Abstract
There is general agreement that DNA synthesis in the single-copy and amplified dihydrofolate reductase (DHFR) loci of CHO cells initiates somewhere within the 55-kb spacer region between the DHFR and 2BE2121 genes. However, results of lagging-strand, early-labelling fragment hybridization (ELFH), and PCR-based nascent-strand abundance assays have been interpreted to suggest a very narrow zone of initiation centered at a single locus known as ori-β, while two-dimensional (2-D) gel analyses suggest that initiation can occur at any of a large number of potential sites scattered throughout the intergenic region. The results of a leading-strand assay and two intrinsic labelling techniques are compatible with a broad initiation zone in which ori-β and a second locus (ori-γ) are somewhat preferred. To determine how these differing views are shaped by differences in experimental manipulations unrelated to the biology itself, we have applied the lagging-strand, ELFH, neutral-neutral, and/or neutral-alkaline 2-D gel assays to CHOC 400 cell populations synchronized and manipulated in the same way. In our experiments, the lagging-strand assay failed to identify a template strand switch at ori-β; rather, we observed a gradual, undulating change in hybridization bias throughout the intergenic spacer, with hybridization to the two templates being approximately equal near a centered matrix attachment region. In the ELFH assay, all of the fragments in the 55-kb intergenic region were labelled in the first few minutes of the S phase, with the regions encompassing ori-β and ori-γ being somewhat preferred. Under the same conditions, neutral-neutral and neutral-alkaline 2-D gel analyses detected initiation sites at multiple locations in the intergenic spacer. Thus, the results of all existing replicon-mapping methods that have been applied to the amplified DHFR locus in CHOC 400 cells are consistent with a model in which two somewhat preferred subzones reside in a larger zone of multiple potential initiation sites in the intergenic region.
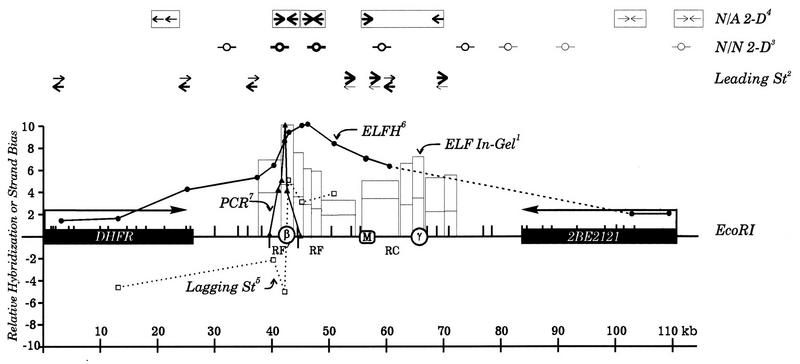
To analyze the replication pattern of a defined mammalian chromosomal locus, we have taken advantage of a methotrexate-resistant CHO cell line (CHOC 400) in which one allele of the early-replicating dihydrofolate reductase (DHFR) locus is amplified ca. 1,000 times (30). The high copy number of the 240-kb repeating unit (amplicon) has facilitated a number of studies designed to identify the replication start site(s) (reviewed in references 8 and 17). By radiolabelling the DNA synthesized at the beginning of the S phase, it was possible to show that replication initiates preferentially in the region lying between the convergently transcribed DHFR and 2BE2121 genes (20) (Fig. 1). In a more quantitative and higher-resolution in-gel renaturation approach, two somewhat preferred initiation regions (termed ori-β and ori-γ) were identified within the intergenic spacer (Fig. 1, ELF in-gel) (26). ori-β and ori-γ are ∼22 kb apart and lie on either side of a prominent matrix attachment region (MAR) (9). In recent years, several additional replicon-mapping techniques with potentially higher resolution and/or sensitivity have been applied to the DHFR locus either in CHOC 400 cells or in parental CHO cells. However, the results from these different approaches do not paint a unified picture.
FIG. 1.
The DHFR initiation locus. A 110-kb region encompassing the convergently transcribed DHFR and 2BE2121 genes and the intergenic region of the DHFR domain is shown. Positions of ori-β, ori-γ, and the MAR on the EcoRI map are indicated by open circles and the square, respectively. The results of seven different replicon-mapping assays are shown diagramatically in the figure. Note that the data from the ELFH, ELF in-gel renaturation, and PCR-based nascent-strand abundance assays have been normalized to one another, with the maximum relative values in each case being set equal to 10 on the scale. The superscript numbers correspond to the following: 1, in-gel renaturation data (26); relative intrinsic radiolabelling of amplified restriction fragments in the indicated 33-kb region in the first 30 min (lower boxes) or 60 min (taller boxes) of the S phase, as quantified by densitometry of labelled amplified restriction fragments after in-gel renaturation; 2, leading-strand data; relative numbers of leading strands moving in the two directions when lagging-strand replication is inhibited by emetine, as estimated by eye from the data in reference 19 (note that the sum of the intensities of the two arrows at any one position was arbitrarily adjusted to be approximately equal, since absolute amounts of labelling to each pair of templates were not assessed in this study); 3, neutral-neutral (N/N) 2-D gel data; relative approximate quantities (indicated by the intensities of the bubbles) of replication bubbles detected in the indicated EcoRI fragments on neutral-neutral 2-D gels in several independent studies (see, e.g., references 10, 11, 13, and 39); note that no bubbles have ever been detected in the DHFR gene; 4, neutral-alkaline (N/A) 2-D gel data; approximate relative quantities of replication forks moving into each end of the indicated fragments, as detected on neutral-alkaline 2-D gels (13, 39); note that the thickness of the arrows corresponds to relative fork numbers at each position; 5, results of the lagging-strand assay (5); hybridization bias of lagging Okazaki fragments to template strands after correction for hybridization to the vector (calculated by dividing the larger of the two values by the smaller); 6, ELFH data (replotted from reference 14) after normalizing the highest value to the highest relative value in the in-gel renaturation data; 7, PCR-based nascent-strand analysis; relative quantities of small nascent strands at different template positions in asynchronous CHO cells, as detected by PCR (replotted from data presented in reference 34 after normalizing the highest value to the highest value in the in-gel renaturation data set). See the original references for experimental details.
One view was suggested by measurements of the template bias of lagging nascent strands, which should switch dramatically at a fixed origin of replication. When this technique was applied to the 11-kb region surrounding ori-β in synchronized CHOC 400 and CHO cells, a pronounced switch in template bias suggested that >80% of initiations occur within a 480-bp fragment centered in the ori-β region (Fig. 1, Lagging St) (5). Thus, ori-β has been designated an origin of bidirectional replication (5). The same result was reported for asynchronous CHOC 400 cells (5), which excluded the possibility of a second active origin in the region of ori-γ; this follows because Okazaki fragments in forks replicating through ori-β from either direction would not change templates at this site, resulting in a much lower switch in template bias than was actually observed. The results of PCR-based nascent-strand abundance assays (34, 38) also argue that ori-β is a highly preferred initiation locus (Fig. 1, PCR) (34). However, neither the lagging-strand nor nascent-strand abundance assay examined the region surrounding ori-γ. Thus, the presence of a preferred initiation site or zone in this region was not formally excluded.
At the other end of the spectrum are results from two-dimensional (2-D) gel analyses of the DHFR locus. A neutral-neutral 2-D gel replicon-mapping method (3) suggests that initiation can occur at virtually any location within the 55-kb intergenic region, with relatively more initiations occurring in the central 30 to 35 kb encompassing ori-β and ori-γ (Fig. 1, N/N 2-D) (10, 11, 13, 39). In apparent agreement, a neutral-alkaline 2-D gel method (33) detects replication forks travelling in both directions within this central region in the early S phase (Fig. 1, N/A 2-D) (11, 13, 39). Two recent variations of the 2-D gel methods that were designed specifically to detect a highly favored start site at ori-β failed to do so (22, 23), lending support to a model in which potential initiation sites are scattered throughout the intergenic region, with ori-β and ori-γ being only somewhat preferred.
