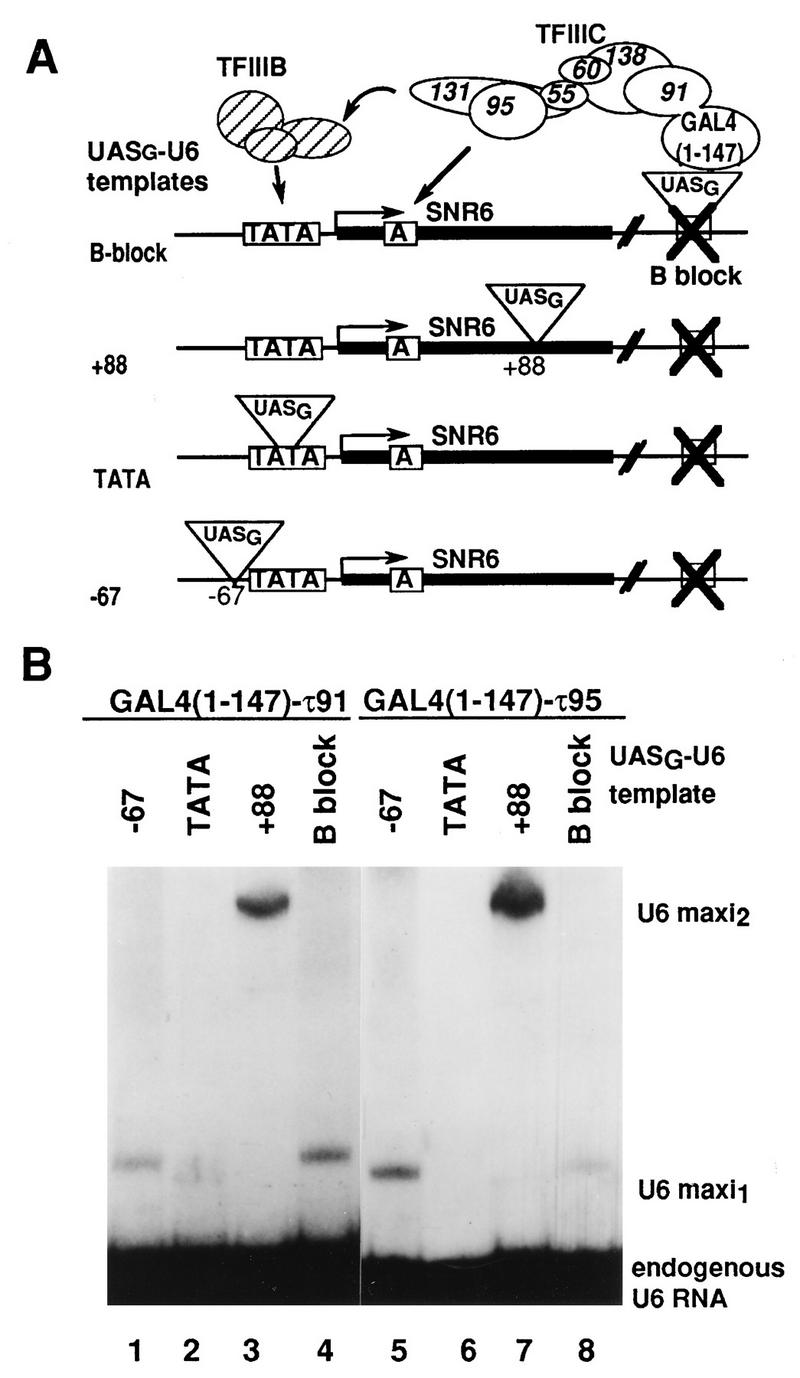
GAL4(1-147)-τ91 activates transcription of a B block-deficient U6 RNA gene.
To gain some insight into the function of the TFC6 polypeptide and assess its involvement in Pol III transcription in vivo, we used a chimeric system in which a B block-deficient U6 RNA gene (SNR6) can be reactivated by fusing the GAL4 DNA binding domain (amino acids 1 to 147) to a subunit of TFIIIC (or TFIIIB) (39). In this system, the extent of SNR6 gene activation depends on the presence of GAL4 binding sites (UASG) at appropriate locations. If TFC6 indeed encoded a subunit of TFIIIC, one would expect that the GAL4(1-147)-τ91 fusion would anchor TFIIIC on the UASG sequence and activate transcription of UASG-U6 RNA genes, as in the case of GAL4(1-147)-τ138 or -τ131 (39). The experimental scheme is shown in Fig. 2A. All genes carry a 24-bp insertion at position +73 to discriminate the transcripts in Northern blots from endogenous U6 RNA. As shown in Fig. 2B, GAL4(1-147)-τ91 activated transcription of UASG-U6 RNA genes that harbored UASG sequences at positions −67, +88 (within the transcribed sequence), and +238 (within the destroyed B block). Similar activation levels were obtained with fusion of GAL4(1-147) to the 95-kDa subunit of TFIIIC (τ95) (Fig. 2B, lanes 5 to 8). In previous work, we reported that a GAL4(1-147)-τ95 fusion was incapable of reactivating the UASG-U6 RNA templates (39). In fact, we found that the plasmid construct used at that time had a stop codon between the GAL4(1-147) and τ95 ORFs that prevented the formation of a hybrid protein. (This mutation went unnoticed because the plasmid was able to complement the deletion of the τ95 gene [39].) Only a faint background signal was obtained when τ91 or τ95 was fused to the GAL4 activation domain or when the cells were transformed with the expression vector (results not shown). These results indicated that TFC6 encoded a component of the Pol III transcription system.
