Abstract
The transcription factor AREB6 contains a homeodomain flanked by two clusters of Krüppel type C2H2 zinc fingers. AREB6 binds to the E-box consensus sequence, CACCTGT, through either the N- or the C-terminal zinc finger cluster. To gain insights into the molecular mechanism by which AREB6 activates and represses gene expression, we analyzed the domain structure of AREB6 in the context of a heterologous DNA-binding domain by transient-transfection assays. The C-terminal region spanning amino acids 1011 to 1124 was identified as a conventional acidic activation domain. The region containing amino acids 754 to 901, which was identified as a repression domain, consists of 40% hydrophobic amino acids displaying no sequence similarities to other known repression domains. This region repressed transcription in vitro in a HeLa nuclear extract but not in reconstituted transcription systems consisting of transcription factor IID (TFIID), TFIIB, TFIIE, TFIIH/F, and RNA polymerase II. The addition of recombinant negative cofactor NC2 (NC2α/DRAP1 and NC2β/Dr1) to the reconstituted transcription system restored the activity of the AREB6 repression domain. We further demonstrated interactions between the AREB6 repression domain and NC2α in yeast two-hybrid assay. Our findings suggest a mechanism of transcriptional repression that is mediated by the general cofactor NC2.
AREB6 is a zinc finger-homeodomain transcription factor whose cDNA was isolated from a HeLa cell expression library with a probe of the Na,K-ATPase α1 subunit gene (Atp1a1) positive regulatory element (ARE). AREB6 regulates the Atp1a1 positively or negatively depending on cell types and in a concentration-dependent manner (55). AREB6 is also known as ZEB, which was identified as a repressor on the immunoglobulin heavy-chain enhancer (14). BZP is a golden hamster homolog of AREB6 and has been reported to change its location from the nucleus to the cytoplasm in response to serum deprivation (12). There is increasing evidence that AREB6 plays important roles in the expression of tissue-specific genes and in various developmental processes. AREB6 works as a negative transcription factor for the interleukin 2 (IL-2) gene to turn off the IL-2 gene transcription just after T-cell activation (56). In anergic T cells, caused by an incomplete T-cell activation, AREB6 plays a key role in the repression of IL-2 gene expression (5). Homozygous AREB6-null mice show severe skeletal defects, such as craniofacial defects, malformation of limbs, lack of invertebral disks, and irregular branching and fusion of ribs (20). Homozygous mice having truncation of the C-terminal zinc finger cluster show defects in early T-cell development (21). These observations suggest that AREB6 regulates various genes by interacting with proteins, including transcription factors in specific tissues and in different developmental stages. Interestingly, AREB6 has three potential DNA-binding domains, i.e., two separated Krüppel type C2H2 zinc finger clusters near the N and C termini and a homeodomain located between them. The homeodomain of AREB6 has no specific DNA-binding activity but interacts with the N-terminal zinc finger domain of AREB6 itself (24).
To understand the molecular basis by which AREB6 activates and represses gene transcription, functional, genetic, and structural studies are indispensable. In our previous study, we demonstrated binding of AREB6 to the consensus E box with the sequence CACCTGT through the N- or C-terminal zinc finger domain (24). We also observed that AREB6 regulates gene transcription either positively or negatively depending on alternative DNA-binding modes, through either the N-terminal or the C-terminal zinc finger domain (24). In the present study, the transcriptional activation and repression domains of AREB6 were identified in the context of a heterologous DNA-binding domain in vivo by transient-transfection assays. The activation domain resides in a glutamic acid-rich region, while the repression domain lies in a hydrophobic region near the C-terminal zinc finger cluster. Transcriptional repression was reconstituted in an in vitro transcription system. We provide evidence for a novel mechanism of transcriptional repression mediated through the general negative transcription cofactor NC2 (39).
MATERIALS AND METHODS
Plasmid constructs.
GAL4 fusion AREB6 domains for cotransfection assays were constructed by subcloning various AREB6 domain-derived sequences into the vector pCMV-GAL4(1–147), which is also known as the GAL4 DNA-binding domain (GDBD). The KpnI-HpaI fragment (nucleotides 1 to 1030), the HpaI-ApaLI fragment (nucleotides 1031 to 2174), and the PmaCI-XbaI fragment (nucleotides 3031 to 3387) of pSVSPORT1/AREB6 (55) were blunt ended and ligated with 8-mer, 12-mer, and 12-mer XbaI linkers, respectively. They were introduced into the XbaI site of pCMV-GAL4(1–147), generating GDBD-AREB6(1–343), GDBD-AREB6(344–726), and GDBD-AREB6(1011–1124), respectively. The ApaLI-PmaCI fragment (nucleotides 2175 to 3030) was blunt ended, ligated with the 12-mer XbaI linker and introduced into the 12-mer XbaI linker-ligated XbaI site of pCMV-GAL4(1–147), generating GDBD-AREB6(726–1010). For 5′- and 3′-deletion mutation constructs between amino acids (aa) 726 and 1010 of AREB6, XbaI-linearized GDBD-AREB6(726–1010) was partially digested with BAL 31 nuclease. The digested ends were filled in with Klenow fragment and coupled with a corresponding length of XbaI linkers to generate an in-frame amino acid junction with GAL4(1–147). Point mutation constructs of GDBD-AREB6(726–1010) were generated by the cassette mutagenesis method and confirmed by sequencing. The mutations were introduced with the following oligonucleotides (the coding sequences of oligonucleotides are described): AREB6(754–901)S762A, serine to alanine at aa 762 (5′-AACAGTGTTTATGCTGTCCAGGAAGAA); AREB6(754–901)N769Q, asparagine to glutamine at aa 769 (5′-AAGAACCCTTGCAGTTGTCTTGCGCA); AREB6(754–901)N885Q, asparagine to glutamine at aa 885 (5′-GTAGAGGATCAGCAGGACTCTGATTCT); AREB6(754–901)K894T, lysine to threonine at aa 894 (5′-ACACCGCCCAAAACGAAAATGCGGAA); AREB6(754–901)N769Q, K894T, asparagine to glutamine at aa 769 and lysine to threonine at aa 894 [the same oligonucleotide as for K894T with a template of AREB6(759–901)N769Q].
Plasmids encoding the histidine-tagged GAL4(1–147) and GAL4 fusion AREB6 proteins were constructed as follows. His6T7-11d (58) was cut with NdeI and BamHI and inserted with NdeI-BamHI fragments from pCMV-GAL4(1–147), GDBD-AREB6(754–901), and GDBD-AREB6(754–901)N769Q. The resulting plasmids were His6GAL4-11d, His6GAL4AREB6(754–901)-11d, and His6GAL4AREB6(754–901)N769Q-11d, respectively. Their expressed proteins were named GAL4(1–147), RD, and RDm, respectively.