The results of other approaches could accommodate either view. In an early-labelling fragment hybridization (ELFH) assay, DNA labelled either in vitro (4, 16) or in vivo (4) in the first 5 to 30 min of the S phase was used to probe immobilized clones from the intergenic region to determine which restriction fragments are the first to be synthesized at the beginning of the S phase. In two studies (4, 16), the resulting hybridization patterns identified a peak of early labelling that encompassed ori-β. However, because of the absence of clones containing and surrounding ori-γ, neither study examined the possibility of a second preferred initiation locus in this region (Fig. 1, ELFH) (16). Indeed, in an in vivo variation of this approach in which psoralen cross-links were used to confine labelling to the region immediately surrounding the origins, a very sharp peak was detected over ori-β and a much broader peak was detected over ori-γ (1). However, the signal from the ori-β region was subsequently shown to arise, at least in part, from two closely spaced copies of the AluI-like repeated sequence family in this region (6, 25), and hybridization could be completely eliminated by inclusion of excess unlabelled CHO DNA in the hybridization solution (25). Thus, although the work described in reference 1 was subsequently cited as evidence for a highly favored initiation site in the region of ori-β (5, 8, 38), the specific labelling of ori-β itself (but not ori-γ, which contains no repeated elements) was indeterminate owing to cross-hybridization with other early-replicating repetitive sequences in the genome.
Finally, determinations of the template bias of leading strands could accommodate either highly preferred sites at ori-β and ori-γ or a much broader zone with ori-β and ori-γ being only somewhat preferred, since the nearest probes lay 5 to 8 kb on either side of the two regions and since there was significant hybridization to both strands at all positions (Fig. 1, Leading St) (19).
There are other inconsistencies among the data sets. As mentioned above, the lagging-strand assay was reported to detect a template switch at the ori-β locus even in asynchronous CHOC 400 cell populations (5). Even if one excludes the possible existence of an ori-γ, this data implies that the DHFR origin is extremely efficient, with initiation occurring in all of the amplicons almost simultaneously in the early S phase in every cell cycle. Thus, at a fork rate of 3 kb/min (21), it should take less than 1 h to replicate most of the 240-kb amplicons. However, intrinsic labelling studies showed that it takes at least 8 h to replicate all of the amplicons (20); furthermore, neutral-neutral and neutral-alkaline 2-D gel methods detected bubbles in the intergenic zone only in the first 2 h of the S phase but single forks persisted for more than 6 h (10, 11, 13, 18, 39). Thus, initiation must occur in only 10 to 15% of the amplicons in any one S phase, with the remainder being replicated passively by forks from distant amplicons. Thus, in an asynchronous culture, a strand switch at ori-β in 10 to 15% of amplicons should be largely obscured by readthrough of this locus in the remaining 80 to 85% that is passively replicated.
Clearly, each replicon-mapping method measures different properties of replicating DNA and each has its own strengths and weaknesses. For example, the neutral-neutral 2-D gel technique can detect initiation events even at minor sites but cannot detect small differences in the relative number of bubbles from one position to the next along a template, owing primarily to differences in restriction fragment sizes along a region of interest (29). The leading- and lagging-strand assays have the advantage that hybridization biases to the two template strands can be reasonably accurately quantified within a given experiment; however, the bias to one strand or the other is never absolute (5, 19, 24, 36) and, unlike 2-D gel approaches, leading- and lagging-strand assays cannot distinguish between minor initiation sites distributed throughout the replicon under study and unavoidable background hybridization.
It is also significant that the indicators in the lagging-strand and ELFH assays are fragments that were labelled in vitro by a 1.5-min pulse of radioactive deoxyribonucleotides (5) and therefore qualify as short-lived replication intermediates whereas 2-D gel methods measure the steady-state in vivo distribution of intermediates and therefore bias toward longer-lived species. Furthermore, the lagging-strand and ELFH assays were performed on cells just as they were entering the S phase (i.e., ∼4 min after release from aphidicolin [5, 16]), while most 2-D gel measurements were performed when initiation in the DHFR locus was maximal (either 20 to 30 min after release from aphidicolin [39] or 80 to 90 min after release from mimosine [10, 11, 13]).
We assume that each assay is valid and accurately reports some aspect of the biological initiation reaction. Therefore, in the present study we attempted to fix several of the experimental variables that could complicate the interpretations of the data. We have applied the lagging-strand, ELFH, neutral-neutral 2-D gel, and/or neutral-alkaline 2-D gel assays to replication intermediates prepared from CHOC 400 cells under the same experimental conditions. Results from these assays appear to be consistent with one another and suggest that there is a broad zone of initiation in the intergenic region of the amplified DHFR domain, with the region encompassing ori-β and ori-γ being somewhat preferred.
(This work fulfills part of the requirement for a Ph.D. in biochemistry from the University of Virginia for S.W.)
MATERIALS AND METHODS
Cell culture and synchronization.
CHOC 400 cells were developed as previously described (30) and were maintained in minimal essential medium supplemented with nonessential amino acids (Bethesda Research Laboratories [BRL]), and 10% Hyclone II (BRL) in an atmosphere of 5% CO2. Synchronization near the G1/S boundary was achieved by first arresting cells in G0 by isoleucine starvation for 45 h and then incubating them in complete medium containing either 10 μg of aphidicolin (Sigma) per ml (12-h incubation) or 400 μM mimosine (Aldrich) (14-h incubation) (10, 27, 32). Drug-containing media were washed out with cold (after aphidicolin) or warm (after mimosine) serum-free medium and replaced with complete medium. Approximately 4 min after removal of aphidicolin or 90 min after removal of mimosine, the cells were harvested for analysis.
In vitro replication reactions and isolation of replication intermediates.
Except where noted, in vitro replication reactions were carried out as previously described (5). Briefly, the cells were washed with ice-cold minimal essential medium, scraped from the plate, and pelleted at 300 × g in an Eppendorf centrifuge for 3 min. A 100-μl aliquot of the cell pellet (∼3 × 107 cells per reaction) was mixed with 120 μl of ice-cold 2× DNA replication cocktail (100 mM HEPES; 0.2 mM each dGTP and dCTP; 0.4 mM each GTP, UTP, and CTP; 8 mM ATP; 20 mM MgCl2; 0.2 mg of bovine serum albumin per ml; 2 mM dithiothreitol; 30% glycerol); note that bromodeoxy-UTP (BrdUTP) was omitted in the present study. Then 10 μl each of [α-32P]dATP and [α-32P]dTTP (Amersham; 3,000 Ci/mmol) and 5 μl of 20% Nonidet P-40 (NP40) were added, and in vitro replication was initiated by incubation at 34°C for 1.5 min. The reactions were quenched by adding 2 ml of 50 mM Tris-HCl (pH 8)–10 mM EDTA–400 mM NaCl–1% sodium dodecyl sulfate (SDS) containing 10 μg of proteinase K (E.M. Laboratories) per ml and 20 μg of RNase A (Qiagen) per ml and incubating the mixture at 37°C for 1 to 2 h. In the pulse-chase experiment, dATP and dTTP were added to 0.1 mM at the end of the labelling period and incubation was continued for 30 min at 34°C before the quenching step.
Isolation of Okazaki and early-labelled fragments.
The lysates received one-third volume of room temperature 5 M NaCl and were quickly and thoroughly mixed. Denatured proteins were precipitated by centrifugation (10,000 rpm at 25°C in an HB-6 rotor [Dupont/Sorvall] for 1 h). The DNA in the supernatant was precipitated with 2 volumes of cold ethanol and centrifuged in the cold at 10,000 rpm in the HB-6 rotor for 1 h, and the pellet was washed several times with 70% ethanol containing 2 M ammonium acetate. For the lagging-strand assay, DNA was dissolved in TE buffer (10 mM Tris [pH 8], 1 mM EDTA) at 100 μl of TE buffer per plate equivalent and subjected to alkaline gel electrophoresis (5). The labelled fraction migrating between 50 and 300 nucleotides (nt) was excised from the gel and recovered by electroelution with a Schleicher & Schuell apparatus. Residual RNA was hydrolyzed by incubation with 0.2 N NaOH for 24 h at 37°C. For the ELFH assay, DNA was dissolved in 0.2 M NaOH–5 mM EDTA and incubated at 37°C for 24 h to degrade residual RNA. The labelled genomic DNA was sonicated for 3 min on ice with a Fisher Scientific Sonic Dismembrator 550 at a setting of 3 with a microtip, and the size of the sonicated DNA was ascertained on an alkaline agarose gel. The solution was neutralized and ethanol precipitated. DNA samples were redissolved in TE buffer and used for hybridization.