FIG. 2.
Activation of the UASG-U6 RNA templates by GAL4(1-147)-τ91 and GAL4(1-147)-τ95. (A) Diagram of UASG-U6 RNA gene activation by GAL4(1-147)-τ91. Each template was named according to the position of the upstream activating sequences (UASG) in each B block-deficient U6 template. TFIIIC is a chimeric τ factor whose wild-type τ91 subunit has been replaced by the GAL4(1-147)-τ91 fusion protein. Chimeric TFIIIC binds the U6 template through the UASG sites, and this allows the factor to bind the A box of the SNR6 gene. TFIIIC then recruits TFIIIB, which directs transcription by Poly III. (B) The UASG-U6 RNA transcripts (U6 maxi2 and U6 maxi1) and the endogenous U6 RNA from cells transformed with pAS91 (lanes 1 to 4) or pAS95 (lanes 5 to 8) were monitored by Northern blot analysis and autoradiography. The UASG binding domain (amino acids 1 to 147) of GAL4 is fused to τ91 in pAS91 or to τ95 in pAS95.
TFC6 encodes the 91-kDa subunit of yeast TFIIIC.
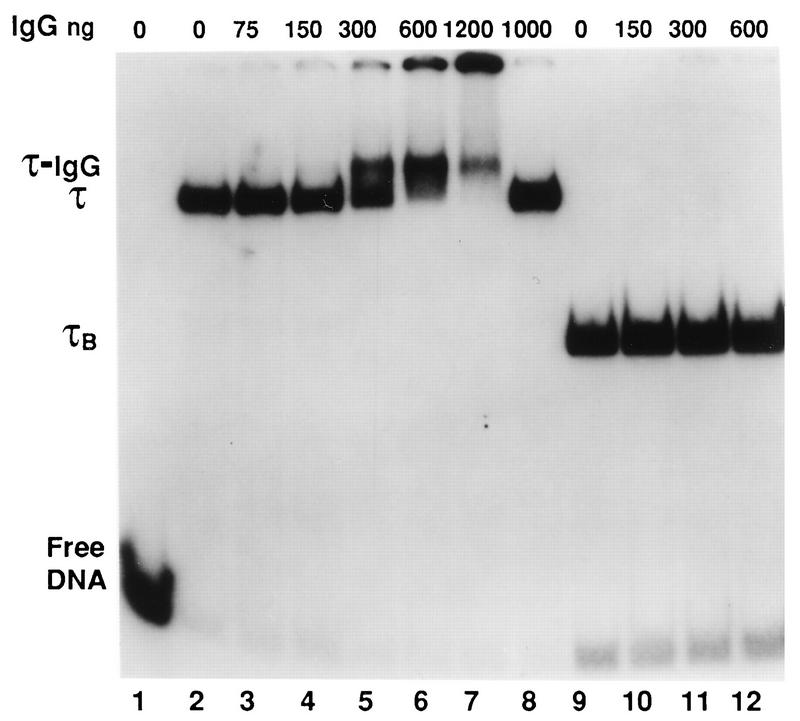
In an early attempt to demonstrate that the 91-kDa polypeptide was a subunit of TFIIIC, we modified the TFC6 gene to fuse the HA epitope at the N-terminal position of the protein (plasmid pYCt91-HA [see Materials and Methods]). The modified gene was functional, since it could replace the wild-type gene in a plasmid shuffling assay in which pYCt91-HA was exchanged for the centromeric plasmid pYc91 (CEN URA3 TFC6). This plasmid exchange, which was done in strain YNRA2, generated strain YNRA3, which lacks the chromosomal copy of TFC6 but survives by expressing the HA-tagged polypeptide (Table 1). We confirmed with Western blots that the HA-tagged TFC6-encoded protein (purified from YNRA3) coeluted with TFIIIC activity throughout three successive chromatographic steps. However, anti-HA monoclonal antibodies did not alter the migration of TFIIIC-tDNA3Glu complexes in gel shift assays and did not interfere with complex formation (results not shown). This negative result was in sharp contrast to those of similar experiments that demonstrated that τ138, τ131, and τ95 were part of TFIIIC (13, 33, 38). The possibility remained, however, that the HA epitope was not accessible to antibodies within TFIIIC-tDNA complex. Therefore, the same experiment was repeated with mouse polyclonal antibodies raised against the recombinant 91-kDa polypeptide. As shown in Fig. 3, these antibodies retarded the migration of preformed TFIIIC-tDNA complexes (lanes 5 to 7), while antibodies from preimmune serum had no effect (lane 8). This experiment confirmed the status of τ91 as a subunit of TFIIIC.