For two-hybrid constructs, pJG4-5/AREB6(726–1010), pJG4-5/AREB6(796–1010), pJG4-5/AREB6(829–1010), and pJG4-5/AREB6(726–829) were made by amplifying the coding region and were inserted into the EcoRI site of the pJG4-5 yeast vector. For pJG4-5/AREB6(754–901) and pJG4-5/AREB6(754–901)N769Q, the fragment from 2260 to 2703 and that from 2260 to 2703 with the N769Q mutation were ligated with an 8-mer EcoRI linker and inserted into the EcoRI site of the pJG4-5 vector. For pEG202/NC2α (LexA-NC2α), the BstEII-BamHI fragment of NC2α (the histone fold region in the N terminus was deleted) was blunt ended and inserted into the blunt-ended BamHI-linearized pEG202. LexA-NC2β was kindly provided by Danny Reinberg.
Cell culture, transient transfections, and reporter gene assays.
The mouse myoblast cell line C2C12 was grown in Dulbecco’s modified Eagle’s medium with a high glucose concentration (4,500 mg/liter) supplemented with 10% fetal calf serum (growth medium). A total of 2 × 105 cells in a 60-mm dish were cotransfected by the calcium phosphate precipitation method as described previously (26), with 4 μg of reporter plasmid and 2 μg of pCMV-GAL4(1–147) or various GDBD-AREB6 plasmids. After 12 h of transfection, the cells were refed with growth medium. The cells were cultivated for a further 36 to 40 h and then harvested. The reporter gene assays for chloramphenicol acetyltransferase (CAT) and luciferase were performed as described previously (26). The reporter plasmid UAS-HTLV1-CAT, which contains the human T-cell leukemia virus type 1 (HTLV-1) promoter harboring five synthetic GAL4 DNA-binding sites (46), was provided by M. Okuda. The other reporter, tk-Galpx3-LUC, which contains the herpes simplex virus thymidine kinase (tk) gene promoter (from −105 to +51) harboring three GAL4 DNA-binding sites, was provided by K. Umezono and was described previously (23). pEF-BOS/βGAL at 0.5 μg (50) was used as an internal control for transfection efficiency. CAT and luciferase activities were normalized with β-galactosidase activity in the same cell lysate.
In vitro transcription reactions.
Preparation of HeLa nuclear extract and phosphocellulose (P11) column fractionation were performed as previously described (7, 40). RNA polymerase II (29) and transcription factor IID (TFIID) (33) were purified from HeLa nuclear extract. The purified TFIID contained substantial amounts of TFIIA (39). As for TFIIH/F, the P11 0.5 M KCl fraction from the HeLa nuclear extract was purified on a DE52 column and pooled fractions containing TFIIH/F (80 to 120 mM KCl) were loaded on a MonoQ column and eluted between 180 and 260 mM KCl. Recombinant TFIIB and TFIIEα/β were expressed and purified as described previously (33). We used 5 μl of HeLa nuclear extract (8 μg of protein per μl) for the transcription reactions with HeLa extract, 2 μl of P11 0.5 M KCl fraction (2.8 μg of protein per μl), and 2 μl of P11 0.85 M KCl fraction (3.8 μg of protein per μl) for the reactions with the P11 fractions. Since the P11 0.5 M KCl fraction contains limiting amounts of TFIIB, we used 10 ng of recombinant TFIIB as a supplement to the P11 0.5 M KCl fraction. In a reconstituted transcription system, we used 10 ng of recombinant TFIIB, 0.8 μl of TFIID (DE52 fraction, 0.35 μg of protein per μl), 10 ng of recombinant TFIIEα, 5 to 10 ng of recombinant TFIIEβ, 0.2 μl of RNA polymerase II (DE52 fraction, 0.5 μg of protein per μl), and 0.8 μl of the TFIIH/F fraction (0.95 μg of protein per μl). For preparation of the heat-treated P11 0.5 M KCl fraction, the P11 0.5 M KCl fraction (2.8 μg of protein per μl) was heat treated at 55°C for 15 min and centrifuged at 12,000 × g for 2 min, and 2 μl of supernatant was used. Recombinant NC2 (NC2α and NC2β/Dr1) was expressed and purified as described previously (16). NC2 standard concentrations (4 ng of NC2α per μl and 30 ng of NC2β per μl) were referred to as 4U as described previously (16). Histidine-tagged GAL4(1–147) and GAL4 fusion AREB6 proteins (RD and RDm) were expressed in Escherichia coli, purified under denaturing conditions on Ni-nitrilotriacetic acid columns (Qiagen), and renatured by differential dialysis. All transcription reactions were performed with 9 ng of linearized plasmid templates, i.e., BstEII-linearized tk-Galp3x-LUC and SmaI-linearized pMRG5. pMRG5 contains the human immunodeficiency virus (HIV) core promoter downstream of five GAL4 DNA-binding sites (33). Transcription reaction mixtures contained 25 mM HEPES-KOH (pH 8.2), 10% glycerol, 4 mM MgCl2, 60 to 65 mM KCl, 5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 500 ng of bovine serum albumin per μl, and 20 U of RNase-Block (Toyobo). UTP, ATP, and GTP (100 μM each), 5 μM CTP, and 0.5 μM [α-32P]CTP (3,000 Ci/mmol) were added for the reaction with tk-Galpx3-LUC, and 100 μM (each) UTP and ATP, 5 μM CTP, 20 μM 3′-o-methyl-GTP, and 0.5 μM [α-32P]CTP (3,000 Ci/mmol) were added for the reaction with pMRG5. As indicated in the figures, 5 or 10 ng of effector proteins [GAL4(1–147), RD, and RDm] was added to the transcription buffer containing the templates and incubated for 10 min at 28°C, followed by the addition of premixed general transcription factors (GTFs). Heat-treated P11 0.5 M KCl fraction or various units of recombinant NC2 were added to the reaction mixture after a 10-min incubation with effector proteins and incubated for 5 min at 28°C before the GTFs were added. For quantification of individual transcripts, dried gels were scanned and quantified with an Instant Imager (Packard).
Yeast two-hybrid interaction assays.
Yeast strains (EGY48) and interaction assays were as previously described (8a). Briefly, the cells were cotransformed with pJG4-5 constructs and pEG202 constructs by the lithium acetate method and selected with Ura−His−Trp− medium. Each double transformant was plated on Ura−His−Trp−Leu− galactose with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates for an interaction assay. An interaction was determined as positive if the transformant became Leu+ and turned blue on X-Gal indicator plates.
RESULTS
Characterization of functional domains of AREB6.
