Abstract
The mei4+ gene of the fission yeast Schizosaccharomyces pombe was cloned by functional complementation. The mei4 disruptant failed to complete meiosis-I but could proliferate normally. mei4+ was transcribed only in meiosis-proficient diploid cells after premeiotic DNA replication. The mei4+ open reading frame encodes a 57-kDa serine-rich protein comprised of 517 amino acids with a forkhead/HNF3 DNA-binding domain in the amino-terminal region. Transcription of spo6+, a gene required for sporulation, was dependent on the mei4+ function. Two copies of the GTAAAYA consensus sequence, proposed as the binding site for human forkhead proteins, were found in the promoter region of spo6+. A gel mobility shift assay demonstrated the sequence-dependent binding of the GST-Mei4 forkhead domain fusion protein to DNA fragments with one of the consensus elements. Deletion of this consensus element from the spo6 promoter abolished the transcription of spo6+ and resulted in a sporulation deficiency. One-hybrid assay of Mei4 which was fused to the Gal4 DNA-binding domain localized the transcriptional activation domain in the C-terminal 140 amino acids of Mei4. These results indicate that Mei4 functions as a meiosis-specific transcription factor of S. pombe.
Meiosis is required for the formation of germ cells which transmit genetic information from generation to generation. This specialized nuclear division is characterized by a reduction in the chromosome number and frequent genetic recombination, both of which have contributed to the evolution of eukaryotes. Although meiosis has basically the same machinery, including spindles, centrosomes, and kinetochores, as mitosis in somatic cells, these two nuclear divisions are different in many aspects. Meiosis-specific gene products must be responsible for the various different features of meiosis, especially those of meiosis-I, such as the synapsis of homologous chromosomes, nondisjunction of sister chromatids, crossing over, and chiasmata formation.
Yeasts are simple eukaryotic organisms which undergo meiosis linked to ascospore formation. Meiosis is induced in diploid cells under conditions of nitrogen starvation, and the haploid tetrads culminate in ascospores. Genetic and cytological studies with the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe have revealed that yeast meiosis consists of a reductional first division and an equational second division with no intervening S phase between them (7, 8, 20, 56), as meiosis in most eukaryotes. Many mutants defective in meiotic events have been isolated and analyzed to identify the meiosis-specific genes that are responsible for the differences between meiosis and mitosis (3, 8).
The meiosis-specific genes are expressed in the germ cells of higher eukaryotes and in the sporulating cells of yeasts. The transcriptional regulation of meiotic genes has been extensively studied with budding yeast (26) and fission yeast (56, 57). The investigation of transcription during early sexual processes of S. pombe has identified some DNA-binding proteins, such as those containing HMG boxes (42), homeobox domains (18), and CREB-like motifs (49).
The mei4+ gene of the fission yeast S. pombe is indispensable for meiosis-I (3, 32, 38). In mei4 mutants, elongated “horsetail” nuclei are at least transiently accumulated (32). This morphology is characteristic of prophase-I nuclei (5, 35). These results strongly suggest that the mei4+ gene products are essential for meiotic prophase-I. Because morphological events unique to meiosis occur mainly during prophase-I, the activity of mei4+ is particularly interesting.
In the present study, we cloned and analyzed mei4+. Nucleotide sequencing suggests that the mei4+ product contains a forkhead DNA-binding domain composed of approximately 120 amino acids which was originally identified as the DNA-binding domain of the hepatocyte-specific transcription factor of rodents (21, 52). More than 60 proteins with this motif in a wide variety of organisms have been compiled in the protein databases. Most of the family members function as tissue-specific transcription factors. Here we present evidence that Mei4 is a meiosis-specific transcription factor in fission yeast.
MATERIALS AND METHODS
Yeast strains and culture conditions.
The S. pombe strains used in this study are listed in Table 1. Cells were grown on YEA complete medium or minimal medium (SD, PM, or EMM2) (9, 25, 29). Mating and sporulation were induced on a malt extract agar or synthetic sporulation medium (SSA or SPA) (9, 29). These media were supplemented with the required nutrients (50 to 100 mg/liter).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| S. pombe haploid strains | ||
| L972 | h− prototroph | U. Leupold |
| L968 | h90 prototroph | U. Leupold |
| C133-1D | h90 mei4-P572 ade6-M216 leu1 ura1 | C. Shimoda |
| C133-4B | h90 mei4-P572 ade6-M216 leu1 | C. Shimoda |
| C206-2A | h90 mei1-B102 ade6-M210 leu1 | C. Shimoda |
| YW917 | h90 mei4::ura4+ (Δ2) ura4-D18 ade6-M216 leu1 | Y. Watanabe |
| JZ878 | h90 ura4-D18 ade6-M216 leu1 | Y. Watanabe |
| S. pombe diploid strains | ||
| CD16-1 | h+/h− ade6-M210/ade6-M216 lys5-391/+ +/cyh1 | C. Shimoda |
| C525 | h90/h90 ura4-D18/ura4-D18 ade6-M216/ade6-M210 leu1/leu1 | C. Shimoda |
| C537 | h90/h90 mei4::ura4+(Δ1)/mei4::ura4+ (Δ1) | C. Shimoda |
| NT-4A | h90/h90 ade6-M216/ade6-M210 ura4-D18/ura4-D18 leu1/leu1 spo6::ura4+/spo6::ura4+ | T. Nakamura |
| JY362 | h+/h− ade6-M210/ade6-M216 leu1/leu1 | Y. Watanabe |
| JZ807 | h+/h− mei4::ura4+ (Δ2)/mei4::ura4+ (Δ2) ura4-D18/ura4-D18 leu1/leu1 ade6-M210/ade6-M216 | Y. Watanabe |
| S. cerevisiae haploid strain SFY526 | MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canr gal4-542 gal80-538 URA3::GAL1-lacZ | Clontech |
Synchronous meiosis was attained basically as described by McLeod and Beach (25). Diploid cells cultured in PM minimal medium to mid-log phase were suspended in a starvation medium, PM lacking ammonium chloride and glucose, and then shaken at 28°C for 12 to 15 h. Meiosis was initiated by adding glucose and glycerol to the starvation culture at final concentrations of 0.1 and 1%, respectively.
Yeast transformation was carried out by means of a highly efficient lithium acetate method (31).
Cloning of mei4+.
mei4+ was cloned by complementation of the mei4-P572 mutation. A homothallic strain, C133-4B (h90 mei4-P572) was transformed by an S. pombe genomic library containing partially digested Sau3A DNA fragments constructed in a multicopy plasmid, pDB248′ (2). The transformants on a sporulation medium (SSA) were stained with iodine vapor, which turned sporulated colonies brown (9). Several brown colonies were microscopically inspected for sporulation ability. A few sporulation-proficient transformants whose suppression activity proved to be plasmid borne were analyzed further.
The plasmid DNA carried by one transformant was rescued in Escherichia coli DH5. This plasmid, named pDB(mei4)1, carried a 13-kb insert (Fig. 1A). The complementation activity was localized on the 2.8-kb KpnI/HindIII fragment by subcloning. pDB(mei4)2, carrying this fragment, complemented mei4-P572 (Fig. 1A).
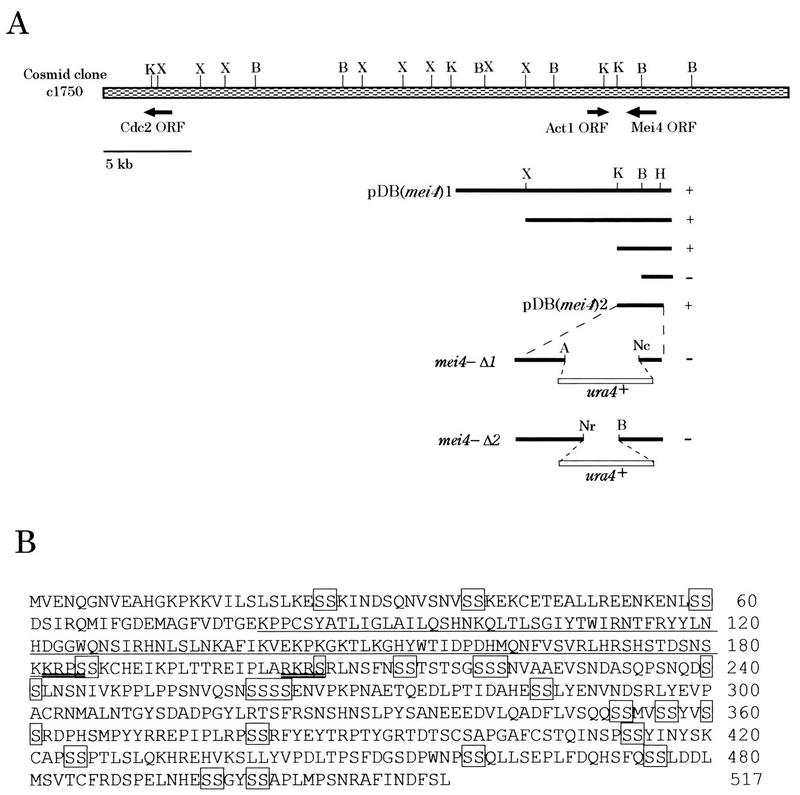
FIG. 1.