FIG. 3.
TFC6 encodes a subunit of factor τ. DNA affinity-purified factor TFIIIC was complexed with the tRNA3Glu gene, and, when indicated, the complex was digested with 10 ng of α-chymotrypsin for 10 min at 25°C to generate the fast-migrating complex τB-tDNA. Complexes were then incubated with increasing amounts of polyclonal antibodies directed against τ91 for 45 min at 25°C. Protein-tDNA complexes were analyzed by electrophoresis. τ, position of τ-tDNA complexes; τ-IgG, band retarded upon antibody binding; τB, protease-resistant domain of TFIIIC bound to tDNA. Lanes 1 to 8, no protease; lanes 9 to 12, α-chymotrypsin treatment. Lane 1, no TFIIIC factor, lanes 2 and 9, no antibody, lanes 2 to 7 and 9 to 12, anti-τ91 immunoglobulin G (IgG). Lane 8, control mouse antibodies.
The TFIIIC factor is made of two large DNA binding domains, τA and τB, of about 300 kDa each, that can be split by limited proteolysis (17, 40). The protease-resistant τB domain forms stable complexes with the B block that can be visualized in gel shift assays. Anti-τ138 antibodies retard the migration of such τB-tDNA complexes (17). We investigated whether τ91 was part of the protease-resistant τB domain by incubating preformed τB-tDNA complexes with the anti-τ91 polyclonal antibodies. Under the conditions that supershifted all of the TFIIIC-tDNA complexes, the migration of the τB-tDNA complex was unaffected by the antibodies (Fig. 3, lanes 9 to 12). This negative result does not, however, exclude the possibility that τ91 belongs to τB together with τ138, as discussed below.
A dominant mutation (TFC6-E330K) suppresses the tfc3-G349E mutation affecting the τ138 subunit.
tsv 115 is a recessive, UV-induced, thermosensitive mutation in the TFC3 gene that causes a G349E substitution in the subunit τ138 (21, 34). This mutation is referred to in this paper as tfc3-G349E. It affects both the level of TFIIIC and its affinity for tDNA (34). A screen for suppressor mutants (see Materials and Methods) yielded 15 colonies that had regained the ability to grow at 37°C after UV mutagenesis of strain D135-3c (MATα tfc3-G349E). Backcrosses to D135-2c (MATa tfc3-G349E) showed that these suppressors were all due to dominant mutations with a monogenic 2:2 segregation. Further crosses showed that four of them were genetically linked to tfc3-G349E. The remaining 11 extragenic suppressors were classified into six distinct linkage groups (presumably corresponding to one gene per group) by crossing them to each other and measuring the frequency of temperature-sensitive (i.e., nonsuppressed) tfc3-G349E segregants in the offspring. One representative of each group was crossed to the tester strain SRY15-5d (MATa ura3-52 trp4 leu2-3 ade2 tfc3-G349E) bearing the trp4− marker, which maps very close to TFC6 (these genes are separated by about 12 kb on chromosome IV). Two suppressor mutations were found to map very close to TFC6 as shown by their linkage to trp4−. To demonstrate that they were allelic to TFC6, we constructed diploid strains that are homozygous for tfc3-G349E and heterozygous for trp4 and for the suppressor allele, and we introduced the tfc6-Δ::HIS3 deletion in these diploids by standard gene disruption. About half of the disruptants regained the temperature-sensitive character of the nonsuppressed tfc3-G349E allele, indicating that they had lost suppression upon inactivation of the TFC6 copy brought by the suppressor strain. The lethal tfc6-Δ::HIS3 deletion cosegregated with the closely linked trp4 marker present in the suppressor strain.