Preparation of plasmids and RNA transcripts.
Restriction fragments from 16 different locations in the 110-kb region shown in Fig. 1 were cloned into a pBS(+) vector (Stratagene); the insert sizes were as follows: 103, 1.2 kb; HCC, 0.8 kb; 3, 0.9 kb; 38, 0.5 kb; 8, 0.9 kb; DGK, 1.2 kb; 14, 0.6 kb; 9, 0.5 kb; 10, 0.7 kb; and 206, 0.7 kb. pBS clones containing fragments HCB and HCC were gifts from Howard Cedar (Hebrew University), and the SK(−) clone DGK was obtained from David Gilbert (Syracuse University). To confirm the absence of repeated sequences, each clone was labelled with [32P]dCTP (3,000 Ci/mmol; Amersham) by random priming (15) and used to probe a restriction digest of genomic CHO DNA separated on a 0.8% agarose gel and transferred to HyBond N+ (Amersham).
The T3 and T7 promoters flanking the polylinker cloning site in the pBS(+) vector were used to prepare full-length transcripts of the plus and minus strands of each insert. Plasmid DNA was linearized at the appropriate restriction site, and the linear form was isolated by agarose gel electrophoresis and recovered with a GeneClean kit (Bio 101). Transcription was carried out with a Promega T3/T7 kit. Template DNA was digested with RNase-free DNase (Promega). An RNeasy kit (Qiagen) was used to purify RNA products and to eliminate unincorporated nucleoside triphosphates. The RNA concentrations were determined spectrophotometrically.
Dot-blotting and hybridization procedures.
A 1-μg portion of each pair of RNA samples or 1 μg of each double-stranded plasmid was dotted onto HyBond N+ with a 96-well Schleicher & Schuell manifold as described in reference 35. The filters were hybridized as described by Church and Gilbert (7) in 5 ml of hybridization fluid containing either ∼ 2 × 106 cpm of 32P-labelled Okazaki fragments or (6 to 10) × 106 cpm of sheared total DNA from the in vitro replication reactions. The membranes were washed for 30 min each in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2% SDS and 0.2× SSC–0.2% SDS at 65°C, and hybridization signals were detected and quantified with a Molecular Dynamics PhosphorImager.
2-D gel electrophoresis.
The cells were harvested at the indicated times in the same way as for in vitro replication assays and were incubated in replication cocktail for 1.5 min, at which time the nuclei were diluted into cell lysis buffer containing digitonin. The matrix-attached replication intermediates were then purified as previously described (12). Briefly, the nuclei were incubated with isotonic lithium diiodosalicylate to remove histone and soluble nonhistone chromosomal proteins (31). The resulting DNA halo was separated from matrix-attached replication intermediates with EcoRI, and the two fractions were separated by centrifugation and finally enriched by benzoylated naphthoylated DEAE-cellulose chromatography (28). The matrix fraction was analyzed by 2-D gel electrophoresis as previously described (3, 10, 12, 13). The digests were transferred to HyBond N+ and were hybridized with the fragments indicated above, which were labelled with [32P]dCTP by random priming (15).
RESULTS
Experimental design of lagging-strand and ELFH assays.
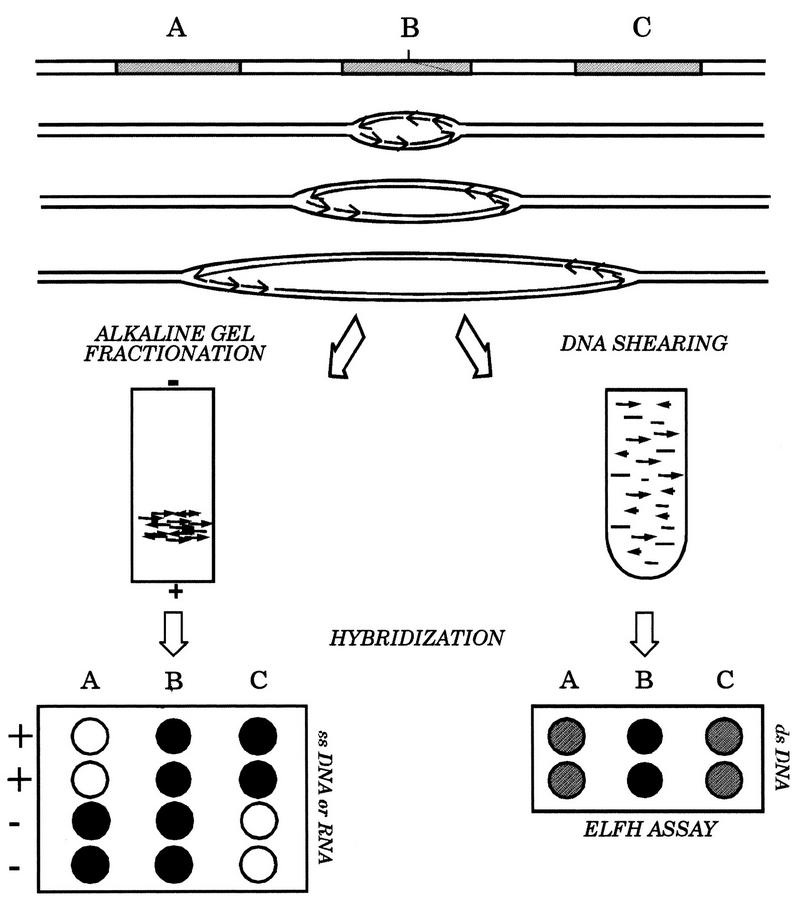
The purpose of this study was to compare the results obtained by lagging-strand, ELFH, and 2-D gel assays as applied to cells synchronized and manipulated under the same conditions. We have had extensive experience with 2-D gel assays, but since the lagging-strand and ELFH assays were new to our laboratory, we devoted considerable effort to standardizing these methods to avoid conditions that could artificially influence the outcomes. The theory and overall experimental design for the two assays are shown in Fig. 2.
FIG. 2.