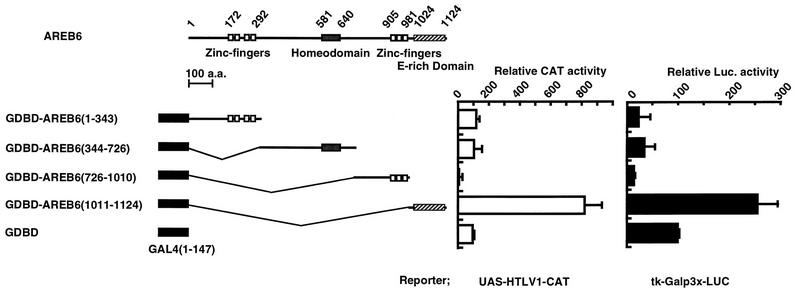
AREB6 is organized in a unique structure of multiple functional domains, containing two zinc finger clusters separated by a homeodomain (24, 55). To identify the domains required for transcriptional activation and repression, respectively, various regions of AREB6 were fused to the GDBD (Fig. 1). GDBD consists of a DNA-binding domain (aa 1 to 90 of GAL4) and a cryptic activation domain (aa 90 to 147) (36). GDBD-AREB6(1–343) contains the N-terminal zinc finger cluster composed of three C2H2-type and one C2HC-type zinc fingers. GDBD-AREB6(344–726) contains the homeodomain. GDBD-AREB6(726–1010) contains the C-terminal zinc finger cluster composed of three C2H2-type zinc fingers. GDBD-AREB6(1011–1124) contains the glutamic acid-rich region. These fusion proteins were tested for their ability to activate or repress gene expression by using reporter plasmids in C2C12 myoblast cells. We chose myoblast cells because we observed that the AREB6 mRNA is abundant in skeletal muscle (55) and the AREB6 protein is produced in myoblast cell lines (25). All associated factors required for the expression of AREB6 regulatory function should exist in these cells. We used two different reporter plasmids, one which contains the HTLV-1 promoter harboring five GAL4 DNA-binding sites fused with the CAT gene (UAS-HTLV1-CAT) and one which contains the tk gene promoter harboring three GAL4 DNA-binding sites fused with the luciferase gene (tk-Galp3x-LUC). As shown in Fig. 1, cotransfection of the GDBD-AREB6(1011–1124) stimulated the activities of the HTLV-1 promoter and the tk promoter about 8-fold and 2.5-fold, respectively, with GDBD as a standard. The region from positions 1011 to 1124 of AREB6 contains 46% acidic amino acids (39% glutamic acid). Cotransfection of plasmids GDBD-AREB6(1–343) and GDBD-AREB6(344–726) had little effect on HTLV-1 promoter activity (Fig. 1, left panel) and moderately repressed the tk promoter (right panel). On the other hand, GDBD-AREB6(726–1010) inhibited the HTLV-1 promoter to 10% and the tk promoter to 15% of the original levels. Since GDBD-AREB6(726–1010) contains the DNA-binding domain which recognizes the E box (CANNTG) (24), it cannot be ruled out that repression is mediated through binding to E-box sequences present in the reporter plasmids. Therefore, we examined reporters lacking GAL4 DNA-binding sites. GDBD-AREB6(726–1010) had little effect (data not shown) on these HTLV-1 and tk promoters, arguing against a direct involvement of the AREB6 DNA-binding region. These results indicate that the region of AREB6(726–1010) functions as a repression domain which is dependent on tethering to the promoter through the heterologous GDBD.
FIG. 1.
Identification of the activation domain and the repression domain of AREB6. Transient-transfection assays were performed in C2C12 cells with the UAS-HTLV1-CAT (left graph) and tk-Galp3x-LUC (right graph) reporter plasmids. The effector plasmids, indicated on the left, express proteins of various portions of the AREB6 fused to the GDBD or the GDBD alone (aa 1 to 147). A plasmid encoding β-galactosidase was included as an internal control for normalizing transfection efficiency. A schematic representation of the structure of the AREB6 protein is displayed at the top. Zinc finger domains (open boxes), a homeodomain (shaded box), and a glutamic acid-rich domain (E-rich, striped box) are shown. Values are represented as relative CAT or luciferase (Luc.) activity with respect to the activity of GDBD, which was set at 100. All transfection assays were repeated three to five times in duplicate.
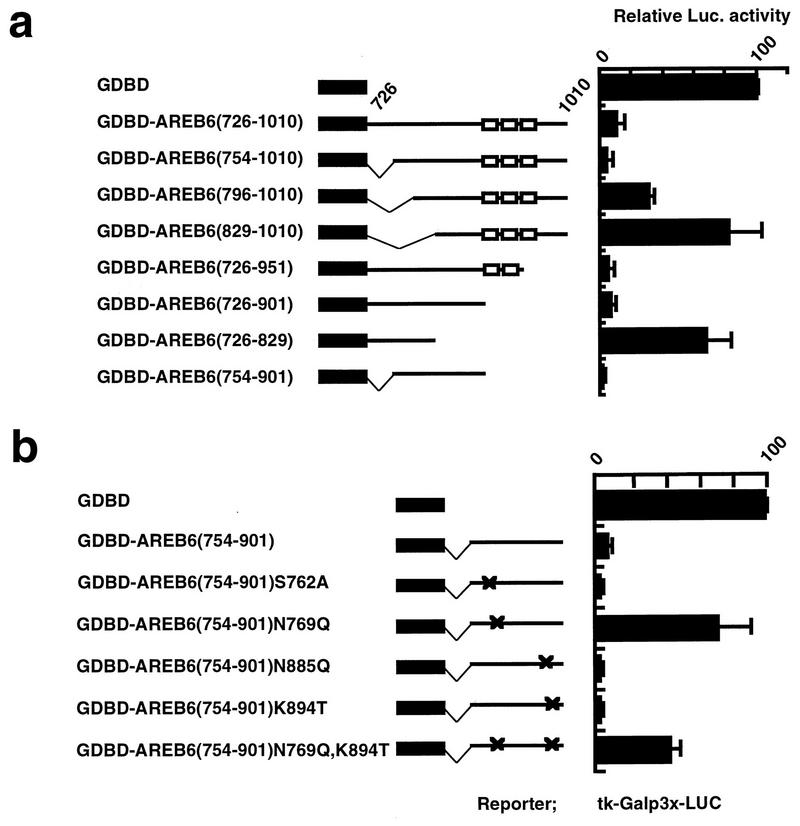
To map the precise location of the repression domain, we constructed fine-deletion and point mutations and tested them for their ability to repress reporter gene expression (Fig. 2). A 5′ deletion to aa 754 and a 3′ deletion to aa 901 had little effect on repression activity, while a 5′ deletion to aa 796 and a 3′ deletion to aa 829 impaired repression activity. The minimal repression domain was revealed to reside within the region spanning aa 754 to 901 of AREB6 (Fig. 2a). This region consists of 40% hydrophobic amino acids and is relatively rich in proline (11%). It has no sequence similarities to other known hydrophobic repression domains, such as the thyroid hormone receptor and retinoic acid receptor α (4) and the proline-rich domain of RGM1 (9). However, sequence comparisons of AREB6(754–901) with the homologous proteins of other species revealed a high degree of identity to mouse (97%), golden hamster (96%), and chicken (93%) proteins. On the other hand, sequence comparison of the activation domain of AREB6(1011–1124) showed lower identities to mouse (69%), golden hamster (57%), and chicken (51%) sequences.