The two suppressor mutations were first grossly localized between positions +484 and +1338 of the TFC6 ORF by a method based on integrative transformation of the suppressor strains with URA3+ integrative plasmids harboring nonfunctional N-terminal or C-terminal fragments of TFC6 (45). After PCR amplification and sequencing of the corresponding DNA region, the two suppressors were found to bear the same dominant mutation (TFC6-E330K) due to a G-A transition at nucleotide 988 of the coding sequence. The TFC6-E330K suppressor is remarkably strong, since the doubling time of the tfc3-G349E/TFC6-E330K diploid cells is the same as that of the wild-type strain at 37°C (130 min). When separated from tfc3-G349E by genetic crosses, the suppressor mutation causes no detectable growth phenotype per se. The fact that an amino acid substitution in τ91 could suppress the temperature-sensitive phenotype of the tfc3-G349E mutant suggested that τ91 might contribute to DNA binding together with τ138.
TFC6-E330K alleviates the DNA binding defect of mutant TFIIIC harboring the temperature-sensitive mutation tfc3-G349E.
To examine the effect of the TFC6-E330K suppressor in vitro, TFIIIC was partially purified from wild-type, tfc3-G349E, and suppressor strains and tested under the same conditions in DNA binding and transcription assays (Fig. 4). The DNA binding activity of mutant TFIIIC harboring the tfc3-G349E mutation is very sensitive to mild heat treatments and is inhibited at moderate salt concentrations (36) (Fig. 4). The mutant factor lost most of its DNA binding activity after 10 min of preincubation at 30°C and was totally inactivated at 35°C, in contrast with the wild-type factor preparation, which resisted up to 40°C. Remarkably, the double-mutant form of TFIIIC (tfc3-G349E/TFC6-E330K) had an intermediate behavior and retained a significant level of DNA binding activity at 35°C (Fig. 4A and B). Mutant, wild-type, and suppressor TFIIIC-tDNA complex formation also showed different salt sensitivities at 25°C. With the same amount of protein fraction used in all three cases, the optimal salt concentration for complex formation with the tRNA3Glu gene was 135 mM KCl. At 175 mM KCl, complex formation with wild-type, suppressor, and mutant factors dropped to 80, 30, and 18% of their maximal values, respectively. At 205 mM KCl, this residual binding activity was further decreased to 15, 3, and <1%, respectively (results not shown). These observations strongly suggested that the τ91 subunit somehow contributed to TFIIIC-DNA binding.
FIG. 4.
Temperature sensitivities of mutant, suppressor, and wild-type TFIIIC. Ultrogel-heparin-purified mutant (Mut) (tfc3-G349E) (•), suppressor (Sup) (tfc3-G349E/TFC6-E330K) (▪), and wild-type (WT) (▴) TFIIIC fractions (∼0.7 μg in 2 μl) were preincubated in standard binding or transcription buffer at the indicated temperatures for 10 min before their DNA binding or in vitro transcription activity was tested. (A) DNA binding activities of TFIIIC mutants. TFIIIC fractions were incubated with a tRNA3Glu gene probe at 25°C for 15 min, and the complexes were analyzed by gel retardation electrophoresis. (B) Complex formation was quantified in a PhosphorImager and is shown as the percentage of complexes formed after preincubation at 20°C. (C and D) Transcription activities of TFIIIC mutants. The tRNA3Leu gene from plasmid pGE2 was transcribed in vitro at 25°C for 45 min in the presence of TFIIIC, recombinant TFIIIB70 and TBP, B", and Pol III. Transcripts were separated in a polyacrylamide gel (C) and quantified in a PhosphorImager (D). In panel D, data are shown as percentages of tRNA3Leu synthesis after preincubation of TFIIIC at 25°C.
The suppressor activity of τ91-E330K was also detectable in transcription assays in vitro. Partially purified preparations of wild-type, mutant, or suppressor TFIIIC were preincubated at different temperatures and added to a transcription system reconstituted with the yeast tRNA3Leu gene, recombinant TBP and TFIIIB70, B" fraction, and purified Pol III. As shown in Fig. 4D, the mutant factor lost half of its transcription activity at 30°C, while the wild-type and suppressor factors remained fully active. At 35°C, the mutant factor was inactivated, but the suppressor retained at least 50% of its activity. The wild-type factor retained activity even up to 40°C. When comparing the temperature-response curves in the DNA binding and transcription assays, it was clear that the partial suppression of the transcription defect by τ91-E330K was well accounted for by the improvement in TFIIIC-DNA binding. The fact that the transcription factor activity of TFIIIC was somewhat less sensitive to heat treatment than its DNA binding activity probably reflects the stabilization of factor-DNA complexes by TFIIIB.
The interaction between τ91 and τ138 subunits, suggested by protein-DNA cross-linking experiments (9) and the present genetic evidence, was further investigated by using the yeast two-hybrid system (12). Possible interactions between τ91 and all of the other class III transcription components (14 Pol III subunits and the components of TFIIIB and TFIIIC) were also analyzed. τ91 was fused to the DNA binding or activation domain of GAL4 and assayed against all the reciprocal class III protein fusions. No interaction could be seen, although the τ91 fusions were well expressed in this yeast system (Fig. 2 and results not shown).