Structure of mei4+ and gene disruption. (A) Restriction map, subcloning, and gene disruption of mei4+. The shaded box indicates the S. pombe cosmid clone c1750, which contained the mei4+ ORF. Solid bars represent S. pombe genomic DNA fragments isolated. pDB(mei4)1 is the mei4+-carrying plasmid which was cloned by complementation from a genomic library. Arrows show the directions and sites of the ORFs for Cdc2, Act1, and Mei4. Complementation of mei4-P572 by each subclone: +, complements; −, does not complement. Restriction enzymes: K, KpnI; X, XbaI; B, BglII; A, AccI; Nc, NcoI; Nr, NruI. (B) Amino acid sequence of Mei4 deduced from the nucleotide sequence, shown in one-letter notation. The forkhead DNA-binding domain is underlined. Possible phosphorylation sites of protein kinase A are double underlined. Serine duplexes, triplexes, and tetraplexes are boxed.
Gene disruption of mei4+.
The mei4::ura4+ null allele was produced by a one-step gene disruption method (36). A BglII/NruI fragment of 470 bp was replaced by a 1.6-kb ura4+ cassette; this allele was designated mei4::ura4+(Δ1). A diploid strain (C525) was transformed with the KpnI/HindIII fragment having this disrupted mei4 allele, and stable Ura+ transformants were isolated. Disruption was confirmed by genomic Southern hybridization (data not shown) and tetrad analysis. A Ura+ segregant which was defective in meiosis was used as a haploid mei4 null mutant. We also constructed another disrupted allele, mei4::ura4+(Δ2), in which the 1.5-kb NcoI/AccI fragment was replaced by the ura4+ cassette. The phenotypes of these disruptants were identical.
DNA sequencing.
The 2.8-kb HindIII/KpnI fragment containing mei4+ was recloned into pUC118/119. A series of nested deletions were produced with exonuclease III and mung bean nuclease. Nucleotide sequences of both strands were determined by the dideoxy termination method (37, 58) with a commercial T7 DNA polymerase sequencing kit (Stratagene). The nucleotide sequence was analyzed with a Genetyx software package (SDC Co.). A similarity search for the amino acid sequence of Mei4 was carried out with proteins in the databases by using the BLAST algorithm.
Southern and Northern analysis.
Genomic DNA was prepared from S. pombe strains basically as described by Hereford et al. (10). Restriction fragments were fractionated on a 0.8% agarose gel and transferred onto a nylon membrane (Biodyne A; Pall Co.). For Northern analysis, total RNA was prepared from S. pombe cultures by the method of Jensen et al. (16). The 32P-labeled riboprobes were prepared by in vitro transcription with T7 RNA polymerase by using a 0.35-kb BglII/XhoI fragment on a pBluescript vector as a template. Hybridization was performed in 50% formaldehyde at 42°C (45). Ethidium bromide staining of rRNAs was used for a loading control. Hybridization with the S. pombe calmodulin gene (cam1) probe was used as an internal reference.
Site-directed mutagenesis.
Oligonucleotide-directed mutagenesis of three forkhead consensus regions was carried out by heteroduplex-PCR protocols according to the instructions of the manufacturer (Takara Shuzo Co.). The oligonucleotides used for generating three different mei4 mutant alleles (mei4-K81Q, -F115D, and -W125S) are as follows: mei4-K81Q, GGTGAAAAT(A)G(C)CAT(C)CGTGTTCTTA; mei4-F115D, AACAAAGCCG(T)A(T)TATCAAAGT; and mei4-W125S, ATGGTGGTTC(G)GCG(A)AAATAGC. The substituted nucleotides are underlined, and the corresponding wild-type nucleotides are in parentheses. The mutagenized sequences were designed to generate the new restriction sites EcoT22I (for mei4-K81Q), EcoRV (for mei4-F115D), and NruI (for mei4-W125S). The amino acid sequences should be changed as follows: KPP (amino acids [aa] 81 to 83) to QAS in mei4-K81Q, F (aa 115) to D in mei4-F115D, and WQ (aa 125 and 126) to SR in mei4-W125S. Correct mutated nucleotides were confirmed by DNA sequencing. The mutated mei4 DNA fragments were inserted into a multicopy plasmid, pAU-KS, and introduced into the mei4 disruptant YW917 and a wild-type strain (JY878).
Construction of the GST-Mei4 fusion gene.
An approximately 360-bp DNA segment containing the forkhead domain (aa 71 to 182) of mei4+ was amplified by PCR and cloned into the pCRII vector (Invitrogen Co.). The EcoRV site which was derived from the forward primer was cut and ligated to BamHI linkers. The BamHI/EcoRI fragment carrying the mei4 forkhead domain was inserted into the BamHI and EcoRI sites located at the 3′ terminus of the glutathione S-transferase (GST) gene of pGEX-2T (Pharmacia Biotech) to construct pGEX(mei4).
An E. coli strain, XLI-Blue, was transformed with pGEX(mei4). Expression of the GST-Mei4 fusion protein was induced by IPTG (isopropyl-β-d-thiogalactopyranoside) in Luria-Bertani medium. Cells were homogenized in buffer containing 30 mM Tris-HCl (pH 7.5) and 30 mM NaCl at 0°C. After centrifugation at 16,000 × g for 15 min at 4°C, most of the fusion protein was recovered in the supernatant fraction (data not shown).
Gel mobility shift assay.
FLEX-U (CTTGAATCAAGTAAATATATATTTTCT), FLEX-D (AAATATTTGAGTAAACAAACAAAATCA), and a mock oligonucleotide (CCCTCTTTCTTTGTTCCTTAT) were labeled with [α-32P]dATP by use of Klenow enzymes with random primers. A standard reaction mixture (20 μl) contained 24 ng of radiolabeled double-stranded oligonucleotide probe, an E. coli crude extract containing 9 ng of protein, and 2 μg of poly(dI-dC) in binding buffer (100 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 60 mM KCl, 1 mM spermidine, 0.1% Nonidet P-40, 7 mM β-mercaptoethanol, and 10% glycerol). In some assays, 4.2 μg of salmon sperm DNA per ml was included in the reaction mixture. The reaction mixture was placed on ice for 60 min and then immediately loaded onto 4% native polyacrylamide gels in TGE buffer, containing 0.6% Tris-HCl (pH 8), 0.078% EDTA, and 2.9% glycine. Polyacrylamide gels were electrophoresed at 15 mA in TGE buffer at 4°C until free probes reached the bottom of the gel. They were fixed with 7% acetic acid and then exposed to X-ray film (Fuji NIF-RX film) for 12 to 18 h at −80°C.
Deletion of the FLEX-D from the spo6 promoter.
The spo6 promoter sequence containing FLEX-D was amplified by PCR. Two different forward primers containing the XhoI site were used: spo6-X (GAGCTCGAGAAAATATTTGAGTAAACAAACAAAA) and spo6-DF (GAGCTCGAGAAAATATTTGAAACAAAATC). The latter sequence lacked the FLEX core heptamer, GTAAACA. The wild-type spo6+ gene was cloned into the multicopy plasmid pAL-KS to give pAL(spo6+). The amplified DNA was digested with XhoI and SalI, and the fragment was then inserted into pAL(spo6+) to replace the corresponding region. The plasmids were designated pAL(spo6)X and pAL(spo6)DF, respectively.
One-hybrid analysis.
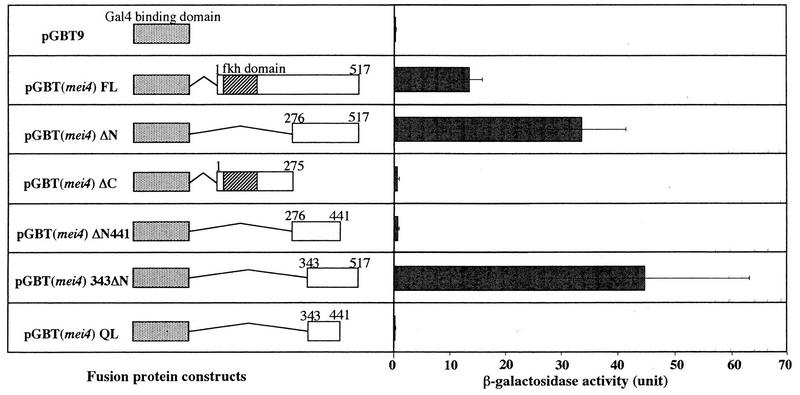
A putative transcriptional activation domain of Mei4 was determined by a one-hybrid assay. The full-length Mei4 open reading frame (ORF) was inserted into pGBT9 (Clontech) so that Mei4 was fused to the carboxyl terminus of the S. cerevisiae Gal4 DNA-binding domain (see Fig. 10). Similar constructs having several truncated mei4 fragments were also made (see Fig. 10). These plasmids were transformed into S. cerevisiae SFY526.
FIG. 10.