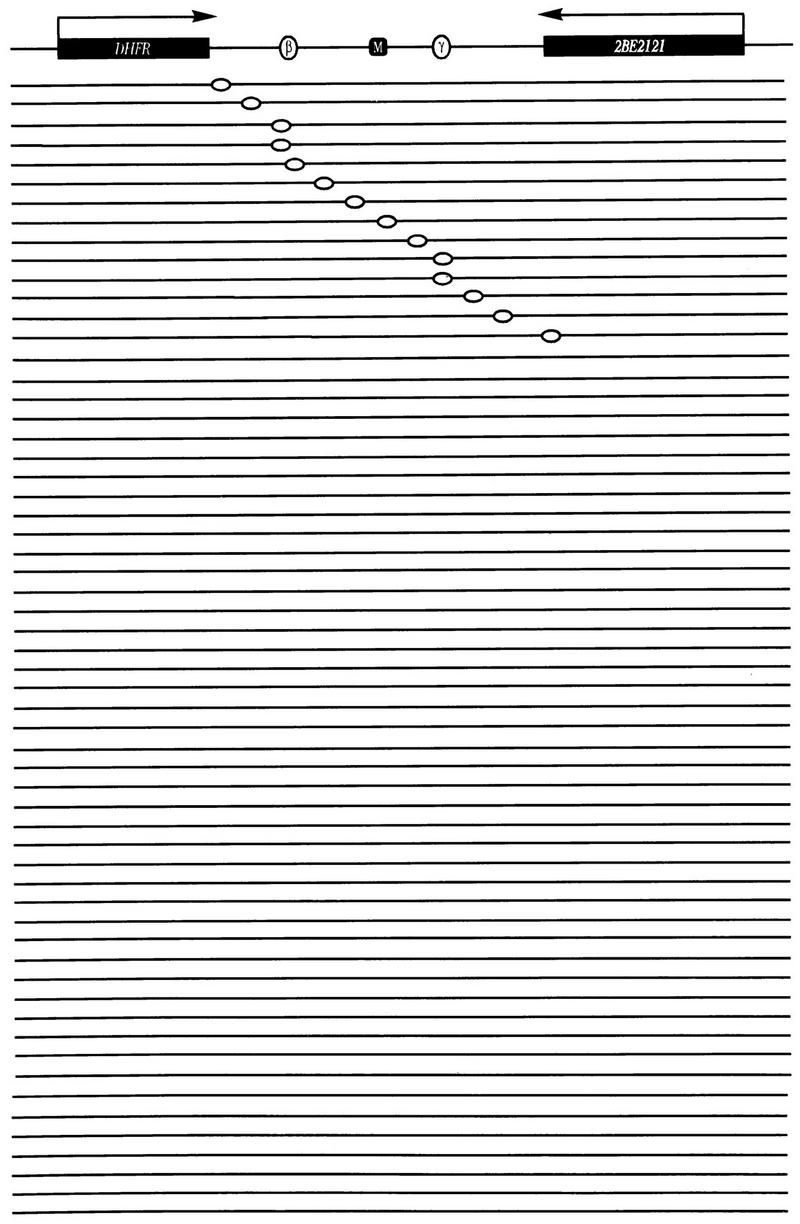
Principles of the ELFH and lagging-strand assays. Synchronous or asynchronous cultures are harvested and in vitro replication reactions are carried out in the presence of radioactive deoxynucleotides as described in the text. A single origin of replication is shown either in the unreplicated state or at three time points after initiation has occurred, to illustrate the distribution of leading- and lagging-strand Okazaki fragments and/or the distribution of intrinsic radioactivity at the time the reaction is quenched; three different restriction fragments (A, B, and C) near the origin are indicated, and nascent (labelled) DNA is indicated by gray lines. Gel-fractionated Okazaki fragments or sheared total DNA were used as hybridization probes on immobilized plus- and minus-strand templates of fragments A to C (represented by single-stranded DNA or RNA transcript equivalents) or on double-stranded subclones for the lagging-strand and ELFH assays, respectively.
In the lagging-strand approach of Burhans et al. (5), CHO or CHOC 400 cells were released from aphidicolin-induced G1/S arrest for ∼4 min and were permeabilized and incubated for 1.5 min in a replication cocktail containing (among other ingredients) BrdUTP and [32P]dATP. Labelled Okazaki fragments were purified first by size and then by precipitation with anti-BrdU and were hybridized to immobilized pairs of plus and minus M13 clones containing selected genomic DNA fragments. Since only six pairs of clones were used and, with the exception of a probe for the DHFR gene, five were within 11 kb of ori-β, the analysis was limited to a small part of the intergenic region. Large corrections in hybridization values were also required owing to hybridization of labelled CHO or CHOC 400 DNA with the M13 vector itself; in the critical case of the two clones between which a template switch was recorded, this correction changed biases of only 2:1 on either side of the midpoint to more than 5:1 (5).
In the present study, both [32P]dATP and [32P]dTTP were included in the replication cocktail and BrdUTP was omitted, to avoid any potential bias generated by an uneven distribution of thymidines between the two complementary strands (24). To give a relatively complete picture of the initiation reaction in the DHFR locus, 16 different indicator fragments from the intergenic region and the two flanking genes were subcloned into a pBS vector. The insert sizes of these probes are given in Materials and Methods. Ten of these probes were used in the lagging-strand assay, and all 16 were used in the ELFH assay. The smaller number of probes used in the lagging-strand assay was dictated by the capacity of the dot blot apparatus (96 wells), the requirement for duplicate samples of both plus- and minus-strand transcripts, and the requirement for an identical blot to hybridize with the sheared radioactive control DNA prepared from asynchronous cells. In most experiments, 12 different probes were used, but only 10 of these were common to all experiments. The results obtained with these probes are presented below.
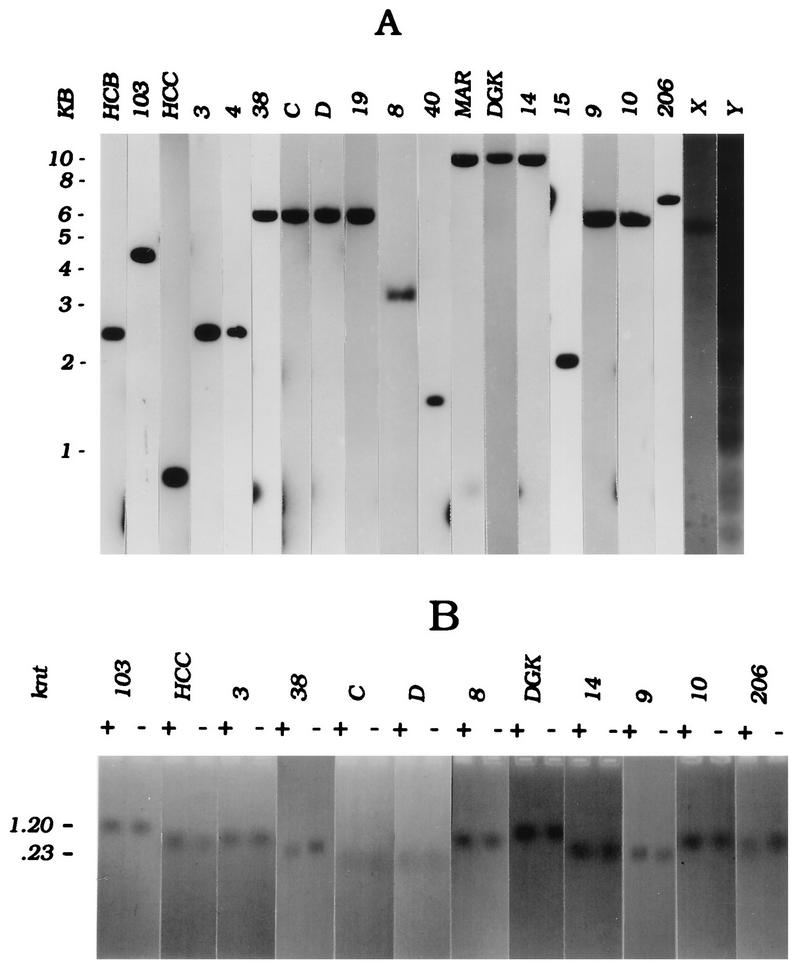
Each insert was shown to be free of repetitive DNA by hybridization to genomic blots of CHO DNA by using the single-copy probe 103, which is derived from an exon of the DHFR gene, as an internal standard (Fig. 3A). The patterns obtained with two fragments containing moderately and highly repeated elements (X and Y, respectively) are included for comparison.
FIG. 3.
Quality control of RNA and cloned templates. (A) CHO genomic DNA was digested with EcoRI, and 15-μg aliquots were run in multiple wells of a 0.8% agarose gel. After blotting to Hybond N+, individual lanes were hybridized with the double-stranded insert from each pBS clone labelled with [32P]dCTP by random priming (1 × 107 to 2 × 107 cpm per blot), and the blots were washed as described in Materials and Methods. For comparison, X and Y are examples of fragments containing moderately or highly repetitive sequence elements, respectively. (B) T3 and T7 transcripts (1 μg each) from each linearized pBS clone were separated on a 1.5% formamide agarose gel; a negative of an ethidium bromide-stained gel is shown. Note that probe 38 yields the smallest transcripts of the collection, resulting in rather diffuse ethidium bromide-staining bands.
A modification of the lagging-strand technique (24), in which the two templates are represented by opposite-strand T3 and T7 RNA transcripts of the inserts in the pBS vector, was used. Thus, nonspecific hybridization to M13 vector sequences was avoided. Figure 3B shows that the RNAs transcribed from these templates were predominantly full-length (Fig. 3B; note that the transcripts from probes C and D are much smaller than the others, resulting in more diffuse bands on the agarose gel). In the ELFH assay, total labelled genomic DNA from the in vitro replication assay was sheared and used as a probe on dot-blots of these double-stranded subclones to identify the earliest labelled fragments (16). Hybridization of total labelled DNA to the pBS vector was not detectable (data not shown).