As the suppressor mutation partially restored the TFIIIC-DNA interaction, we explored the possibility that τ91 polypeptide could interact directly with DNA. The DNA binding activity of recombinant τ91 protein was first detected in a Southernwestern analysis in which denatured-renatured recombinant His6-HA-tagged τ91 (rτ91) bound strongly a 32P-labelled poly(dA · dT) probe (data not shown). There was no equivalent DNA binding when nonrenatured rτ91 or other recombinant His6-HA-tagged TFIIIC subunits, such as rτ95 or rτ55 (36), were used. The DNA binding properties of τ91 were further analyzed in gel retardation assays. The recombinant τ91 polypeptide was purified to apparent homogeneity in three steps (Ni2+ affinity chromatography in a fast protein liquid chromatography Hitrap column, chromatography on a Smart Heparin-HyperD column, and then gel filtration through a Smart Superdex-75 column) and used in gel shift assays with different nucleic acid probes (Fig. 5). No band shift could be detected after incubation of rτ91 with the full-length tRNA3Glu gene (lanes 7 to 9). However, under the same conditions, rτ91 interacted strongly with the same heat-denatured DNA probe. Three complexes of decreasing mobility were observed (Fig. 5, lanes 4 to 6). They probably corresponded to the binding of one, two, or three molecules of rτ91 per DNA strand, as suggested by protein titration experiments (results not shown). The protein retarded the migrations of the two separated DNA strands with about the same efficiency, which was further indicative of a nonspecific interaction. A nonspecific interaction with single-stranded nucleic acids may reflect a role in DNA or RNA binding during the transcription process. When incubation was with a labelled tRNAMet probe, a weak complex was detected (Fig. 5, lanes 1 to 3). This interaction might point to a role of τ91 in binding the nascent RNA transcript. This possibility was not further investigated.
FIG. 5.
Nucleic acid binding activity of rτ91 protein. About 2 ng of highly purified rτ91 protein was incubated with the different nucleic acid probes in 15-μl mixtures containing 20 mM Tris-HCl (pH 8.0), 120 mM KCl, 20 μg of bovine serum albumin, 10% glycerol, 10 to 20 fmol (10,000 cpm) of 32P-5′-end-labelled nucleic acid probe, and, when indicated, 2.5 mM MgCl2. After 15 min of incubation at 25°C, rτ91-nucleic acid complexes were separated from free DNA or tRNA by nondenaturing electrophoresis on an 8% polyacrylamide gel. The nucleic acid probes were 20 fmol of native spleen tRNAMet (tRNA) (lanes 1 to 3), 20 fmol of 200-nucleotide-long single-stranded tRNA3Glu gene (ssDNA) (lanes 4 to 6), and 10 fmol of 200-bp double-stranded tRNA3Glu gene (ds DNA) (lanes 7 to 9). The arrowhead indicates the rτ91-tRNAMet complex, and arrows point to the rτ91–single-stranded tDNA complexes.
DISCUSSION
Yeast TFIIIC is a multisubunit transcription factor that displays a remarkable adaptation to multiple DNA sequences and protein targets, leading to the formation of stable preinitiation complexes. Pursuing the analysis of this factor, we report here the isolation and characterization of TFC6, the gene encoding the 91-kDa polypeptide of TFIIIC, and present biochemical and genetic evidence that τ91 is an essential subunit of TFIIIC which participates with τ138 in TFIIIC-DNA interaction.
τ91 is an essential subunit of TFIIIC.
Highly purified preparations of TFIIIC obtained in different laboratories consistently contained a 91-kDa polypeptide which was initially disregarded as a potential TFIIIC subunit because it was weakly stained by silver and was present in variable ratios with respect to other subunits (3, 17, 42). Nevertheless, it comigrated with TFIIIC components in nondenaturing gel electrophoresis (3, 16, 17, 42), coimmunoprecipitated with τ138, τ131, and τ95 subunits in absence of target DNA (13), and, most remarkably, was specifically located at the 3′ end of the 5S RNA genes in TFIIIC-TFIIIA-DNA complexes (9). We isolated the gene encoding τ91 by using the amino acid sequence information obtained from the gel-purified polypeptide. Mouse polyclonal antibodies directed to the recombinant protein supershifted TFIIIC-tDNA complexes in gel shift assays, thus confirming the stable association of τ91 within the factor-DNA complexes in vitro. To assess the implication of τ91 in class III gene activation in vivo, we used a chimeric system in which a B block-deficient SNR6 gene harboring GAL4 binding sequences (UASG) is reactivated when the cells express the τ138 or τ131 subunit of TFIIIC fused to the GAL4 DNA binding domain (39). Similarly, expression of the GAL4(1-147)-τ91 fusion activated UASG-containing SNR6 genes to various extents, depending on the position of the UASG sequences. This result indicated that τ91 contributed to the formation of the preinitiation complexes in vivo by anchoring TFIIIC on the UASG template and confirmed its status as a subunit of TFIIIC. The fact that TFIIIC remained functional (i.e., able to properly assemble TFIIIB) in vivo when anchored on the DNA through either one of four distinct subunits (τ138, τ131, τ95, and τ91) illustrates the extraordinary flexibility of the TFIIIC-SNR6 interaction.