FIG. 2.
Identification of the repression domain by fine-deletion mutation and point mutation constructs. (a) Luciferase activities of GDBD-fused 5′- and 3′-deletion mutation constructs of AREB6(726–1010) relative to that of GDBD, which was set as 100. tk-Galp3x-LUC was used as a reporter. (b) Luciferase activities of GDBD-fused various point mutation constructs of AREB6(754–901) relative to that of GDBD, which was set as 100. Positions of point mutations are indicated by crosses. tk-Galp3x-LUC was used as a reporter. All transfection assays were repeated three times in duplicate.
To identify important amino acids that transmit repression activity, we constructed various point mutations at the positions of potential N-glycosylation sites and potential casein kinase II phosphorylation sites (Fig. 2b). We found that the mutations of AREB6(754–901)N769Q and double mutation AREB6(754–901)N769Q,K894T, which contain mutations of asparagine at 769 in the N-glycosylation site (NLS) to glutamine, abolished repression. Other point mutation constructs showed almost the same repression activity as the wild-type GDBD-AREB6(754–901). Approximately the same amounts of the fusion proteins were expressed in the transfected cells as demonstrated by gel retardation assays with a probe with the GALY DNA binding site (data not shown). To examine whether the glycosylation is important for the repression activity of GDBD-AREB6(754–901), a construct that contains a mutation of serine at 771 (NLS) to alanine, which is also expected to abolish glycosylation, was used. The repression activity was not diminished by this mutation (data not shown). Addition of the glycosylation inhibitor tunicamycin to the culture medium of transfected cells had no effect on repression (data not shown), also arguing against the involvement of glycosylation. These results indicate that AREB6(754–901) acts as a repression domain and that the mutation of the asparagine at 769 to glutamine causes the loss of the repression activity of AREB6.
The repression domain of AREB6 inhibits transcription in vitro.
The activation domain of AREB6 resembles those of well-known acidic activators like GAL4, GCN4, and VP16 (22, 47, 52). On the other hand, the repression domain of AREB6 may have a novel structure that has not been found in other transcription repressors. Therefore, the molecular mechanism by which the repression domain functions was analyzed in vitro. We tested the effects of bacterially expressed AREB6(754–901) on the transcription activity in HeLa nuclear extract. Figure 3a shows the effects of RD (which contains aa 754 to 901 of AREB6 fused to GAL4 aa 1 to 147) and RDm (which contains the mutation of an asparagine to a glutamine at position 769 of RD) with tk-Galpx3-LUC as a template. The basal transcription was not detected (lane 1), while the addition of GAL4(1–147) stimulated transcription from the accurate start site (lane 2). Compared to GAL4(1–147), RD repressed the transcription activity and gave rise to additional bands above the accurate transcript (lane 3). On the other hand, RDm showed higher activity than GAL4(1–147) (lane 4).
FIG. 3.
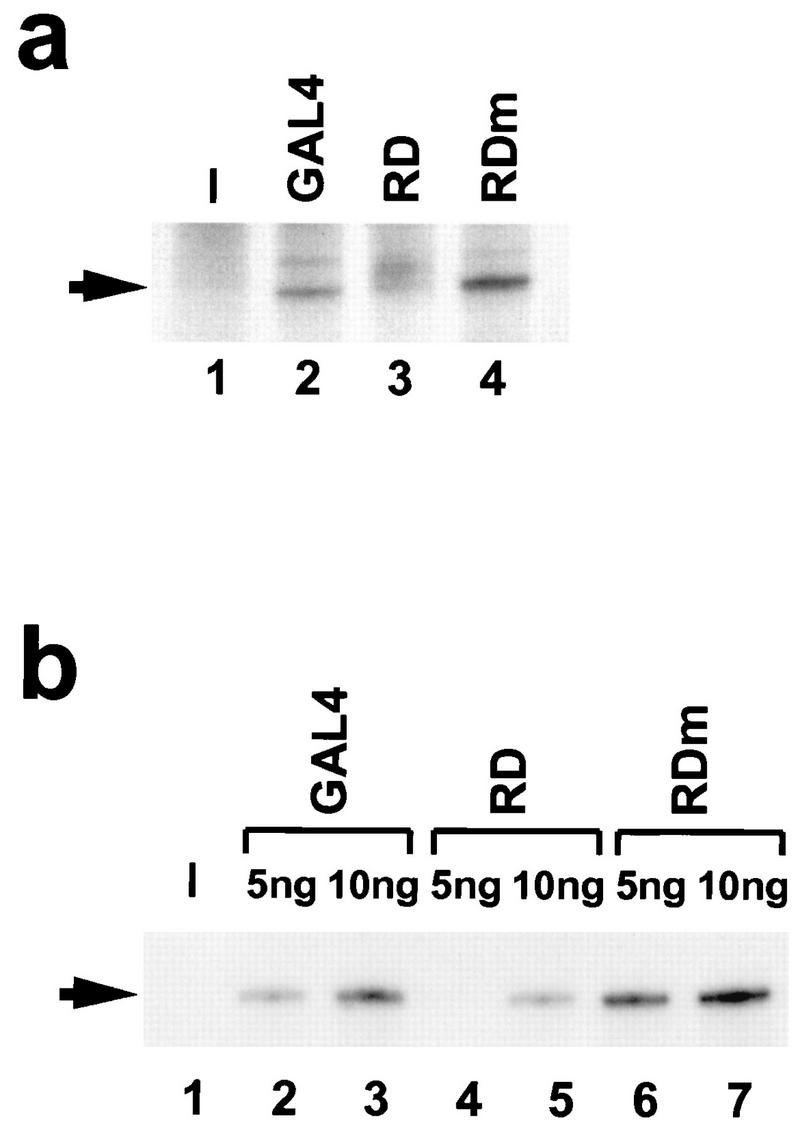
RD of AREB6 but not RDm represses transcription in vitro. In vitro transcription assays were performed with HeLa nuclear extract. We used 9 ng of linearized plasmids tk-Galp3x-LUC (a) and pMRG5 (b). (a) Transcription reactions were performed in the absence (lane 1) or presence of 10 ng of bacterially expressed GAL4(1–147) (lane 2), RD (lane 3), and RDm (lane 4). (b) Transcription reactions were performed in the absence (lane 1) or presence of GAL4 (lanes 2 and 3), RD (lanes 4 and 5), and RDm (lanes 6 and 7). The amounts of proteins added are indicated above (5 or 10 ng). The arrows indicate the positions of accurate transcripts.