One-hybrid analysis to localize the activation domain of Mei4. (Left panel) Construction of fusion proteins between a Gal4 DNA-binding domain and Mei4 proteins. Numerals represent the positions of amino acid residues from the N terminus. S. cerevisiae SFY526 was transformed with the indicated plasmids. fkh, forkhead. (Right panel) β-Galactosidase activities, with standard deviations, for three independent transformants.
β-Galactosidase activity was assayed as follows (1). A single colony of yeast transformants was grown in SD-Trp liquid medium at 28°C to the early stationary phase. Cells were washed with and resuspended in Z buffer. The optical density at 600 nm (OD600) was determined as a measure of cell density. The cells were permeabilized by being vortexed in 0.8 ml of Z buffer containing 0.04 ml of 0.1% sodium dodecyl sulfate and 0.04 ml of chloroform, and then 0.16 ml of a 4-mg/ml o-nitrophenyl-β-d-galactoside solution was added. The reaction mixture was incubated at 30°C. The reaction was terminated by adding 3 volumes of 1 M Na2CO3. The OD420 of the supernatant was measured. One unit of β-galactosidase activity was defined as (OD420 × 1,000)/(OD600 × T [minutes] × V [milliliters]), where T is the reaction time and V is the volume of cell suspension used in the assay.
Fluorescence-activated cell sorter (FACS) analysis.
Cellular DNA content was determined by flow cytometry basically as described by Watanabe and Yamamoto (48). Cells were fixed with 70% ethanol and stained with propidium iodide, and then the fluorescence intensity was measured with a flow cytometer (model EPICS-C; Coulter).
DAPI staining.
S. pombe cells were fixed with 3.7% formaldehyde at 28°C for 30 min. The nuclear chromatin region was stained with 4′,6-diamidino-2-phenylindole (DAPI) at 1 μg/ml. Stained cells were observed under a fluorescence microscope (Olympus BHS-RFK).
RESULTS
Cloning and sequencing of mei4+.
mei4+ is essential for the progression through meiotic prophase-I. For further analysis of mei4-mediated steps, we isolated a 13-kb genomic DNA fragment which complemented the mei4-P572 mutation. Subcloning localized the complementation activity on a 2.8-kb KpnI/HindIII fragment (Fig. 1A).
Sequencing of this fragment (2,788 bp) identified an uninterrupted ORF composed of 1,551 nucleotides. Our sequence was identical to the sequence in the cosmid clone c1750, the nucleotide sequence of which was determined recently in the S. pombe genome sequence project. The mei4+ gene has been mapped in the vicinity of cdc2+ (0.6 centimorgan) on chromosome II (38). This cosmid clone (38 kb) also contained the ORF for Cdc2, indicating that our cloned gene was likely mei4+ itself (Fig. 1A). This assumption was further verified by genetic crosses between the disruptant strain and the original mei4-P572 mutant. The deduced mei4+ gene product is a 57-kDa serine- and threonine-rich protein composed of 517 amino acids (Fig. 1B).
Phenotypes brought about by the disruption and overexpression of mei4+.
The chromosomal mei4+ gene in a diploid strain (C525) was disrupted. The disrupted diploid strain was then sporulated, and the tetrads were dissected. Most asci produced four viable spore clones, indicating that the mei4 null mutation did not confer lethality to the cells. There were no differences in growth and cell size between the wild-type strain and the mei4Δ mutant (data not shown).
A homothallic haploid strain harboring mei4Δ was able to mate, but the resultant diploid zygotes were asporogenic. DAPI staining revealed that many zygotes contained one horsetail or a rounded nucleus (Table 2 and Fig. 2A), indicating that both mutants were arrested before meiosis-I. There were no significant differences in meiotic phenotypes between the disrupted null mutant and the mei4-P572 mutant.
TABLE 2.
Mating and sporulation of mei4-defective and mei4-overexpressing strains
| Expta | Strain | Relevant genotype | Zygotes (%) | Asci (%)b
|
Nuclear type of zygotes (%)c
|
||||
|---|---|---|---|---|---|---|---|---|---|
| 4 spore | 2 spore | 1N(r) | 1N(ht) | 2N | 3-4N | ||||
| 1 | JZ878 | h90 mei4+ | 10 | 60 | 0 | 4 | 5 | 1 | 90 |
| YW917 | h90 mei4Δ | 65 | 0 | 0 | 76 | 24 | 0 | 0 | |
| YW917 | h90 mei4Δ [pREP(mei4+)] | 13 | 47 | 8 | 13 | 5 | 2 | 80 | |
| 2 | YW917 | h90 mei4Δ [pAL19] | 69 | 0 | 0 | 28 | 71 | 1 | 0 |
| YW917 | h90 mei4Δ [pAL(mei4+)] | 25 | 29 | 5 | 16 | 32 | 20 | 32 | |
| YW917 | h90 mei4Δ [pAL(mei4-K85Q)] | 56 | 0 | 0 | 38 | 61 | 1 | 0 | |
| YW917 | h90 mei4Δ [pAL(mei4-W115S)] | 63 | 0 | 0 | 40 | 60 | 0 | 0 | |
| YW917 | h90 mei4Δ [pAL(mei4-F125D)] | 55 | 0 | 0 | 37 | 62 | 1 | 0 | |
In experiment 1 cells were cultured on SSA medium for 2 days; in experiment 2 cells were incubated in PM liquid medium lacking N for 12 h.
Two- and four-spore asci were differentially counted.
1N(r), mononucleate zygotes containing a round nucleus; 1N(ht), mononucleate zygotes with a horsetail nucleus; 2N, binucleate zygotes; 3-4N, tri- or tetranucleate zygotes.
FIG. 2.
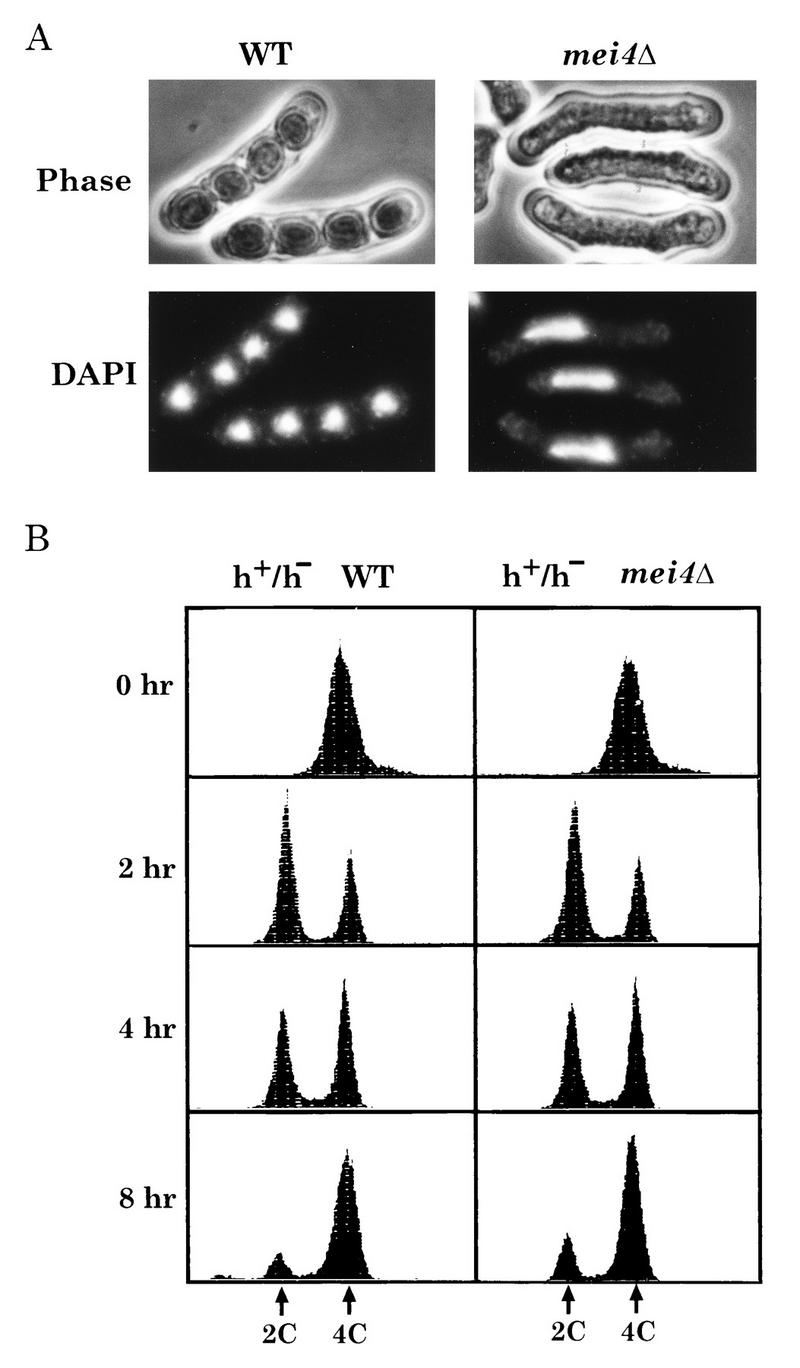
Phenotypes of the mei4Δ strain. (A) Nuclear morphology. The wild-type (WT) strain (L968) and the mei4 disruptant (YW917) were cultured on malt extract agar for 2 days. (B) Premeiotic DNA synthesis complete in a diploid mei4Δ strain. A wild-type diploid strain (JY362) and a homozygous mei4Δ mutant (JZ807) were cultured in nitrogen-free medium, PM lacking N, for the indicated times, and samples were processed for FACS analysis. Fluorescence intensities corresponding to 2C and 4C DNA contents are indicated.