It is remarkable that all the components of the yeast Pol III transcription system characterized to date (14 subunits of Pol III, the 3 components of TFIIIB, and 3 subunits of TFIIIC) are encoded by unique and essential genes. The τ91 subunit of TFIIIC does not depart from this rule, since haploid cells with TFC6 deleted are not viable.
τ91 cooperates with τ138 in TFIIIC-tDNA complexes.
Starting from the tfc3-G349E mutation, which alters τ138 and affects TFIIIC-DNA binding, we isolated a suppressor mutation causing an E330K substitution in τ91 that restored cell growth at nonpermissive temperatures. This genetic interaction between τ138 and τ91 suggested that τ91 contributes to DNA binding along with τ138. Indeed, the double-mutant form of TFIIIC (tfc3-G349E/TFC6-E330K) has significantly improved DNA binding and transcription activities compared to the single-mutant TFIIIC (tfc3-G349E). This observation suggested that τ91 stabilized the τ138-DNA interaction. Whether τ91 participates directly in DNA binding remains an open question. The sequencing of τ91 did not shed much light on the possible function of the polypeptide, but the central cluster of 13 Cys and His residues suggests a metal binding site and a possible role in DNA binding. The suppressor mutation itself lies in the Cys-His cluster. It was previously noted that the formation of TFIIIC-tDNA and τB-tDNA complexes was inhibited by the zinc chelator 1,10-phenanthroline, while EDTA had no effect (17). Since no obvious zinc binding domain has been detected in the yeast TFIIIC subunits cloned so far (τ138, τ131, and τ95, τ91 could be the target of the chelator.
In mobility shift experiments, rτ91 displayed a strong affinity for single-stranded DNA, weakly bound tRNA, but did not interact detectably with a full-length double-stranded tRNA gene. The significance of this nucleic acid binding activity is unclear. RNA interaction might point to a role of τ91 in tRNA transcript release or tRNA processing. RNA-protein cross-linking experiments with Pol III transcription complexes have suggested contacts between human TFIIIC and the nascent transcript (5). Competitor single-stranded DNA was found to affect TFIIIC-tDNA interaction, primarily at the level of the A block (40, 52). These observations suggested that the single-stranded DNA binding component belongs to the τA domain and raise the question of the location of τ91 within the TFIIIC-tDNA complexes. A polypeptide the size of τ91 was located at the 3′ end of the 5S RNA gene by site-directed photo-cross-linking (9) and was clearly distinct from the closely migrating 95-kDa polypeptide (τ95), which itself was cross-linked in the vicinity of the A block in tRNA genes or at a similar location in the 5S RNA gene (3, 9). τ95 and τ91 have very close calculated molecular masses (∼74 kDa), but it seems unlikely that the residual cross-linked nucleotides inverted the migrations of these two polypeptides in SDS gels. In addition, a β-galactosidase–τ95 fusion incorporated into TFIIIC was located in the τA domain by electron microscopy (13). By inference, and in agreement with the present genetic evidence, τ91 is very likely part of the τB domain together with τ138. Nevertheless, our attempts to demonstrate the presence of τ91 in the protease-resistant τB-tDNA complex have failed. Anti-τ91 polyclonal antibodies shifted the migration of TFIIIC-tDNA but not τB-tDNA complexes. The proteolysed complex may have lost the τ91 epitopes recognized by the antibodies (as well as the single-stranded-DNA binding activity). Indeed, τ91 appears to be very sensitive to proteolysis within TFIIIC-tDNA complexes, because anti-τ91 antibody reactivity (as detected in supershift assays) was totally lost under limited proteolysis conditions that did not generate τB-tDNA complexes (results not shown). The protease sensitivity of τ91 may also explain the frequent presence of the τ91-related 75-kDa polypeptide in purified TFIIIC fractions.
Relationship between yeast TFIIIC and human TFIIIC.