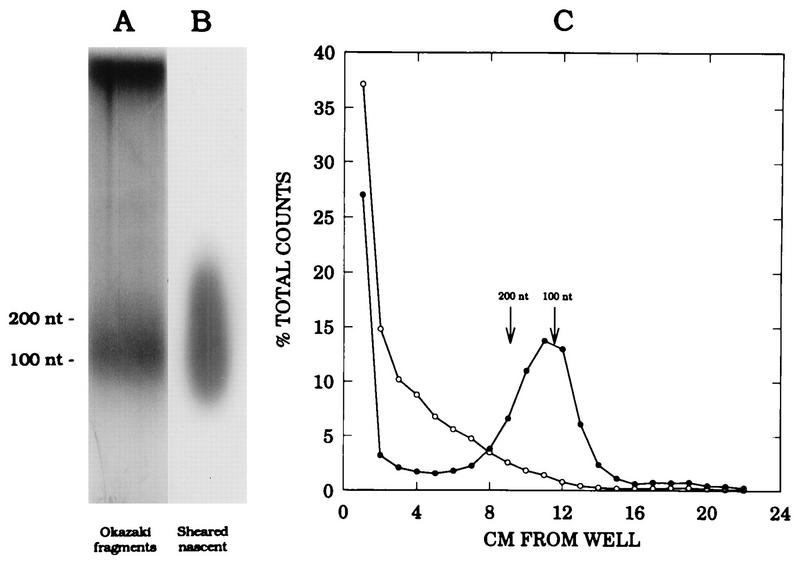
To label replication intermediates synthesized in the very early S phase, exactly the same synchronization and in vitro replication method was used as that described by Burhans et al. (5): cells were released from the aphidicolin block for 4 min and incubated for 1.5 min in the in vitro replication cocktail (in the original study [5], no bias was observed in samples harvested prior to 3 min after release, probably because the sizes of the leading strands at this time would not differ significantly from those of the lagging strands). After quenching the reaction, Okazaki fragments and total genomic DNA were isolated for use in the lagging-strand and ELFH assays, respectively. In Fig. 4A and B, agarose gel separations of the Okazaki fragments, which migrate between ∼50 and 300 nt, as well as the total labelled DNA fraction after sonication, are shown. The data in Fig. 4C show that the labelled Okazaki fraction can be chased into high-molecular-weight DNA by a 30-min incubation with unlabelled nucleotides. We have also demonstrated that the average size of the majority of the Okazaki fraction is reduced by 8 to 12 nt after treatment with RNase to remove RNA primers and separation on an acrylamide gel (data not shown).
FIG. 4.
Characterization of Okazaki fragment preparation and sheared genomic DNA. (A and B) Fractions of either the Okazaki fragment preparation or the sheared total genomic DNA labelled in vitro for 1.5 min were separated on a 1.8% alkaline agarose gel and subjected to autoradiography; sizes were determined by comparison to a 100-nt ladder (BRL). (C) In two in vitro reactions, nuclei were labelled as in panel A and incubated for an additional 30 min in either the absence (open circles) or presence (solid circles) of unlabelled dATP and dTTP to determine whether small, rapidly labelled fragments could be chased into high-molecular-weight DNA. The DNA was then purified and separated as in panel A; the gel was cut into 0.5-cm slices, and the radioactivity in each slice was determined by liquid scintillation counting.
The lagging-strand assay suggests a broad zone of potential initiation sites in the intergenic region.
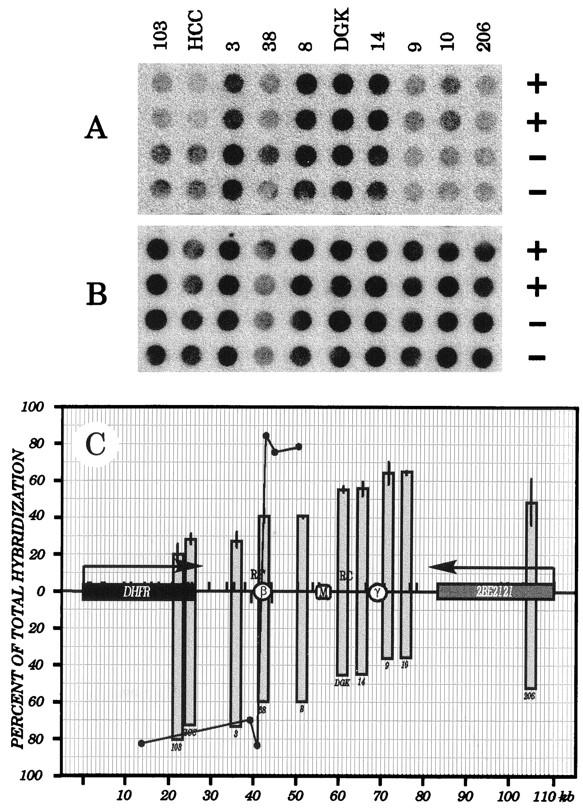
Labelled Okazaki fragments isolated from very early S-phase cells were used to probe dot blots containing immobilized plus- and minus-strand transcripts of 10 different fragments from the DHFR locus (Fig. 5A). Hybridization values to each of the template strands were determined with a PhosphorImager. These hybridization values were normalized to values obtained when duplicate dot blots were probed with total sheared DNA isolated from in vitro-labelled asynchronous CHOC 400 cultures (Fig. 5B), which corrects for any potential intrinsic hybridization bias. Normalized hybridization values of early S-phase Okazaki fragments to plus and minus strands (expressed as a percentage of the total hybridization to both strands) are plotted as a function of map position in Fig. 5C. For comparison, the results of Burhans et al. (5) are also presented (Fig. 5C) but were transformed from fold-of-bias to the percentage of total counts hybridizing to each strand. Note that the size range of inserts is 0.4 to 1.2 kb and that equal amounts (1 μg) of DNA were loaded into each well. Thus, the signals will be lower for the smallest fragments (e.g., probe 38). Also note that there is some variation among individual experiments (reflected by the error bars in Fig. 5C and observed by others [5, 24]); thus, the observed biases in the example shown in Fig. 5A may differ somewhat from those measured in the other two experiments and, as a consequence, from the mean of the three experiments reflected in the graph.
FIG. 5.
Lagging-strand template bias changes gradually throughout the intergenic region in early-S-phase CHOC 400 cells. (A) CHOC 400 cells were released from an aphidicolin block for 4 min, and DNA was labelled in vitro for 1.5 min; 2 × 106 cpm of the labelled Okazaki fraction was hybridized to duplicate dot blots containing 1 μg each of the indicated plus- or minus-strand RNA equivalents; relative hybridization values were determined on a PhosphorImager (three individual experiments). (B) Asynchronous CHOC 400 cultures (three experiments) were labelled in vitro for 1.5 min, the total genomic DNA was sheared, and 6 × 106 cpm was hybridized with a blot identical to that in panel A. (C) In each of three independent experiments, values for each duplicate determined in panel A were divided by the averaged value for the same dots in panel B and are expressed as a percentage of the total hybridization to both templates for each clone; these normalized values are plotted as a function of map position. Standard errors of the mean are indicated. The solid line connecting the solid circles represents the data of Burhans et al. (5), which was transformed from fold-of-bias to the percentage of total hybridization. For simplicity, only hybridization to the preferred strand at each location is shown.
In close agreement with earlier analyses of the DHFR locus by the lagging-strand assay (5, 16), a relatively strong minus-strand hybridization bias (>80%) was detected in the DHFR gene in the very early S phase. This result is expected if forks move mainly outward through the gene from start sites within the intergenic region, as suggested by all previous studies on this locus. Within the intergenic region itself, hybridization to the minus-strand template gradually decreases from ∼75% of total at the position of probe 3 to ∼60% near ori-β (probe 38) to ∼35% near probe 10. This result is predicted for a broad initiation zone (note that probe 38 is the smallest cloned insert in the series tested in the experiment depicted in Fig. 5; thus, the hybridization signal is correspondingly small). To the right of probe 10, an apparent transition occurs, resulting in a 50:50 distribution of hybridization to the two templates in the 5′ end of the 2BE2121 gene. This data is compatible with the results of neutral-neutral and neutral-alkaline 2-D gel analyses, in which a low level of initiation was shown to occur within and to the right of 2BE2121 (11, 13).