We also tested another template plasmid containing the HIV promoter harboring five GAL4 DNA-binding sites (pMRG5). In transient-transfection assays with HeLa cells, the HIV promoter activity was also repressed by GDBD-AREB6(754–901) but not by GDBD-AREB6(754–901)N769Q (data not shown). As shown in Fig. 3b, the basal transcription of pMRG5 was not detected (lane 1). GAL4(1–147) activated transcription above background levels in a dose-dependent manner (lanes 2 and 3), as did RDm (lanes 6 and 7), while RD repressed transcription (lanes 4 and 5). These results indicated that the repression domain of AREB6 can inhibit transcription of the tk and the HIV promoters through upstream GAL4 DNA-binding sites in in vitro transcription systems.
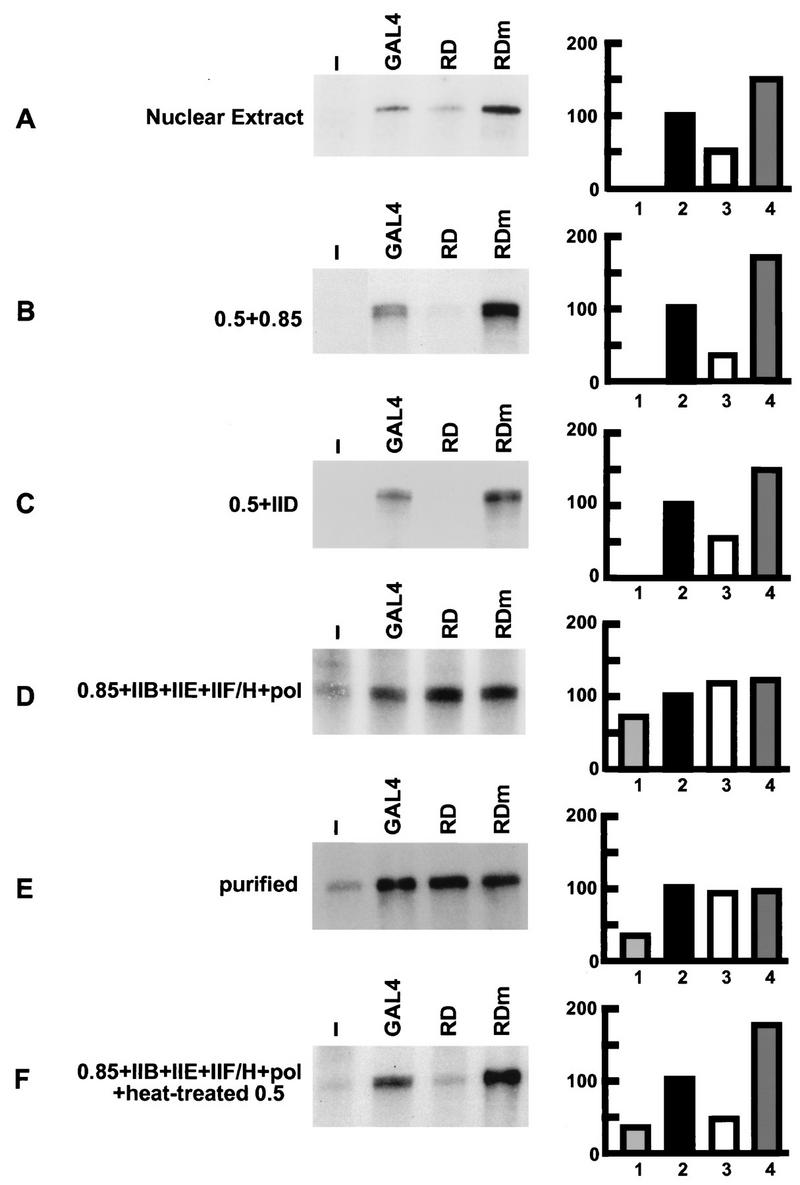
To characterize the factors which mediate the repression by the RD, we examined the expressed proteins in partially enriched and more purified reconstituted transcription systems (Fig. 4). RD exhibited almost the same repression potential in a transcription system partially enriched for general transcription factors (phosphocellulose P11 0.5 M KCl and P11 0.85 M KCl fractions derived from HeLa nuclear extract) as in the HeLa nuclear extract (Fig. 4A and B). The P11 0.5 M KCl and P11 0.85 M KCl fractions contain several positive and negative cofactors in addition to the general transcription factors. To analyze if cofactors are essential for repression by RD or if RD directly influences general transcription factors, more purified transcription systems lacking cofactors were used. We found that RD still exhibited repression activity when the P11 0.85 M KCl fraction was substituted by purified TFIID (Fig. 4C). In contrast, the repression activity of RD was not observed when the P11 0.5 M KCl fraction was substituted by purified TFIIB, TFIIE, TFIIH/F, and RNA polymerase II (Fig. 4D) or when only purified general transcription factors were used (Fig. 4E). This indicates that direct interactions between RD and the components of the basal transcription machinery are not sufficient for repression. Rather, some factors in the P11 0.5 M KCl fraction are essential for repression activity of RD.
FIG. 4.
Identification of fractions which contain possible factors mediating the repression activity of RD. Transcription reactions were performed with 9 ng of linearized pMRG5, using HeLa nuclear extract (A); the P11 0.5 M KCl and P11 0.85 M KCl fractions (B); the P11 0.5 M KCl fraction and TFIID (C); the P11 0.85 M KCl fraction, TFIIB, TFIIE, TFIIH/F, and RNA polymerase II (D); and TFIID, TFIIB, TFIIE, TFIIH/F, and RNA polymerase II (E). The heat-treated P11 0.5 M KCl fraction was added to the reaction mixture containing the P11 0.85 M KCl fraction, TFIIB, TFIIE, TFIIH/F, and RNA polymerase II (F). A 5-ng portion of GAL4(1–147) which is indicated as GAL4, RD, or RDm was added as the effector protein. The graphs indicate the amounts of individual transcripts quantified by the Instant Imager (Packard). Values are represented as activities relative to the activity of GAL4, which was set as 100. Transcription activities of basal (without any effectors) (lane 1), GAL4 (lane 2), RD (lane 3), and RDm (lane 4) are shown.
The RD of AREB6 represses transcription through NC2.