Four subunits of yeast TFIIIC (τ138, τ131, τ95, and τ91) have now been cloned. No significant sequence similarity was found between τ138 or τ91 and other protein sequences in current data banks, including the known subunits of human TFIIIC2, TFIIICα and -β (30, 35, 51). The B block binding subunit, TFIIICα, being almost twice as large as τ138, may conceivably correspond to a fusion of two or more polypeptides, but this appears to be unlikely as TFIIICα shows no significant similarity to τ91, τ138, or the two smallest subunits of yeast TFIIIC, τ60 and τ55 (36). This evolutionary divergence need not mean that the basic mechanisms of Pol III gene activation are different in yeast and human cells. It remains possible that τ91 (or even τ138) has a human counterpart in TFIIIC2 components yet to be cloned or in the TFIIIC1 fraction, which extends the footprint of TFIIIC2 to downstream sequences over the termination region (57). It is nevertheless remarkable that τ131 (the TFIIIB-assembling subunit of TFIIIC) and τ95 are, instead, evolutionarily conserved, as shown by the existence of sequence homologs in the C. elegans genome, in data banks of human cDNA (τ131), and in the S. pombe genome (τ95). Human homologs of these polypeptides presumably belong to TFIIIC2 (57). There is also a clear sequence similarity between the human TFIIIB90 and yeast TFIIIB70/BRF1 (56). Furthermore, a functional interchangeability of human and yeast TFIIIB components was recently demonstrated in vitro, pointing to the existence of a human homolog of yeast TFIIIB90/B" (54). Finally, the yeast Pol III subunits (C34 and C31) involved in TFIIIB recognition and chain initiation (2, 28, 31, 55, 59) are conserved in human Pol III (58). Thus, the complex protein-protein interactions involved in TFIIIB assembly and Pol III recruitment have clearly been conserved among eucaryotes, while the recruitment of TFIIIC on the B block element may involve fairly divergent polypeptide structures.
ACKNOWLEDGMENTS
We are grateful to Anny Ruet and Yveline Frobert for help in raising mouse polyclonal antibodies, to Janine Huet and Emmanuel Favry for recombinant TBP, recombinant TFIIIB70, and Pol III preparations, and to Martin Lanzendörfer for advice on recombinant protein purification and single-stranded DNA binding. We thank Michel Riva and Françoise Bouet for their help in peptide microsequence determination. We thank Christian Marck for pointing out to us the presence of τ131 and τ95 homologs in data banks and for helpful discussions, and we thank Carl Mann for improving the manuscript.
This work was supported by a grant from the European Union BIOTECH program (to A.S.). R.A. was supported by a long-term postdoctoral FEBS fellowship. N.M. and S.R. were supported by a fellowship from the French Ministére de l’Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Baker R E, Hall B D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984;3:2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomew B, Durkovich D, Kassavetis G A, Geiduschek E P. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartholomew B, Meares C F, Dahmus M E. Photoaffinity labelling of RNA polymerase III transcription complexes by nascent RNA. J Biol Chem. 1990;265:3731–3737. [PubMed] [Google Scholar]
- 6.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulanger P A, Yoshinaga S K, Berk A J. DNA-binding properties and characterization of human transcription factor IIIC2. J Biol Chem. 1987;262:15098–15105. [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Braun B R, Bartholomew B, Kassavetis G A, Geiduschek E P. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J Mol Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 10.Burnol A-F, Margottin F, Huet J, Almouzni G, Prioleau M-N, Méchali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993;362:475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- 11.Burnol A F, Margottin F, Schultz P, Marsolier M C, Oudet P, Sentenac A. Basal promoter and enhancer elements of yeast U6 snRNA gene. J Mol Biol. 1993;233:644–658. doi: 10.1006/jmbi.1993.1542. [DOI] [PubMed] [Google Scholar]
- 12.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 13.Conesa C, Swanson R N, Schultz P, Oudet P, Sentenac A. On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/τ. J Biol Chem. 1993;268:18047–18052. [PubMed] [Google Scholar]
- 14.Dean N, Berk A J. Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol Cell Biol. 1988;8:3017–3025. doi: 10.1128/mcb.8.8.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean N, Berk A J. Separation of TFIIIC into two functional components by sequence specific DNA affinity chromatography. Nucleic Acids Res. 1987;15:9895–9907. doi: 10.1093/nar/15.23.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabrielsen O S, Huet J. Magnetic DNA affinity purification of yeast transcription factor. Methods Enzymol. 1993;218:508–525. doi: 10.1016/0076-6879(93)18038-e. [DOI] [PubMed] [Google Scholar]
- 17.Gabrielsen O S, Marzouki N, Ruet A, Sentenac A, Fromageot P. Two polypeptide chains in yeast transcription factor τ interact with DNA. J Biol Chem. 1989;264:7505–7511. [PubMed] [Google Scholar]