Thus, the strand bias observed near the ori-β region differs from the data of Burhans et al. (5). In that study, an equal and opposite template bias of ∼85% was detected between their probes C and D, which correspond approximately to the two halves of our probe 38 (Fig. 5C). Based on the magnitude of the observed shift in template bias, it was estimated that more than 80% of all initiations in the DHFR locus occur within ±240 bp of the center of probe 38 (5). This proposal predicts that hybridization to the two different template strands of probe 38 itself should be approximately equal. However, in our experiments, hybridization to probe 38 displays a 60% hybridization bias toward the minus-strand template while equal hybridization to the two strands occurs approximately at the position of the MAR, which lies ∼14 kb downstream (Fig. 5C).
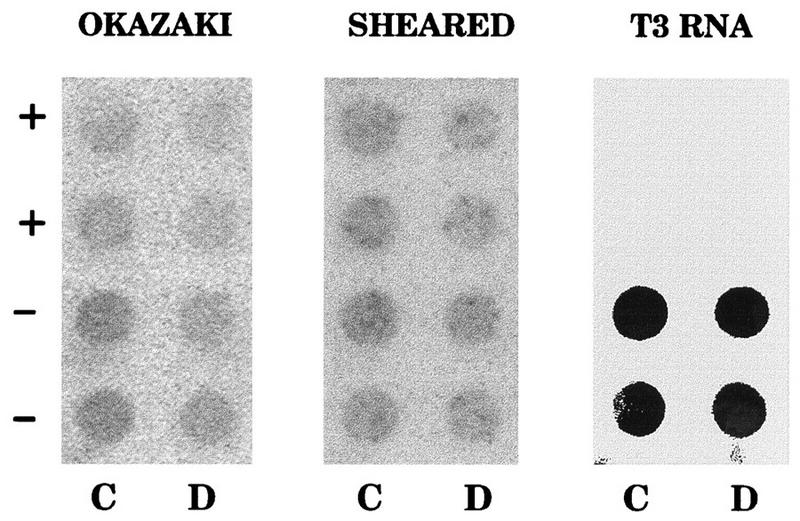
We have tested directly whether there is a shift in template bias in the center of probe 38. The two halves of this fragment (probes C and D in reference 5) were subcloned into a pBS vector and were again probed with Okazaki fragments synthesized in vitro in very-early-S-phase nuclei. In seven independent experiments, Okazaki fragments appeared by eye to hybridize about equally to the minus-strand templates in both halves of probe 38 (Fig. 6, left panel). However, since each half of probe 38 is only ∼250 bp long, the signals were not strong enough to assign accurate template biases. The same blot was stripped and reprobed with total sheared genomic DNA from an asynchronous CHOC 400 culture, and the results show that approximately equal amounts of each RNA template were loaded onto the filter (Fig. 6, center panel). Finally, we confirmed that the sense of the plus and minus strands was correctly assigned by stripping the blot and reprobing with a transcript of the entire probe 38 insert (Fig. 6, right panel).
FIG. 6.
A lagging-strand switch in template bias is not detected at ori-β. The two halves of probe 38 (representing probes C and D in reference 5) were subcloned into the pBS vector, transcripts were prepared from the two templates, and ∼1 μg of each RNA was blotted in duplicate onto four different filters (the sense of each template equivalent is denoted by the + and − symbols). The blots were then hybridized with Okazaki fragments labelled for 1.5 min in vitro 4 min after the release from aphidicolin (left panel). The blot was stripped and reprobed with total sheared DNA from an asynchronous culture labelled in vitro (center panel) or with a radioactive T3 transcript of probe 38 itself (right panel), which encompasses both fragments and represents the lower-strand template as pictured in Fig. 1. Note that the strand assignments of probes C and D were also confirmed by sequencing and comparison to the sequence of the ori-β region (25).
Only minimal hybridization biases are detected in the DHFR locus when Okazaki fragments are isolated from asynchronous CHOC 400 cells.
In the original application of the lagging-strand assay to the DHFR locus, it was reported that even in asynchronous CHOC 400 cell cultures, a pronounced switch in hybridization bias was detected between the two halves of the ori-β-containing fragment (probe 38) (5). In the absence of fixed termination sites between amplicons and/or between ori-β and ori-γ, this outcome requires that initiation in the DHFR locus occur very synchronously in the early S phase in the great majority of amplicons, since passive readthrough of ori-β from the ori-γ region or from neighboring amplicons would decrease the switch in template bias in proportion to the percentage of inactive amplicons. However, both neutral-neutral and neutral-alkaline 2-D analyses (11, 13, 18), as well as intrinsic labelling data (20), show clearly that only 10 to 15% of the 1,000 DHFR amplicons in CHOC 400 cells actually sustain active initiation events, with the remaining 85 to 90% of amplicons being replicated by forks moving through passively later in the S phase.
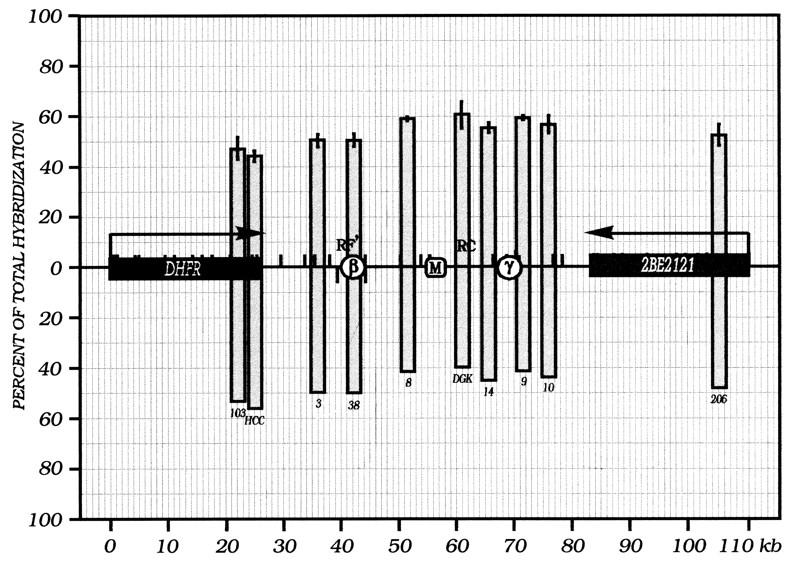
As shown in Fig. 7, Okazaki fragments synthesized in vitro in nuclei prepared from asynchronous cell populations displayed only minor differences in hybridization bias to the two template strands at different positions within the examined region; this is consistent with a minority of amplicons initiating somewhere within the intergenic zone, with the majority being read through passively and therefore exhibiting nearly a 50:50 distribution of template bias to the two strands.
FIG. 7.
Very little lagging-strand template bias can be detected in the DHFR locus in DNA prepared from asynchronous CHOC 400 cell cultures. In vitro reactions (four separate experiments) were carried out on asynchronous CHOC 400 cell populations, and ∼3 × 106 cpm of labelled Okazaki fragments was hybridized to identical dot blots to those described in the legend to Fig. 5. The data were normalized to duplicate blots that were hybridized with in vitro-labelled total sheared DNA from asynchronous cultures. Standard errors of the mean are indicated.
Results of the ELFH assay on early-S-phase cells are consistent with a broad zone of initiation encompassing both ori-β and ori-γ.
Three different in vivo intrinsic labelling studies suggested the presence of a second, somewhat preferred initiation site or zone other than ori-β within the intergenic region of the DHFR domain (termed ori-γ [Fig. 1]) (1, 19, 26). In the absence of a strong terminus somewhere near the MAR, for which there is no compelling evidence from neutral-neutral 2-D gel analyses, the presence of a second preferred initiation region near ori-γ is not compatible with a strong lagging-strand switch at ori-β: in amplicons in which only ori-γ is active, ori-β should be read through passively and therefore would not display a template switch.