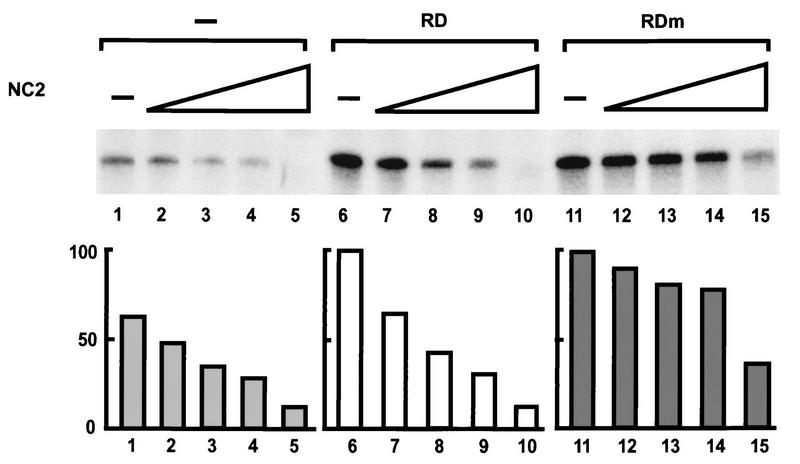
Heat treatment of the P11 0.5 M KCl fraction denatures most proteins in the fraction, among them all the general transcription factors, but not the positive cofactor PC5 (17) and the negative cofactor NC2 (15). To examine the possibility that these heat-stable factors are involved in transcriptional repression of RD, we added the heat-treated P11 0.5 M KCl fraction to the reaction mixture consisting of the P11 0.85 M KCl fraction, TFIIB, TFIIE, TFIIH/F, and RNA polymerase II. Indeed, transcriptional repression by RD was recovered (Fig. 4F), indicating that heat-stable factors in the P11 0.5 M KCl fraction mediate repression. The negative cofactor NC2 has been shown to repress basal transcription by direct interaction with the TATA-binding protein (TBP) (27). To directly address whether NC2 can also mediate repression of RD, we added E. coli-expressed and purified NC2α and NC2β/Dr1, instead of the heat-treated P11 0.5 M KCl fraction, to the purified system (Fig. 5). The transcription activities with RD and RDm in the absence of NC2 were comparable (Fig. 4E and 5, lanes 6 and 11), which was not the case in the presence of NC2. With 0.08 U of NC2, the transcription in the presence of RDm was 90% (lane 12) while the transcription in the presence of RD was repressed to 66% (lane 7) of the activities in the absence of NC2 (lanes 6 and 11). With the addition of 0.4 U of NC2, the activity with RDm was decreased only to 80% whereas the activity with RD was repressed to 42% (lanes 8 and 13). With 0.8 U of NC2, the transcription activity in the presence of RD was repressed to 31% (lane 9) while the activity in the presence of RDm was 79% (lane 14). When saturating amounts of NC2 (4 U) were added to the reaction mixture, the activity in the presence of RD was 12% (lane 10) and the transcription in the presence of RDm was 31%. The repression by NC2 in the absence of RD or RDm should be interpreted as repression of basal transcription, as reported previously (27, 39). The basal transcription activity was 65% (lane 1) without NC2, and the activities were decreased to 49, 33, 30, and 13% by the addition of 0.08, 0.4, 0.8, and 4 U of NC2, respectively (lanes 2 to 5). Transcription repression in the presence of RD was more pronounced than the basal transcription repression. These results showed that RD but not RDm actively represses transcription in the presence of NC2.
FIG. 5.
Effects of recombinant NC2 (NC2α and NC2β/Dr1) on transcription in the absence of any effectors (lanes 1 to 5), in the presence of 5 ng of RD (lanes 6 to 10) or in the presence of 5 ng of RDm (lanes 11 to 15). Transcription reactions were performed with TFIID, TFIIB, TFIIE, TFIIH/F, and RNA polymerase II. The transcripts without NC2 (lanes 1, 6, and 11) or in the presence of 0.08 U (lanes 2, 7, and 12), 0.4 U (lanes 3, 8, and 13), 0.8 U (lanes 4, 9, and 14), or 4 U (lanes 5, 10, and 15) of NC2 are shown. The definition of units is given in Materials and Methods. Values are represented as activities relative to that of RD without NC2 (lane 6), which was set as 100.
Direct interaction between RD and NC2 in yeast two-hybrid assay.
The above results suggested that there was a specific interaction between NC2 and RD. To test this hypothesis, we analyzed RD-NC2 interaction in various assays. We did not observe binding of NC2 to immobilized glutathione S-transferase (GST)-RD or GST-RDm, nor could we detect NC2-TBP-RD ternary complex formation on a TATA-containing DNA by gel mobility shift assays (data not shown). Therefore, we used a more sensitive yeast two-hybrid assay and observed binding of RD to NC2α (Table 1). Surprisingly, not only RD but also RDm interacted with NC2α, which lacks the histone fold domain in the construct (16). Neither RD nor RDm interacted with NC2β. On the other hand, AREB6(726–1010), which contains RD (Fig. 1), and AREB6(726–829), which showed little repression activity (Fig. 2a), interacted with both NC2α and NC2β.
TABLE 1.
Interaction between various domains of AREB6 and NC2α/βa
| Product expressed by pJG4-5 construct | pEG202 construct expression of:
|
|
|---|---|---|
| NC2α | NC2β | |
| AREB6(754–901); RD | + | − |
| AREB6(754–901)N769Q; RDm | + | − |
| AREB6(726–1010) | + | + |
| AREB6(796–1010) | − | − |
| AREB6(829–1010) | − | − |
| AREB6(726–829) | + | + |
Yeast strain EGY48 was cotransformed with pJG4-5 constructs (rows) and pEG202 constructs (columns). An interaction was determined as positive (+) if the transformant became Leu+ and turned blue on X-Gal indicator plates and negative (−) if it did not. The regions contained in pJG4-5 AREB6 constructs are shown in Fig. 2a.
DISCUSSION
AREB6 contains a highly negatively charged activation domain.
It is well known that acidic domains and glutamine- or proline-rich domains act as transcriptional activation domains. Some activation domains can interact directly with a number of components of the general transcription machinery in vitro (reviewed in reference 49). TFIID, TFIIB, TFIIH, and RNA polymerase II are known to play a central role in transcription activation by acidic activation domains in vitro (30, 37, 42). Through the interactions with TFIIB, the activation domains stimulate the formation of a preinitiation complex. Activators can also stimulate transcription indirectly by reversing the inhibitory effects of chromatin. Anionic regions rich in aspartic acid and glutamic acid residues are characteristic of many proteins that interact with chromatin, such as nucleoplasmin (8) and HMG1 (53). The acidic activator Gal4 can displace a nucleosome from the GAL1 promoter in vivo (2). The activation domain of AREB6 (aa 1011 to 1124) is highly negatively charged. We dissected this activation domain into four parts and found that the full transcription activation was achieved by all four subregions in an additive manner (25). This observation suggests that the net negative charge of the domain is important, rather than specific protein-protein interactions through critical amino acids. The low degree of amino acid sequence conservation among different species (see Results) is consistent with this notion.
AREB6 contains an active repression domain.
It was previously reported that δEF1 (the chicken homolog of AREB6) (13) antagonizes the action of MyoD family proteins through E-box binding-site competition (45). It has also been reported that ZEB (AREB6) acts as a repressor by competing with other basic helix-loop-helix proteins for binding to the E box in the immunoglobulin heavy-chain enhancer (14). However, we showed that AREB6 contains a potent repression domain by using fusion proteins of AREB6 with a heterologous GDBD in cotransfection experiments. Such transcriptional inhibition via transferable repression domains has been termed active repression, since it is not mediated simply by steric hindrance or by competition with other DNA-binding proteins. The AREB6 repression domain is highly conserved among species, and a change of asparagine to glutamine at 769, which has subtle effects on the conformations, abrogates activity. This might suggest that an interaction between cofactors and RD through aa 769 of AREB6 is important for its repression activity.