We show below that mei4+ is transcribed only in meiotic cells (see Fig. 3). Overexpression of mei4+ in nutrient and sporulation media was examined. Ectopic expression of mei4+ was attained by placing it under control of the inducible nmt1 promoter on the pREP1 plasmid. This construct, named pREP(mei4), was introduced into mei4Δ null mutants. When mei4+ was ectopically expressed by transferring cells to SSA medium without thiamine, it complemented the mei4 mutation completely (Table 2). We noticed that a small fraction (at most 10%) of the population formed two-spored asci. Untimely and/or excess expression of mei4+ may disorder the meiotic process. Overproduction of mei4+ in vegetative cells in rich growth medium, however, did not cause meiosis and sporulation.
FIG. 3.
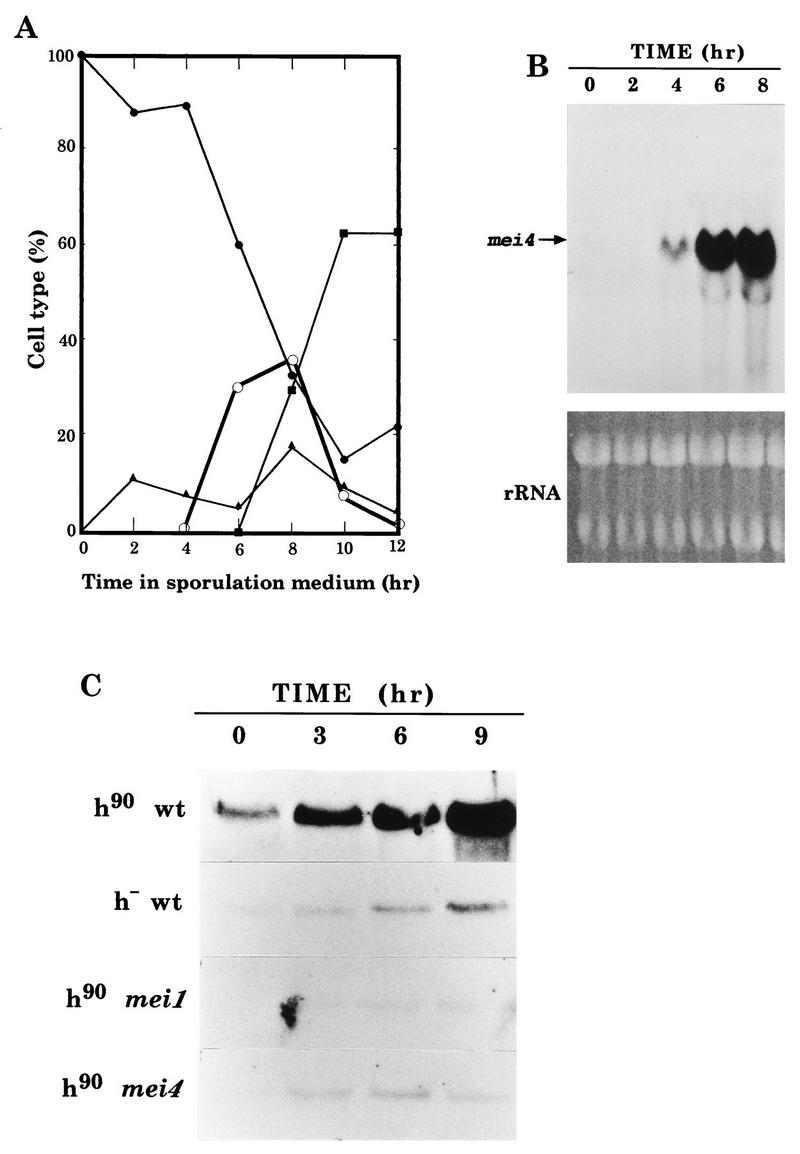
Transcriptional regulation of mei4+. (A) Kinetics of meiosis in a heterothallic diploid strain, CD16-1. Cells were cultured for 15 h in a modified minimal medium lacking both nitrogen and carbon sources (PM lacking N and C), and then glucose and glycerol were added to induce meiosis. A portion of this synchronously sporulating culture was withdrawn, fixed, and stained with DAPI. Frequencies of mononucleate cells with a rounded nucleus (closed circles), mononucleate cells with a horsetail nucleus (open circles), binucleate cells (triangles), and tetranucleate cells (squares) were determined. The other portion of the synchronous culture was subjected to RNA extraction. (B) Northern analysis. An autoradiogram of Northern analysis for mei4 mRNA is shown. Gel staining with ethidium bromide to assess loading abundance is presented. (C) Northern analysis of mei4+ in meiosis-deficient strains. Log-phase cultures grown in a minimal medium (PM with N) were incubated in PM lacking N for the indicated times. Samples were subjected to Northern analysis with a mei4-specific probe. To assess the loading abundance, ethidium bromide staining of the gels was done (data not shown). Strains: L968 (h90 wild type [wt]), L972 (h− wild-type), C206-2A (h90 mei1-B102), and C133-1D (h90 mei4-P572).
Premeiotic DNA replication in the mei4Δ strain.
To see whether the premeiotic S phase was completed, nitrogen-starved mei4Δ cells were subjected to FACS analysis (Fig. 2B). At 2 h after the nutritional shift-down, a discrete G1 (2C) peak appeared, which then shifted to a G2 (4C) peak between 4 and 8 h, indicating normal execution of premeiotic DNA replication in mei4Δ cells. Changes in the DNA content of mei4Δ cells were very similar to those of the wild-type strain. We conclude that mei4Δ cells undergo premeiotic DNA replication normally.
Transcriptional regulation of mei4+.
The fact that mei4+ is required only for meiosis-I prompted us to examine whether its expression is restricted to the meiotic process. Synchronous meiosis was induced in a wild-type diploid strain, CD16-1, by a shift-down to a starvation medium as has been reported previously (25). As shown in Fig. 3A, after 4 h of incubation in a nitrogen-free medium, cells entered prophase-I and so-called horsetail nuclei began to accumulate. Cells which finished meiosis-II appeared at 6 h, and mature asci were observable at 10 h.
Cells were harvested at 2-h intervals, and total RNA was subjected to Northern blot analysis with a mei4-specific probe. As Fig. 3B shows, the mei4 transcripts (2.3 kb) were hardly detected in vegetative cells (at 0 h). Remarkable induction of mei4+ mRNA was observed after 4 h of incubation. These results indicate that mei4+ was transcriptionally regulated, and abrupt induction occurred before or at the onset of prophase-I. Similar observations have been reported by Iino et al. (14) based on the pat1-driven synchronous meiosis.
The mei4+ transcript level in nitrogen-free sporulation medium was severely reduced in both a heterothallic haploid strain and the mutant harboring mei1-B102, which is allelic to mat2-Pm (Fig. 3C). Because meiosis is blocked before the premeiotic S phase in these strains, the mei4+ transcription is dependent upon meiosis rather than upon nitrogen starvation. Interestingly, the transcription level was also reduced in mei4-P572 cells, suggesting a positive autoregulatory mechanism (Fig. 3C). These results indicate that transcription of mei4+ is induced during early meiosis and probably before prophase-I.
Characteristics of the predicted Mei4 protein.
The predicted amino acid sequence of Mei4 was examined for similarity to known proteins in databases by using the BLAST program. The amino-terminal domain of Mei4 (aa 81 to 172) shows prominent sequence homology with an array of proteins of the forkhead/HNF3 family, also called the winged helix family (6), including the Drosophila nuclear protein forkhead (53) and the murine HNF3, which is the hepatocyte-specific transcription factor (21) (Fig. 4). This domain may function as the sequence-specific DNA-binding domain. The third α-helix of HNF3, corresponding to aa 122 to 135 of Mei4, makes major-groove contact with DNA (6). The primary structure of this domain, composed of approximately 100 amino acids, was compared for Mei4 and 67 forkhead proteins in the databases. Its phylogenetic tree suggested that S. cerevisiae Hcm1 is the closest member and that Mei4 and Hcm1 seem to constitute a subfamily (data not shown). The HCM1 gene was cloned as a high-copy suppressor of calmodulin mutants (59). The molecular function of Hcm1 has not yet been elucidated.
FIG. 4.
Mei4 contains a forkhead DNA-binding motif. A comparison of the amino acid sequences of forkhead domains in a few typical forkhead proteins is shown. Amino acids identical in Mei4 and the others are shown in white against black. Mei, Mei4 (S. pombe); Hcm, Hcm1 (S. cerevisiae), HNF, HNF3α (rat); FKH, forkhead (fly); FRE, FREAC-1 (human).