However, previous applications of the lagging-strand and ELFH assays to the CHOC 400 DHFR domain focused attention only on the ori-β region and concluded that it constituted the major nascent-strand start site in this domain (5, 16). Neither of these studies examined the region of ori-γ directly. Furthermore, both studies examined replication intermediates prepared only a few minutes after entry into the S phase, whereas the studies that detected ori-γ focused either on log-phase cells (19) or on cells that had been in the S phase for at least 30 min after the release from aphidicolin (1, 26). Thus, if ori-γ were to fire slightly later than ori-β, it could have escaped detection in these studies.
The ELFH assay was therefore repeated with 16 clones covering the intergenic region as well as the two flanking genes. CHOC 400 cells were synchronized at the G1/S boundary with aphidicolin as described above, released into the S phase for 4 min, and harvested and incubated in the in vitro replication cocktail for 1.5 min. Total DNA was isolated, sheared to ∼150 bp in length, and used as a hybridization probe on dot blots containing the immobilized double-stranded subclones.
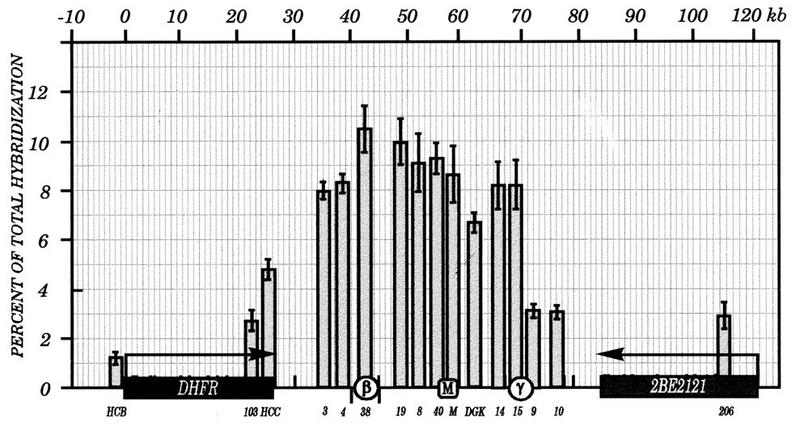
As shown in Fig. 8, at this very early time point after the release from aphidicolin, the entire intergenic region, including both ori-β and ori-γ, is labelled substantially, with a slight bias toward the ori-β half of the region and a somewhat lower labelling index near the DGK probe. These data are quite similar to those obtained by intrinsic labelling approaches (1, 26) and by 2-D gel analyses (39) performed on cells 30 min after the release from aphidicolin, when initiation in this locus is maximal. Furthermore, they are quite similar to those obtained by Gilbert et al. (16), with the exception of the region encompassing ori-γ, which was not examined in their study (Fig. 1, dotted line).
FIG. 8.
The ELFH assay suggests a broad zone of initiation in very-early-S-phase CHOC 400 cells. In vitro replication reactions were carried out for 1.5 min on cells harvested 4 min after the release from aphidicolin. Total sheared DNA was used as a hybridization probe on the indicated immobilized double-stranded pBS clones. Relative hybridization values were normalized for loading and intrinsic hybridization efficiency by normalizing to the values obtained by hybridizing with genomic DNA from asynchronous cells labelled under the same in vitro replication reaction conditions. Standard errors of the mean of three independent in vitro reactions are shown.
2-D gel methods detect multiple nascent-strand start sites in the very early S phase.
Both the lagging-strand and ELFH assays, which have been interpreted to suggest a very circumscribed initiation zone in or near ori-β, were performed on cells just after entry into the S phase (i.e., 4 min after removal of aphidicolin) (5, 16). In contrast, most of the 2-D gel analyses that suggest a broad zone of initiation throughout the intergenic region were performed on cells at the peak initiation period (i.e., 30 min after release from aphidicolin or 90 min after release from mimosine) (10, 11, 13, 39). (Note that there is a 50- to 60-min lag after mimosine removal before substantial amounts of replication begin [10, 32].) Therefore, differences in sampling times could yield different results.
To some extent, this issue has already been addressed in the lagging-strand and ELFH assays summarized in Fig. 5 and 8: when probes representing the entire intergenic zone were included in these analyses, the resulting hybridization patterns were more consistent with initiations occurring at any of multiple sites distributed throughout the intergenic region in the first few minutes of the S phase rather than with initiations occurring at a very circumscribed region around ori-β.
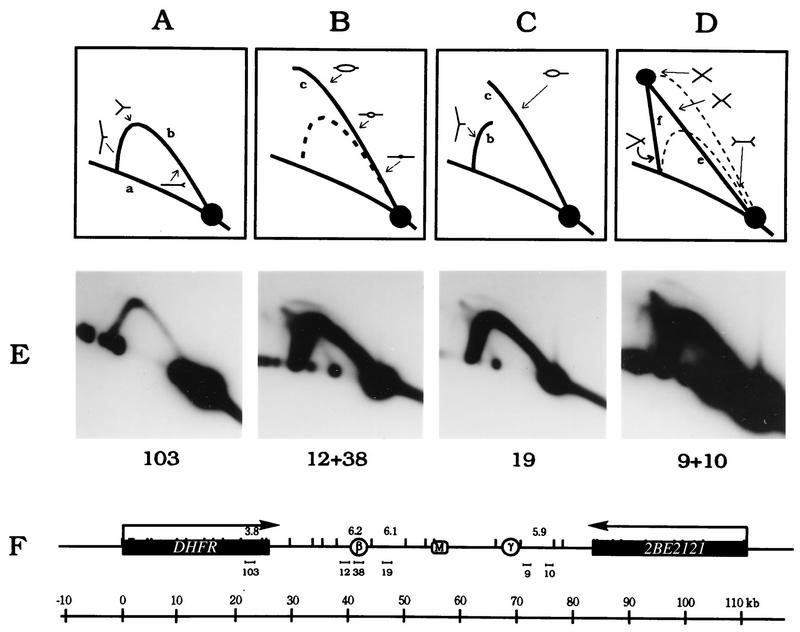
However, we have not previously determined the 2-D gel patterns of replication intermediates in DNA sampled and isolated under the same conditions as those used for the lagging-strand and ELFH assays. Therefore, CHOC 400 cells were collected at the G1/S boundary with aphidicolin, released for 4 min, permeabilized with NP40, and incubated at 34°C for 1.5 min in the in vitro replication cocktail. Replication intermediates were then isolated by our standard method (12), using EcoRI to digest the DNA, and were analyzed by the neutral-neutral 2-D gel replicon-mapping method (3). The principle of this assay is diagrammed in Fig. 9A to D, and the results are shown in Fig. 9E.
FIG. 9.
Neutral-neutral 2-D gel analysis on very-early-S-phase cells detects initiation in multiple intergenic fragments. (A to D) Principle of the neutral-neutral 2-D gel technique (3). (E) CHOC 400 cells were released from aphidicolin for 4 min. After a 1.5-min incubation in the in vitro reaction, replication intermediates were prepared and separated on a 2-D gel as previously described (10, 12). A transfer of the gel was hybridized successively with probes 12 plus 38, 19, 9 plus 10, and 103, with a 30-min incubation with 0.4 N NaOH at 42°C to remove each probe. Because of the low level of replication intermediates at this very early time point, each blot had to be exposed to film for 14 days on average, which made it difficult to normalize the signal strengths of the 1n spots with each probe.
When a transfer of the 2-D gel was hybridized with probes 12 plus 38, which are specific for the 6.2-kb EcoRI fragment that contains ori-β, a typical composite pattern consisting of a faint bubble arc and a strong single fork arc was observed. Therefore, at this very early time point, this fragment sometimes sustains internal initiation events but most often is replicated passively by forks originating from outside of the fragment. Hybridization of the blot with probe 103, which detects the DHFR gene, detects very few replication intermediates at this very early time point, consistent with the ELFH data in Fig. 7 and confirming that most of the observed replication forks in the intergenic zone must have initiated from within the zone itself and would not have had time to enter the gene from neighboring amplicons. This suggestion is supported by the presence of both bubble arcs and strong single-fork arcs in the 6.1- and 5.9-kb intergenic fragments, which are detected with probes 19 and probes 9 plus 10, respectively (Fig. 9E). Thus, by the criterion of neutral-neutral 2-D gel analysis of very-early-S-phase cells, initiation sites appear to be chosen from a broad region encompassing a large part of the intergenic zone. (Note that the bubble arcs in all three intergenic fragments are weak relative to those that would be observed in these fragments in the early S phase in vivo. This result is predicted since no new initiations occur in the in vitro reaction; therefore, only bubbles that have not yet matured beyond the bounds of the restriction fragment in which they initiated in vivo are detected.)