Active repression through the negative cofactor NC2.
Recently, there has been progress toward identifying target molecules of active repression domains of DNA-binding transcription repressors (reviewed in references 19, 28, and 43). Many eukaryotic transcription repressors have been reported to interact with general transcription factors in vitro. For example, the unliganded thyroid hormone receptor interacts with TFIIB (3) as well as with TBP (10), resulting in inhibition of preinitiation complex formation (11). The repression domain of the homeodomain protein even-skipped (18, 51), which is encoded by a Drosophila segment polarity gene, interacts with TBP (54) and may prevent TFIID binding to a promoter (1). Another homeodomain protein, Krüppel (Kr), which is encoded by a Drosophila gap gene, also contains the repression domain (35, 38). The interaction between Kr and TFIIEβ results in transcriptional repression (44). A murine homeodomain transcription factor, Msx-1, represses transcription by interacting with a protein complex composed of TBP and TFIIA (DA complex) or with one composed of TBP, TFIIA, and TFIIB (DAB complex) (6). The adenovirus oncoprotein E1A interacts with TBP and represses transcription, but the repression is reversed by TFIIB (48). Through interactions with general transcription factors, these repressor proteins are thought to sterically block the assembly of subsequent proteins, freeze the assembled transcription initiation complex, or disassemble the preinitiation complex. Another target of repressors in the general transcription machinery has been found by genetic experiments with yeast. SRB10 and SRB11, which are members of the C-terminal domain interacting polylpeptides in yeast RNA polymerase II, are required for full repression by the SSN6/TUP1 repressor (34). AREB6 is the first transcription factor which targets one of the general negative cofactors, NC2. NC2 was discovered by its ability to bind stably to TBP and to repress basal transcription (27, 39). NC2 consists of two subunits, NC2α (16, 41) and NC2β/Dr1 (27). This complex has also been defined as the repressor-corepressor complex Dr1-DRAP1 (41). Binding of NC2 to the TBP-promoter complexes prevents the assembly with TFIIA (31, 39). NC2 may also affect the conformation of the DNA-TBP complex, since it weakens the association of TFIIB with the complex (16, 27). The inhibitory effects of Dr1 are counteracted by the viral immediately-early activator (32) and some cellular transcriptional activators (57). NC2 has been thought to control the overall basal activity of genes in cells and has been reported to be released by upstream transcription factors to potentiate transcriptional activation. AREB6 is the tissue-specific transcriptional repressor which binds to and functions in conjunction with NC2. Thus, it appears likely that NC2 not only controls basal transcription but also functions as a mediator of tissue-specific transcription factors.
Interaction with NC2 is not sufficient for repression.
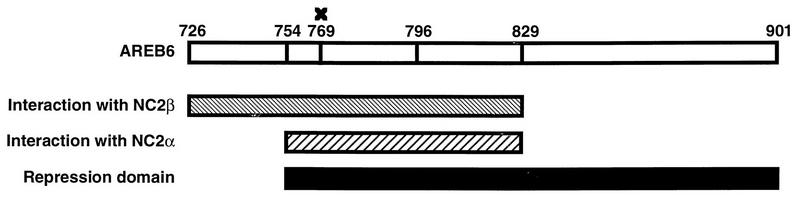
As shown in Table 1, the C-terminal region of NC2α interacts with AREB6(754–901), AREB6(754–901)N769Q, AREB6(726– 1010), and AREB6(726–829) but not with AREB6(796–1010) or AREB6(829–1010). Thus, the interaction is specific, and the region from aa 754 to 829 of AREB6 is sufficient for interaction with NC2α. However, the mutation of aa 769, which releases repression in vivo, does not abolish interaction with NC2α (see below). NC2β interacts with AREB6(726–1010) and AREB6(726–829) but not with AREB6(754–901), AREB6(796–1010), or AREB6(829–1010). This indicates that the region from aa 726 to 829 of AREB6 is sufficient for interaction with NC2β. However, interaction with NC2β seems not to be essential for the repression activity [Fig. 2a, GDBD-AREB6(754–901)]. This does not necessarily mean that NC2β is not required for repression, since AREB6 binds NC2α outside of the histone fold and NC2α can recruit NC2β through the histone fold (16).
As shown in Fig. 6, interaction of NC2α with AREB6 is essential but not sufficient for the repression activity in vivo [Fig. 2a, GDBD-AREB6(726–829)]. We supposed that interaction with one or more corepressors, which are contained in the TFIIF/H and RNA polymerase II fractions, might be necessary for NC2 repression activity through aa 769 and through the region from positions 829 to 901 of AREB6. It could be also possible that introduction of the mutation at aa 769 caused a new interaction with some positive cofactors. To clarify this point, we used the most highly purified system, consisting of TFIID, TFIIB, TFIIE, more purified TFIIF, more purified TFIIH, and more purified RNA polymerase II, and titrated NC2 in the presence of RD or RDm. Even in the most highly purified system, we observed almost the same results as we did with the purified system in the experiment whose results are shown in Fig. 5. From these results, we suppose that aa 769 of AREB6 plays a key role in the direct interaction between AREB6 and GTFs or in the interaction between NC2 and GTFs.
FIG. 6.
Summary of the identified domains of AREB6. The region from aa 754 to 829 and that from aa 726 to 829 of AREB6 are sufficient for interaction with NC2α and NC2β, respectively (Table 1). The RD was identified at aa 754 to 901 in transient-transfection assays (Fig. 2a). The mutation at aa 769 (indicated by a cross) abolished transcriptional repression in vivo but not the interaction with NC2α.
We have previously reported that AREB6 binds to DNA through either the N- or the C-terminal zinc finger domain, depending on the promoter context. AREB6 can regulate promoter activity positively or negatively by the alternative DNA-binding modes. What is the molecular basis of the activator-repressor switch of AREB6? Here we have shown that AREB6 has both an acidic activation domain and an active RD and that these bipartite functional domains may act through distinct factors on the transcription machinery. To further reveal the relationship between alternate binding modes and the positive/negative regulatory functions, it will be necessary to examine transcription regulation on natural target promoters through the DNA-binding domains of AREB6.
ACKNOWLEDGMENTS
We thank K. Kaiser and A. Goppelt for providing materials and for discussions. We also thank K. Umezono for tk-Galp3x-LUC, M. Okuda for UAS-HTLV1-CAT and pCMV-GAL4(1-147), J. Fujisawa for His6T7-11d, and D. Reinberg for LexA-NC2β.