Another prominent feature of Mei4 was the abundance of serine and threonine residues, which constitute 25 and 8%, respectively. Serine duplex, triplex, and tetraplex motifs appeared 19 times (Fig. 1B), although their meaning is unclear. Two possible sites, KRPS and RKRS, for phosphorylation by the cyclic AMP-dependent protein kinase were found following the C terminus of the forkhead domain (aa 182 to 185 and aa 203 to 206).
Mutations in the forkhead domain abolish Mei4 function.
In order to examine whether the forkhead domain is essential for the function of Mei4, highly conserved amino acids in this domain were altered by site-directed mutagenesis. Mutated mei4 alleles on a multicopy plasmid were transformed into a mei4 disruptant. As Table 2 shows, none of the three different mutant alleles could complement the mei4 null mutation, indicating that these amino acid substitutions severely affected the mei4 gene function. They did not interfere with meiosis or sporulation in a wild-type strain (data not shown), indicating that the mutations in the forkhead domain represented a dominant-negative phenotype.
Transcription of the meiotic spo6+ gene depends on Mei4.
We next searched for meiosis and sporulation genes whose transcription was dependent on the putative transcription factor Mei4. Transcription of genes known to be responsible for meiosis, sporulation, or recombination was examined in the mei4Δ strain. The spo6+ gene is essential for meiosis-II and sporulation (3, 11, 12) and is transcribed to generate two mRNA species that are different in size and expression pattern (30). The 2.1-kb species was constitutively expressed at a low level in both vegetative and meiotic cells, while the smaller, 1.4-kb species is absent in vegetative cells and highly induced in meiotic cells (30). These two transcripts have different transcriptional start points (30). We found that the 1.4-kb meiosis-specific spo6+ mRNA was almost absent in the mei4Δ mutant (Fig. 5). In addition, overexpression of mei4+ stimulated the transcription of spo6 even in a nutrient medium, indicating that spo6+ may be a target gene of Mei4.
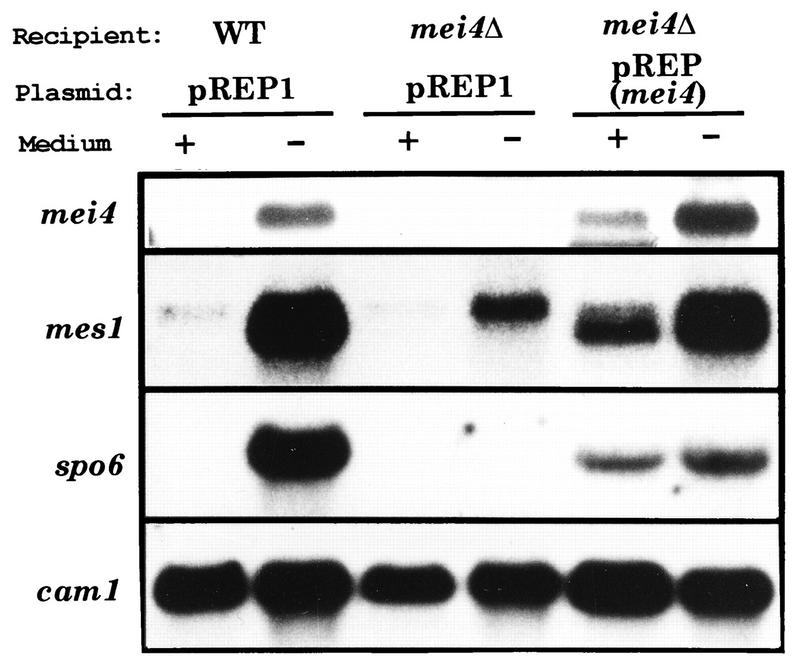
FIG. 5.
Northern blot analysis indicating that transcription of spo6+ is dependent on mei4+ function. Strains: WT, a wild-type diploid strain (JY362); mei4Δ, a diploid strain harboring the mei4Δ null allele homozygously (JZ878). These strains were transformed by either pREP1 vector plasmid or pREP1(mei4). Transformants were incubated in PM with (+) or without (−) 10 mM NH4Cl. Specific radioactive probes were for mei4, mes1, spo6, and cam1. Although the spo6 probe hybridizes with two mRNA species (2.1 and 1.4 kb), only the meiosis-specific 1.4-kb mRNA is shown.
The mes1+ gene, responsible for meiosis-II, carries one short intron, and splicing of this intron is accomplished only during meiosis (19). As shown in Fig. 5, both transcription and splicing of mes1+ occurred in the wild-type strain cultured in nitrogen-free medium. However, transcription of mes1+ was reduced in mei4Δ cells. In contrast, mes1+ was induced in a mei4Δ strain overexpressing mei4+, even when cells were cultured in nitrogen-rich medium. In addition, splicing of the mes1 intron was highly dependent on the mei4 function. These observations suggest that transcription of mes1 and splicing of its intron are regulated by mei4+.
The Mei4 forkhead domain binds to the FLEX sequence.
We addressed the question of whether Mei4 could bind to a specific sequence. In the case of the human forkhead proteins called FREAC, the recognition sequence was GTAAAYA, which seemed to be a consensus core sequence for forkhead proteins in general (34). In this article, this heptamer sequence will be designated the core heptamer. Interestingly, two sequences which are identical to the FREAC core heptamer were found in the possible 5′ regulatory region of spo6+ (Fig. 6A). Therefore, we examined the binding of Mei4 to two kinds of core heptamer-containing sequences of 27 nucleotides, as presented in Fig. 6B. These sequences were designated FLEX-U and FLEX-D (for FREAC-like consensus element of spo six), where the suffixes U and D represent the upstream and downstream elements, respectively.
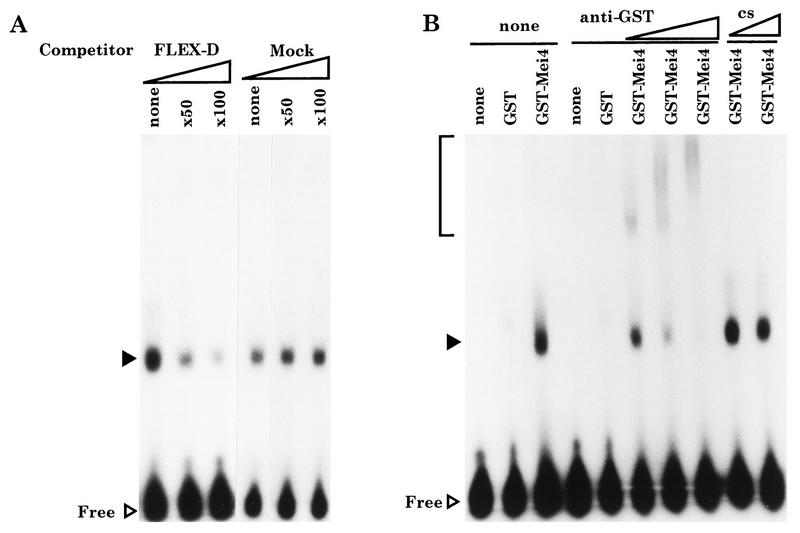
FIG. 6.
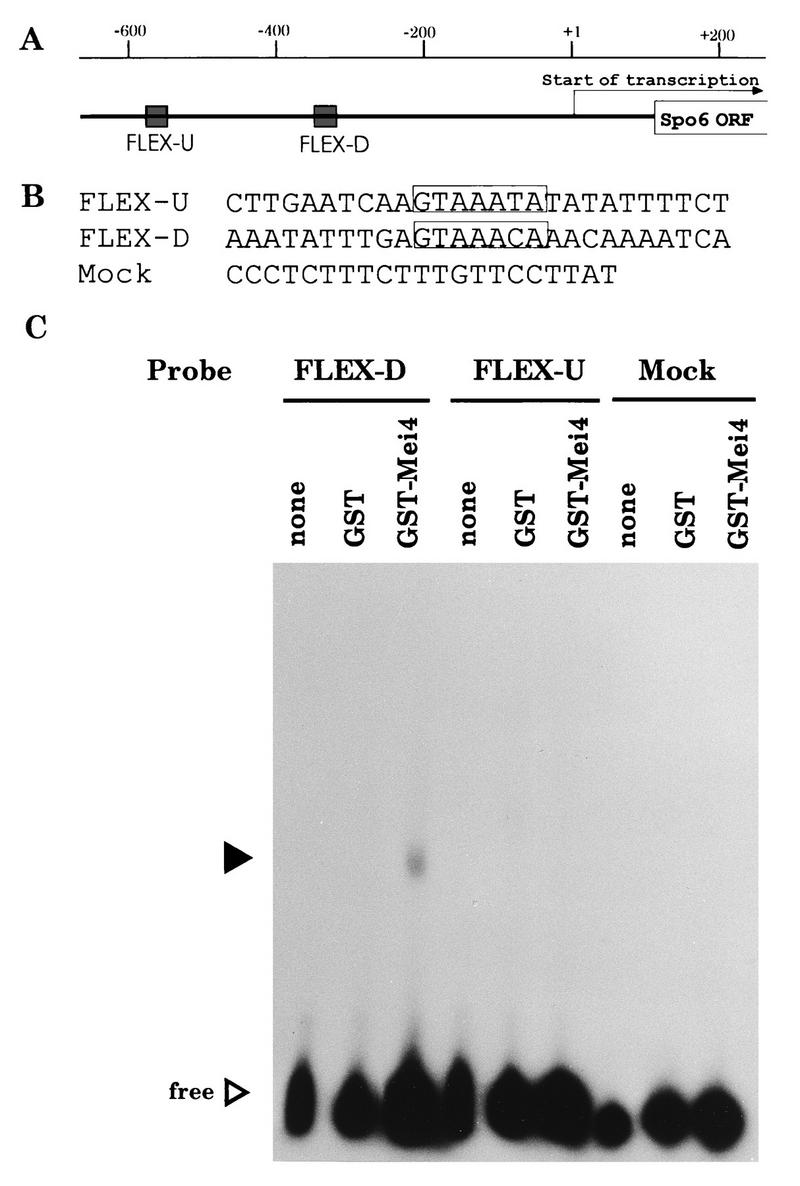
Gel mobility shift assay with a recombinant Mei4 protein. (A) Map of the 5′ promoter sequence of spo6+. Shaded boxes called FLEX-U and FLEX-D indicate the putative recognition sequences containing the core heptamer motif. An arrow represents the direction of transcription and the site of the start point. (B) Sequences of oligonucleotides used for probes. The mock oligonucleotide is the recognition site for Ste11. (C) Gel shift analysis. A whole-cell extract from E. coli expressing the GST-Mei4 forkhead fusion protein was incubated with labeled oligonucleotide probes. Closed and open triangles indicate shifted bands and free probes, respectively.