- 18.Gabrielsen O S, Oyen T B. The requirement for the A block promoter in tRNA gene transcription in vitro depends on the ionic environment. Nucleic Acids Res. 1987;15:5699–5713. doi: 10.1093/nar/15.14.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 20.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 21.Harris S D, Pringle J R. Genetic analysis of Saccharomyces cerevisiae chromosome I: on the role of mutagen specificity in delimiting the set of genes identifiable using temperature-sensitive-lethal mutations. Genetics. 1991;127:279–285. doi: 10.1093/genetics/127.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf D H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 23.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 24.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 25.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 107–126. [Google Scholar]
- 26.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 29.Kovelman R, Roeder R G. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267:24446–24456. [PubMed] [Google Scholar]
- 30.Lagna G, Kovelman R, Sukegawa J, Roeder R G. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol Cell Biol. 1994;14:3053–3064. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lannutti B, Persinger J, Bartholomew B. Probing the protein-DNA contacts of a yeast RNA polymerase III transcription complex in a crude extract: solid phase synthesis of DNA photoaffinity probes containing a novel photoreactive deoxycytidine analog. Biochemistry. 1996;35:98212–9831. doi: 10.1021/bi960525x. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence C W. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- 33.Lefebvre O, Carles C, Conesa C, Swanson R N, Bouet F, Riva M, Sentenac A. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor TFIIIC. Proc Natl Acad Sci USA. 1992;89:10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefebvre O, Rüth J, Sentenac A. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J Biol Chem. 1994;269:23374–23381. [PubMed] [Google Scholar]
- 35.L’Etoile N D, Fahnestock M L, Shen Y, Aebersold R, Berk A J. Human transcription factor IIIC box B binding subunit. Proc Natl Acad Sci USA. 1994;91:1652–1656. doi: 10.1073/pnas.91.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manaud, N., E. Deprez, R. Arrebola, and C. Conesa. Unpublished results.
- 37.Marck C. “DNA strider”: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marck C, Lefebvre O, Carles C, Riva M, Chaussivert N, Ruet A, Sentenac A. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeat and basic-helix-loop-helix motifs. Proc Natl Acad Sci USA. 1993;90:4027–4031. doi: 10.1073/pnas.90.9.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsolier M-C, Chaussivert N, Lefebvre O, Conesa C, Werner M, Sentenac A. Directing transcription of an RNA polymerase III gene via GAL4 sites. Proc Natl Acad Sci USA. 1994;91:11938–11942. doi: 10.1073/pnas.91.25.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marzouki N, Camier S, Ruet A, Moenne A, Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor τ. Nature. 1986;323:176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- 41.Parsons M C, Weil P A. Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J Biol Chem. 1992;267:2894–2901. [PubMed] [Google Scholar]
- 42.Parsons M C, Weil P A. Purification and characterization of Saccharomyces cerevisiae transcription factor TFIIIC. Polypeptide composition defined with polyclonal antibodies. J Biol Chem. 1990;265:5095–5103. [PubMed] [Google Scholar]
- 43.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 44.Rameau R, Puglia K, Crowe A, Sethy I, Willis I M. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol Cell Biol. 1994;14:822–830. doi: 10.1128/mcb.14.1.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozenfeld, S., and P. Thuriaux. Unpublished results.
- 46.Rüth J, Conesa C, Dieci G, Lefebvre O, Düsterhöft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz P, Marzouki N, Marck C, Ruet A, Oudet P, Sentenac A. The two DNA-binding domains of yeast transcription factor τ as observed by scanning transmission electron microscopy. EMBO J. 1989;8:3815–3824. doi: 10.1002/j.1460-2075.1989.tb08559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sethy I, Willis I M. Recessive mutations in the second largest subunit of TFIIIC suggest a new step in RNA polymerase III transcription. Gene Exp. 1995;5:35–47. [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinn E, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of a TFIIIC2 subunit (TFIIICβ) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- 52.Stillman D J, Caspers P, Geiduschek E P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985;40:311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- 53.Swanson R N, Conesa C, Lefebvre O, Carles C, Ruet A, Quemeneur E, Cagnon J, Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor τ (TFIIIC) Proc Natl Acad Sci USA. 1991;88:4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teichmann M, Dieci G, Huet J, Rüth J, Sentenac A, Seifart K. Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J. 1997;16:4708–4716. doi: 10.1093/emboj/16.15.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Roeder R G. TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Roeder R G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 59.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 60.Willis I, Oksman A, López-De-León A. The PCF1-1 mutation increases the activity of the transcription factor (TF) IIIB fraction from Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:3725–3730. doi: 10.1093/nar/20.14.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 62.Yoshinaga S K, Boulanger P A, Berk A J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci USA. 1987;84:3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshinaga S K, L’Etoile N D, Berk A. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]