This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Austin R J, Biggin M D. A domain of the even-skipped protein represses transcription by preventing TFIID binding to a promoter: repression by cooperative blocking. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelrod J D, Reagan M S, Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993;7:857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- 3.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M-J, O’Malley B W. Interaction of human thyroid hormone receptor β with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baniahmad A, Köhne A C, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker J C, Brabletz T, Kirchner T, Conrad T, Bröcker E-B, Reisfeld R A. Negative transcriptional regulation in anergic T cells. Proc Natl Acad Sci USA. 1995;92:2375–2378. doi: 10.1073/pnas.92.6.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catron K M, Zhang H, Marshall S C, Inostroza J A, Wilson J M, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 8.Dingwall C, Dilworth S M, Black S J, Kearsey S E, Cox L S, Laskey R A. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1989;6:69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Donzeau M, Winnacker E-L, Meisterernst M. Specific repression of Tax trans-activation by TAR RNA-binding protein TRBP. J Virol. 1997;71:2628–2635. doi: 10.1128/jvi.71.4.2628-2635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estruch F. The yeast putative transcriptional repressor RGM1 is a proline-rich zinc finger protein. Nucleic Acids Res. 1991;19:4873–4877. doi: 10.1093/nar/19.18.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor α can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 12.Franklin A J, Jetton T L, Shelton K D, Magnuson M A. BZP, a novel serum-responsive zinc finger protein that inhibits gene transcription. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. δ-Crystallin enhancer binding protein δEF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 14.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goppelt, A., and M. Meisterernst. Unpublished data.
- 16.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 17.Halle J-P, Stelzer G, Goppelt A, Meisterernst M. Activation of transcription by recombinant upstream stimulatory factor 1 is mediated by a novel positive cofactor. J Biol Chem. 1995;270:21307–21311. doi: 10.1074/jbc.270.36.21307. [DOI] [PubMed] [Google Scholar]
- 18.Han K, Manley J L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 19.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 20.Higashi, Y. Personal communication.
- 21.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Knodoh H. Impairment of T cell development in δEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda K, Kawakami K. Cis-elements in differential expression of Na+,K+-ATPase α2 subunit gene in muscle differentiation. Biochim Biophys Acta. 1996;1308:67–73. doi: 10.1016/0167-4781(96)00071-1. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, Kawakami K. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem. 1995;233:73–82. doi: 10.1111/j.1432-1033.1995.073_1.x. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda, K., and K. Kawakami. Unpublished data.
- 26.Ikeda K, Nagano K, Kawakami K. Anomalous interaction of Sp1 and specific binding of an E-box-binding protein with the regulatory elements of the Na,K-ATPase α2 subunit gene promoter. Eur J Biochem. 1993;218:195–204. doi: 10.1111/j.1432-1033.1993.tb18365.x. [DOI] [PubMed] [Google Scholar]
- 27.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 28.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 29.Kaiser K, Stelzer G, Meisterernst M. The coactivator p15(PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T K, Roeder R G. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci USA. 1994;91:4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T K, Zhao Y, Ge H, Bernstein R, Roeder R G. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 32.Kraus V B, Inostroza J A, Yeung K, Reinberg D, Nevins J R. Interaction of the Dr1 inhibitory factor with the TATA binding protein is disrupted by adenovirus E1A. Proc Natl Acad Sci USA. 1994;91:6279–6282. doi: 10.1073/pnas.91.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kretzschmar M, Stelzer G, Roeder R G, Meisterernst M. RNA polymerase II cofactor PC2 facilitates activation of transcription by GAL4-AH in vitro. Mol Cell Biol. 1994;14:3927–3937. doi: 10.1128/mcb.14.6.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licht J D, Hanna-Rose W, Reddy J C, English M A, Ro M, Grossel M, Shaknovich R, Hansen U. Mapping and mutagenesis of the amino-terminal transcriptional repression domain of the Drosophila Krüppel protein. Mol Cell Biol. 1994;14:4057–4066. doi: 10.1128/mcb.14.6.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y-S, Carey M F, Ptashne M, Green M R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y-S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 38.Margolin J F, Friedman J R, Meyer W K-H, Vissing H, Thiesen H-J, Rauscher F J., III Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 40.Meisterernst M, Roy A L, Lei H M, Roeder R G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 41.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 42.Roberts S G E, Ha I, Maldonado E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 43.Roberts S G E, Green M R. Dichotomous regulators. Nature. 1995;375:105–106. doi: 10.1038/375105a0. [DOI] [PubMed] [Google Scholar]
- 44.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jäckle H. Control of transcription by Krüppel through interactions with TFIIB and TFIIEβ. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 45.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima Y, Kondoh H. The δ-crystallin enhancer-binding protein δEF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimazaki T, Okazawa H, Fujii H, Ikeda M, Tamai K, McKay R D G, Muramatsu M, Hamada H. Hybrid cell extinction and re-expression of Oct-3 function correlates with differentiation potential. EMBO J. 1993;12:4489–4498. doi: 10.1002/j.1460-2075.1993.tb06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singler P B. Acid blobs and negative noodles. Nature. 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 48.Song C-Z, Loewenstein P M, Toth K, Tang Q, Nishikawa A, Green M. The adenovirus E1A repression domain disrupts the interaction between the TATA binding protein and the TATA box in a manner reversible by TFIIB. Mol Cell Biol. 1997;17:2186–2193. doi: 10.1128/mcb.17.4.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stargell L A, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki-Yagawa Y, Kawakami K, Nagano K. Housekeeping Na,K-ATPase α1 subunit gene promoter is composed of multiple cis elements to which common and cell-type-specific factors bind. Mol Cell Biol. 1992;12:4046–4055. doi: 10.1128/mcb.12.9.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.TenHarmsel A, Austin R J, Savenelli N, Biggin M D. Cooperative binding at a distance by even-skipped protein correlates with repression and suggests a mechanism of silencing. Mol Cell Biol. 1993;13:2742–2752. doi: 10.1128/mcb.13.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 53.Tsuda K, Kikuchi M, Mori K, Waga S, Yoshida M. Primary structure of non-histone protein HMG1 revealed by the nucleotide sequence. Biochemistry. 1988;27:6159–6163. doi: 10.1021/bi00416a050. [DOI] [PubMed] [Google Scholar]
- 54.Um M, Li C, Manley J L. The transcriptional repressor even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe Y, Kawakami K, Hirayama Y, Nagano K. Transcription factors positively and negatively regulating the Na, K-ATPase α1 subunit gene. J Biochem (Tokyo) 1993;114:849–855. doi: 10.1093/oxfordjournals.jbchem.a124267. [DOI] [PubMed] [Google Scholar]
- 56.Williams T M, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher III F J, Kant J A. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 57.Yeung K C, Inostroza J A, Mermelstein F H, Kannabirian C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 58.Yoshimura T, Fujisawa J, Yoshida M. Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J. 1990;9:2537–2542. doi: 10.1002/j.1460-2075.1990.tb07434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]