To demonstrate that Mei4 can bind to the FLEX sequences, a fusion protein composed of GST and the forkhead domain of Mei4 (aa 71 to 182) was subjected to a gel retardation assay with radioactive FLEX-U and FLEX-D as probes. As shown in Fig. 6C, the shifted band was observed only with FLEX-D. The fusion protein caused no mobility shift with either FLEX-U or the mock oligonucleotide. The GST protein without the forkhead region did not recognize any oligonucleotides examined. These gel shift assays indicated that the Mei4 fusion protein could bind to the FLEX-D sequence by the amino-terminal forkhead domain.
We then tested competition of binding with nonlabeled oligonucleotides to examine the sequence specificity. Binding of GST-Mei4 fusion protein to the labeled FLEX-D oligonucleotide was severely inhibited by adding cold competitor of the same sequence, but no such competition was observed with unrelated oligonucleotides (Fig. 7A). Thus, we conclude that the binding was sequence specific.
FIG. 7.
Gel mobility shift analysis indicating the specificity of binding between FLEX-D and the recombinant Mei4 protein. (A) Competition assay. FLEX-D probes were incubated with GST-Mei4. Closed and open triangles indicate shifted bands and free probes, respectively. (B) Supershift experiment using the anti-GST antibody. A different amount of antibody was included in the reaction mixture. The probe used was FLEX-D. The supershifted bands due to anti-GST are indicated by a bracket. cs, control serum.
To confirm that the GST-Mei4 fusion protein itself bound to FLEX-D, a supershift analysis with an anti-GST antibody (Pharmacia Biotech) was carried out. When the antiserum was included in the reaction mixture, the shifted band was further retarded (Fig. 7B), clearly indicating that the GST-Mei4 fusion protein was responsible for the mobility shift.
The GST-Mei4 fusion protein specifically recognizes FLEX-D.
To corroborate that the core consensus sequence of FLEX-D was important for recognition by Mei4, we tested binding of Mei4 to the mutated oligonucleotide in which the central AAA of the core heptamer was replaced by CCC (Fig. 8A). The fusion protein could not bind to this mutated oligonucleotide (FLEX-Dm2) (Fig. 8B).
FIG. 8.
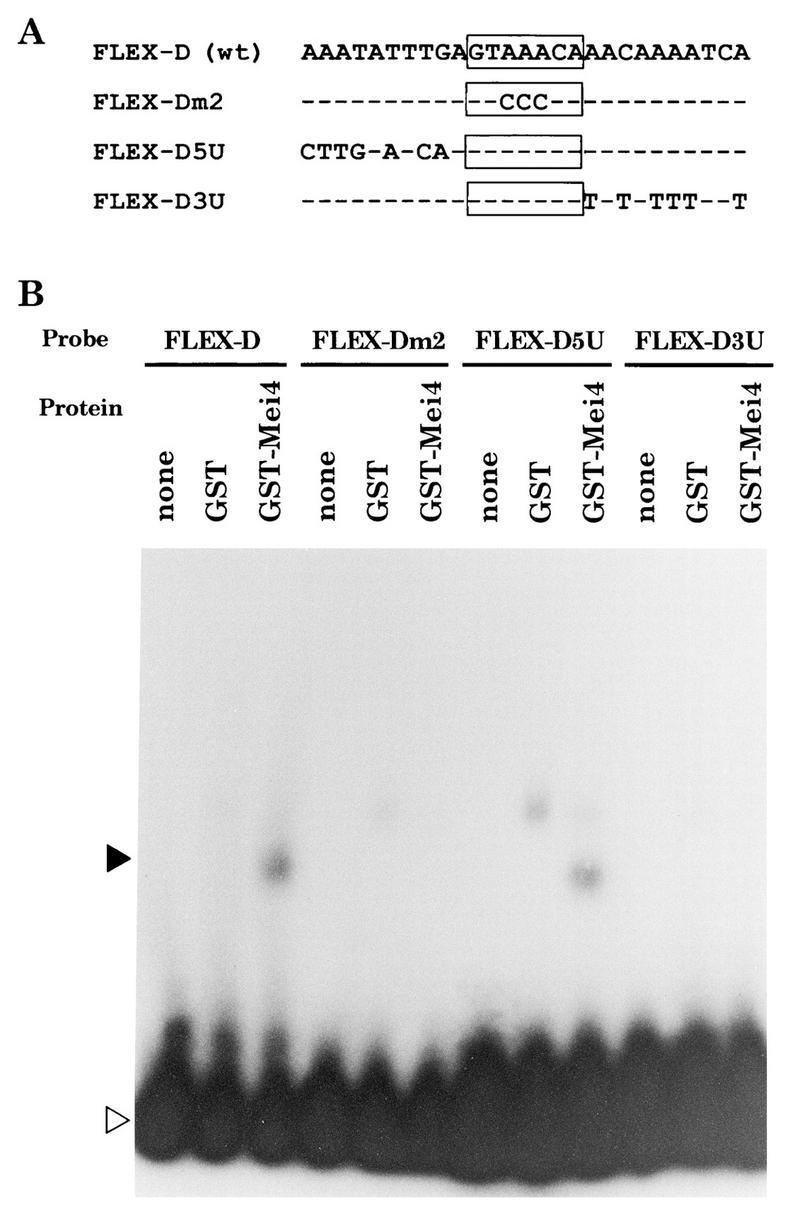
Gel mobility shift analysis with mutated FLEX-D oligonucleotides. (A) Sequences of oligonucleotides used. The core heptamer is boxed, and nucleotides identical to those in FLEX-D are indicated by dashes. wt, wild type. (B) Results of gel shift assay. Closed and open triangles indicate shifted bands and free probes, respectively.
The GST-Mei4 fusion protein recognized FLEX-D but not FLEX-U (Fig. 6C). Only the sixth base was different in the core heptamers of FLEX-U (GTAAATA) and FLEX-D (GTAAACA). This difference, however, did not affect the binding affinity, as the replacement of T by C or vice versa only slightly influenced the band intensity (data not shown). This suggests that the variation in the FLEX core heptamer itself is not the cause for the binding affinity. Thus, we examined the significance of the flanking sequences. The 5′ and 3′ flanking sequences of FLEX-D were replaced with those of FLEX-U as shown in Fig. 8A. The gel shift assay (Fig. 8B) indicated that oligonucleotides in which the 3′ flanking sequence was replaced (FLEX-D3U) could hardly be recognized by the GST-Mei4 fusion protein. A FLEX-D5U probe gave an additional band when mixed with the GST-Mei4 fusion protein. This slower-migrating band might be a nonspecific one, because a band of the same mobility was also found with GST alone. We conclude that the 3′ flanking sequence, in addition to the core heptamer, is important for recognition by Mei4.
FLEX-D is essential for the transcription of spo6+.
The forkhead domain of Mei4 could bind to FLEX-D in vitro. We addressed the question of whether this sequence could function as a transcriptional cis element. The FLEX-D core heptamer was deleted from plasmid-borne spo6+. The plasmid carrying this deleted allele of spo6 (spo6-DF) was transformed into a diploid strain (NT-4A) homozygous for spo6Δ to test its ability to complement the sporulation defect. As shown in Fig. 9A, the spo6-DF allele complemented the spo6 null mutation only very weakly, while the spo6+ and spo6-X genes carrying the core heptamer in the promoter were able to rescue the sporulation defect.
FIG. 9.
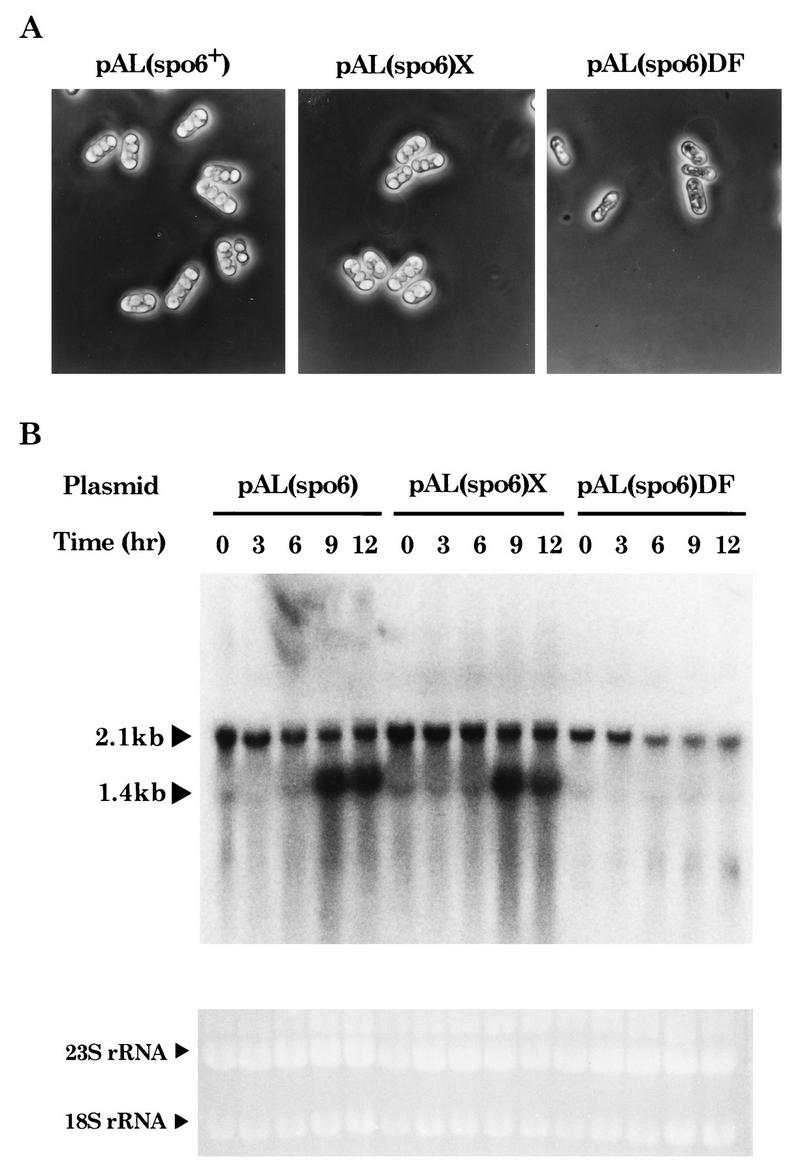
Deletion of the core heptamer in the FLEX-D element from the spo6 promoter reduces sporulation ability and spo6 transcription. (A) Complementation of the sporulation defect of spo6Δ. Plasmid pAL(spo6) is pAL-KS carrying spo6+. pAL(spo6)X and pAL(spo6)DF are pAL-KS containing the spo6 gene with or without the core heptamer of FLEX-D, respectively. A diploid spo6 disruptant, NT-4A, was transformed with the pAL-KS-based plasmids, and then the transformants were sporulated on SSA for 2 days at 28°C. (B) Northern blot analysis showing the reduced expression of spo6 caused by the deletion of the promoter element. A diploid spo6 disruptant strain (NT-4A) transformed with the indicated plasmids was grown in nitrogen-rich medium (EMM2 with N) and shifted to nitrogen-free medium (EMM2 without N). Cells were harvested at the indicated times after the transfer to EMM2 without N. Total RNA was extracted and subjected to Northern analysis with the spo6-specific probe. Two different mRNA species, of 2.1 and 1.4 kb, are indicated. Gel staining with ethidium bromide indicates roughly equal amounts of RNA in each lane.
Transcription of spo6 in these transformants was examined by Northern analysis (Fig. 9B). Two classes of the spo6 transcripts were detected. The meiosis-specific 1.4-kb band was prominent in the spo6+ and spo6-X strains but was almost completely missing in the spo6-DF strain. On the other hand, the 2.1-kb mRNA species was not affected by the mutation. These observations support that FLEX-D serves as the cis-acting element for the Mei4 transcription factor in spo6+ cells.
The activation domain resides in the C-terminal region of Mei4.
To dissect the transcriptional activation activity of Mei4, we used an S. cerevisiae one-hybrid analysis. The full-length Mei4 protein was fused to the S. cerevisiae Gal4 DNA-binding domain on plasmid pGBT9. The resulting plasmid, pGBT(mei4)FL, was transformed into S. cerevisiae SF526, which carried the GAL1 promoter upstream of the lacZ reporter gene. The β-galactosidase activity in the transformants with pGBT(mei4)FL was significantly higher than that in transformants with the control plasmid pGBT9 (Fig. 10), confirming that Mei4 is able to activate transcription.
Deletion analysis localized the activation domain of Mei4 in the C-terminal region from aa 343 to 517. As expected, the N-terminal half containing the forkhead domain did not stimulate transcription. Deletion of the region between aa 441 and 517 completely eliminated the activation activity.
We conclude that Mei4 is a meiosis-specific transcription factor, in which the N-terminal 120 aa (aa 71 to 190) constitute a DNA-binding domain and the C-terminal 175 aa (aa 343 to 517) constitute a transcriptional activation domain (Fig. 4C).
DISCUSSION
Mei4 is a meiosis-specific transcription factor.
The following observations support that Mei4 is a meiosis-specific transcription factor of S. pombe. (i) mei4+ is transcribed only in meiotic cells. (ii) Mei4 contains a forkhead DNA-binding domain in the N-terminal region. Mutations introduced into this domain abolish the mei4 function. (iii) A meiosis-specific gene, spo6+, is not transcribed in mei4 null mutants. Ectopic expression of mei4+ in rich growth medium causes transcription of spo6+. (iv) A recombinant Mei4 protein could bind specifically to the FLEX-D DNA fragment, a putative cis element on spo6+. Deletion of this element totally eliminated the transcription of spo6+. (v) A one-hybrid assay proved the ability of Mei4 to activate transcription. The activation domain was localized in the C terminus.
Consensus cis element recognized by forkhead proteins.
We demonstrated that Mei4 is able to bind to the 27-mer DNA fragment called FLEX-D. It contains the core heptamer GTAAACA, which is identical to the elements required for human forkhead proteins (34). Furthermore, the core sequence of FLEX-D meets the requirement for the binding of HNF3β, (G/A)(T/C)(C/A)AA(C/T)A (33). Our mutational analysis demonstrated that this core heptamer is indispensable for the DNA-protein recognition between FLEX-D and Mei4. The fact that mammalian and yeast forkhead proteins recognize the common cis element suggests that DNA-binding properties have been conserved rather tightly among the forkhead family proteins.
A transcriptional cascade operates to drive meiosis in S. pombe.
Recent studies revealed that several putative transcription factors are integrated into a regulatory cascade leading to the initiation and progression of meiosis in S. pombe. A part of this cascade culminating in the expression of spo6+ and mes1+, which are essential for meiosis and sporulation, is illustrated in Fig. 11.
FIG. 11.
A regulatory cascade including some putative transcription factors, culminating in the expression of spo6+ and mes1+, which are necessary for meiosis and sporulation. Arrowheads and vertical bars indicate stimulatory and inhibitory actions, respectively. Putative transcription factors are boxed. Other gene products: Pka1, a catalytic subunit of A kinase; Pat1, a serine/threonine protein kinase; Mei3, a Pat1 inhibitor; Mei2, an RNA-binding protein.
Sexual differentiation in fission yeast is initiated on nutritionally poor media. Starvation signals are mediated through both the adenylate cyclase-protein kinase A pathway (15, 23, 24, 28) and the Wik1 (Wak1)-Wis1-Sty1 (Spc1) mitogen-activated protein kinase cascade (40, 41, 46). The latter pathway activates the Atf1 (Gad7) transcription factor, which is phylogenetically related to the mammalian transcription factor Atf (17, 54). Those signal transduction pathways may join at the expression of ste11+, which encodes a key transcription factor responsible for a wide variety of genes required for sexual development. The transcription of ste11+ requires Atf1 and is repressed by A kinase (Pka1). Ste11 recognizes the promoter element called the TR box, which is found in the 5′ upstream region of all of the target genes, including mei2+, mat1+, ste6+, and others (13, 18, 42, 47, 55). Most of the target genes are transcribed in response to a nitrogen starvation signal (1, 13, 39, 42, 55). The mat1+ products, Mat1-Pm and Mat1-Mm, cooperatively activate transcription of mei3+ (1, 18), which encodes the inhibitor of the Pat1 protein kinase (25). Ste11 thus promotes meiosis by indirectly enhancing mei3+ transcription (Fig. 11).
The mitotic G1-to-S transition is controlled by a complex of transcription factors, Cdc10-Res1 or Cdc10-Res2, which recognizes the MCB box (4, 22, 27, 43, 44). Two genes necessary for DNA replication, cdc18+ and cdc22+, are the targets of these complexes (22). Another transcription activator, called Rep1, is also included in the complex (43). The complex composed of Rep1, Cdc10, and Res2 (or Res1) primarily supports premeiotic DNA synthesis rather than mitotic DNA synthesis (27, 60). Interestingly, transcription of res2+ requires rep1+, and that of rep1+ requires Ste11 (43). Therefore, the premeiotic S phase is dependent on Ste11.
As mentioned above, transcription of mei4+ is stimulated by nitrogen starvation. Because the transcriptional activation is observed neither in haploid strains nor in an mei1 mutant in which meiosis is blocked before the premeiotic DNA synthesis, the mei4 expression is not a simple response to starvation signals. It is important to identify the factor regulating the transcription of mei4+. One likely candidate is Mei2, which is a crucial inducer of meiosis (48). As shown in Fig. 11, Mei2 is positively regulated by Ste11 and negatively controlled by the Pat1 protein kinase which blocks the initiation of meiosis in haploid cells under growth conditions (42, 50). Heterothallic haploid cells carrying the activated mei2 allele (mei2-SATA) undergo meiosis even in a rich growth medium (50). mei4+ transcription is totally abolished in mei2Δ cells, and full transcription of mei4+ was observed in the mei2-SATA mutant in nitrogen-rich medium (51). These facts indicate that mei4 transcription is dependent upon mei2+ function. From the fact that the mei4 mRNA level decreases in mei4-P572 cells (Fig. 3C), we speculate that there is a positive feedback mechanism for mei4+ transcription by its product. However, the GTAAAYA motif for Mei4-binding sites is not present in the promoter region of mei4+.
Targets of Mei4.
We demonstrated that Mei4 regulates expression of spo6+ and mes1+ at the transcriptional level. Two copies of the consensus core motif GTAAAYA were found in the promoters of both genes. However, the phenotypes of spo6Δ and mes1Δ mutants are not identical to that of the mei4Δ mutant, which is blocked in prophase-I, suggesting that there are certain targets of Mei4 other than spo6+ and mes1+. It is noteworthy that the spliced mature mes1+ mRNA could not be detected in mei4Δ cells. Splicing of mes1 mRNA appears to be regulated by some hypothetical meiosis-specific splicing factor(s). We speculate that the expression of these putative splicing factors may depend on the Mei4 transcription factor.
It is known that cdc13+ and cdc25+ are not fully transcribed in mei4 disruptant strains (14). The reduction of the mRNA levels of these cdc genes may be responsible for the arrest of mei4 null mutants in prophase-I. Genes that are essential for the progression through prophase-I have not yet been reported for S. pombe. It is highly probable that some of these genes are possible targets of Mei4.
ACKNOWLEDGMENTS
This study was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan to C.S., Y.W., and M.Y.
REFERENCES
- 1.Aono T, Yanai H, Miki F, Davey J, Shimoda C. Mating pheromone-induced expression of the mat1-Pm gene of Schizosaccharomyces pombe: identification of signaling components and characterisation of upstream controlling elements. Yeast. 1994;10:757–770. doi: 10.1002/yea.320100607. [DOI] [PubMed] [Google Scholar]
- 2.Beach D, Nurse P. High-frequency transformation of the fission yeast Schizosaccharomyces pombe. Nature. 1981;290:140–142. doi: 10.1038/290140a0. [DOI] [PubMed] [Google Scholar]
- 3.Bresch C, Muller G, Egel R. Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- 4.Caligiuri M, Beach D. Sct1 functions in partnership with cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell. 1993;72:607–619. doi: 10.1016/0092-8674(93)90079-6. [DOI] [PubMed] [Google Scholar]
- 5.Chikashige Y, Hiraoka Y. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark K L, Halay E D, Lai E, Burley S K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 7.Egel R. Mating-type genes, meiosis, and sporulation. In: Nasim A, Young P, Johnson B F, editors. Molecular biology of the fission yeast. New York, N.Y: Academic Press; 1989. pp. 31–73. [Google Scholar]
- 8.Esposito R E, Klapholz S. Meiosis and ascospore development. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 211–287. [Google Scholar]
- 9.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 10.Hereford L, Fahner K, Woolford J, Jr, Rosbash M, Kabak D B. Isolation of yeast histone genes H2A and H2B. Cell. 1979;18:1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- 11.Hirata A, Shimoda C. Electron microscopic examination of sporulation-deficient mutants of the fission yeast Schizosaccharomyces pombe. Arch Microbiol. 1992;158:249–255. doi: 10.1007/BF00245240. [DOI] [PubMed] [Google Scholar]
- 12.Hirata A, Shimoda C. Structural modification of spindle pole bodies during meiosis II is essential for the normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast. 1994;10:173–183. doi: 10.1002/yea.320100205. [DOI] [PubMed] [Google Scholar]
- 13.Hughes D A, Fukui Y, Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990;344:355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- 14.Iino Y, Hiramine Y, Yamamoto M. The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe. Genetics. 1995;140:1235–1245. doi: 10.1093/genetics/140.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 16.Jensen R, Sprague G F, Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci USA. 1983;80:3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells. 1996;1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
- 18.Kelly M, Burke J, Smith M, Klar A, Beach D. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988;7:1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishida M, Nagai T, Nakaseko Y, Shimoda C. Meiosis-dependent mRNA splicing of the fission yeast Schizosaccharomyces pombe mes1+ gene. Curr Genet. 1994;25:497–503. doi: 10.1007/BF00351668. [DOI] [PubMed] [Google Scholar]
- 20.Kupiec M, Byers B, Esposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J B, Jones E W, editors. Molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 21.Lai E, Prezioso V R, Smith E, Litvin O, Costa R H, Darnell J. HNF-3A, a hepatocyte-enriched transcription factor of novel structure, is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- 22.Lowndes N F, McInerny C J, Johnson A L, Fantes P A, Johnston L H. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+ Nature. 1992;355:449–452. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- 23.Maeda T, Mochizuki N, Yamamoto M. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1990;87:7814–7818. doi: 10.1073/pnas.87.20.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda T, Watanabe Y, Kunitomo H, Yamamoto M. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J Biol Chem. 1994;269:9632–9637. [PubMed] [Google Scholar]
- 25.McLeod M, Beach D. A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature. 1988;332:509–514. doi: 10.1038/332509a0. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto M, Tanaka K, Okayama H. res2+, a new member of the cdc10+/SWI4 family, controls the “start” of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13:1873–1880. doi: 10.1002/j.1460-2075.1994.tb06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochizuki N, Yamamoto M. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol Gen Genet. 1992;233:17–23. doi: 10.1007/BF00587556. [DOI] [PubMed] [Google Scholar]
- 29.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1990;194:793–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, T., M. Kishida, and C. Shimoda. Unpublished data.
- 31.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducting vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson L W, Eden U, Egel-Mitani M, Egel R. Asynaptic meiosis in fission yeast? Hereditas. 1978;89:189–199. [Google Scholar]
- 33.Overdier D G, Porcella A, Costa R H. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinow C F. The number of chromosomes in Schizosaccharomyces pombe: light microscopy of stained preparations. Genetics. 1977;87:491–497. doi: 10.1093/genetics/87.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothstein R. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimoda C, Hirata A, Kishida M, Hashida T, Tanaka K. Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1985;200:252–257. doi: 10.1007/BF00425432. [DOI] [PubMed] [Google Scholar]
- 39.Shimoda C, Uehira M, Kishida M, Fujioka H, Iino Y, Watanabe Y, Yamamoto M. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1987;169:93–96. doi: 10.1128/jb.169.1.93-96.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 41.Shiozaki K, Shiozaki M, Russell P. Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol Biol Cell. 1997;8:409–419. doi: 10.1091/mbc.8.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 43.Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SWI4 gene products. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas P S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warbrick E, Fantes P A. Five novel elements involved in the regulation of mitosis in fission yeast. Mol Gen Genet. 1992;232:440–446. doi: 10.1007/BF00266249. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe Y, Iino Y, Furuhata K, Shimoda C, Yamamoto M. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 1988;7:761–767. doi: 10.1002/j.1460-2075.1988.tb02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe Y, Yamamoto M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol. 1996;16:704–711. doi: 10.1128/mcb.16.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, Y. Unpublished data.
- 52.Weigel D, Jackle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990;63:455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- 53.Weigel D, Jurgens G, Kuttner F, Seifert E, Jackel H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson M G, Samuels M, Takeda T, Toone W M, Shieh J C, Toda T, Millar J B, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 55.Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol Cell Biol. 1995;15:4964–4970. doi: 10.1128/mcb.15.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto M. The molecular control mechanism of meiosis in fission yeast. Trends Biol Sci. 1996;21:18–22. [PubMed] [Google Scholar]
- 57.Yamamoto M, Imai Y, Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle J R, Broach J B, Jones E W, editors. Molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1037–1106. [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 59.Zhu G, Muller E G, Amacher S L, Northrop J L, Davis T N. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the forkhead family of DNA-binding proteins. Mol Cell Biol. 1993;13:1779–1787. doi: 10.1128/mcb.13.3.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Y, Takeda T, Nasmyth K, Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]