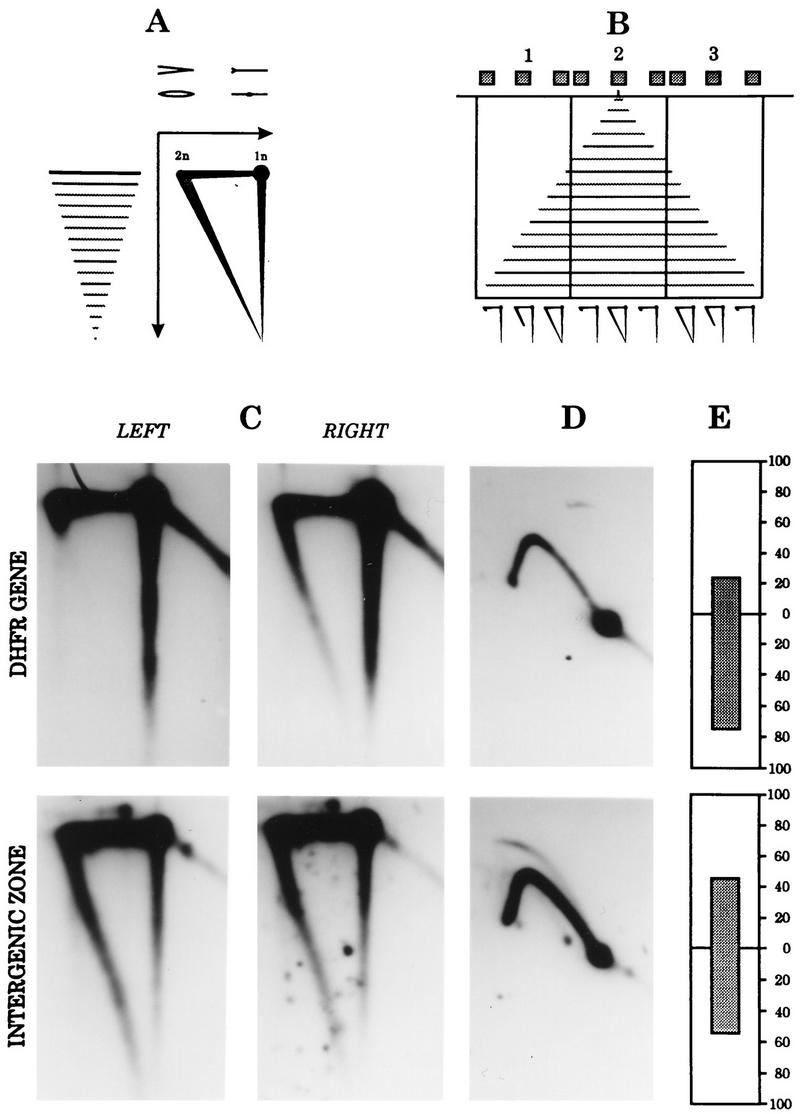
In a second approach, the lagging-strand and neutral-alkaline and neutral-neutral 2-D gel assays were applied to the same preparation of CHOC 400 cells sampled 90 min after the release from mimosine. Most cells have entered the S phase by this time, initiation in the DHFR locus is maximal (10, 11), and most of our recent 2-D gel analyses were performed on material sampled at this time. The principle of the neutral-alkaline 2-D gel replicon-mapping method is diagrammed in Fig. 10A and B, and the results are shown in Fig. 10C and D.
FIG. 10.
Lagging-strand and 2-D gel assays paint a unified picture of initiation 90 min after the release from mimosine at the peak initiation period. (A) Cartoon of the migration pattern of the template (uppermost) and nascent (smaller) strands in a neutral (first-dimension)-alkaline (second-dimension) gel. (B) Hybridization patterns obtained when the ends and center of individual fragments in a region surrounding a relatively fixed origin were probed; vertical lines represent restriction sites, and gray boxes represent hybridization probes. Typical patterns are shown below. (C) Sixteen 15-cm plates of CHOC 400 cells were released from a G0 block into 400 μM mimosine for 14 h, after which the drug was removed and the cells were allowed to proceed into the S phase for 90 min; all the plates were harvested in the cold; 12 plates were used as the source of replication intermediates for neutral-alkaline 2-D gels (A to C) or neutral-neutral 2-D gels (D), and 4 were prepared for in vitro replication reactions (E). The neutral-alkaline 2-D gel transfer was probed successively with radioactive labels for the left and right ends of a 3.8-kb EcoRI fragment in the DHFR gene (C, left and right panels, respectively) and for the 6.1-kb EcoRI fragment just to the right of the ori-β locus in the intergenic zone; the neutral-neutral 2-D gel was hybridized with probes 103 (upper) and 19 (lower). The lagging-strand hybridization data for probes 103 (upper) and 19 (lower) are expressed as in Fig. 5.
In the neutral-alkaline 2-D gel analysis, when a fragment from the DHFR gene was hybridized with probe 103 for the origin-proximal end, a very prominent and complete diagonal was detected (Fig. 10C, upper right panel); subsequent hybridization of the same transfer with probe 100 for the origin-distal end detected only very large nascent strands (upper left panel). This result shows that 90 min after the release from mimosine, replication forks are moving overwhelmingly outward through the DHFR gene from the direction of the intergenic zone. We estimate the bias in the outward direction to be greater than 10-fold in this experiment. However, when the same blot was stripped and reprobed with the two end labels for the intergenic 6.1-kb EcoRI fragment lying just to the right of ori-β (lower panels), strong diagonals were detected with both probes, showing that approximately equal numbers of replication forks enter this fragment from the directions of ori-β and from sites situated to the right of this fragment. Thus, ori-β cannot represent the only (or even the predominant) initiation site in this locus.
The neutral-neutral 2-D gel assay results on the same material are shown in Fig. 10D. Consistent with the results of all earlier 2-D gel analyses on mimosine-synchronized cells sampled at 90 min (10, 11, 13), a probe specific for the DHFR gene detected a relatively weak single fork arc but no replication bubbles (Fig. 10D, upper panel). In the same material, probe 19 for the 6.1-kb EcoRI fragment to the right of ori-β detected a complete bubble arc and a stronger single fork, confirming the delocalized initiation mode in the intergenic zone (lower panel).
Finally, in the same cell preparation sampled 90 min after the release from mimosine, the distribution of Okazaki fragment hybridization to the minus-strand template in the DHFR gene (i.e., probe 103) was approximately 75% in this experiment whereas hybridization to the minus strand in the region of probe 8 was ∼55%. Therefore, the results of the lagging-strand assay agree qualitatively with those of the neutral-alkaline 2-D gel assay on the same material, but the lagging-strand assay appears to be less sensitive to strand bias, based on results with the DHFR gene. Furthermore, the results of the lagging-strand assay are very similar whether the cells are sampled ∼4 min after the release from aphidicolin or 90 min after the release from mimosine.
DISCUSSION
In two previous studies, novel variations of the neutral-neutral and neutral-alkaline 2-D gel methods were developed to determine whether these methods could fail to detect a major initiation site near ori-β (22, 23). Instead, the results supported our previous proposal that nascent-strand start sites are chosen from many potential sites scattered throughout the 55-kb intergenic region, with a concentration of sites in the central 30- to 35-kb encompassing the ori-β and ori-γ regions (10, 11, 13, 39).
In previous reports, results of the lagging-strand (5) and ELFH (16) assays were interpreted to mean that ori-β is the major initiation site in the DHFR locus (5, 16). In the present study, we have adapted these two assays to our laboratory and have modified them in the following ways in an effort to avoid inadvertent bias in the results: (i) BrdUTP was omitted from and [32P]dTTP was included in the in vitro replication cocktail along with [32P]dATP to normalize any bias from an unequal distribution of thymidines on the two strands; (ii) indicator clones were chosen so as to examine the entire intergenic region as well as the flanking genes; and (iii) in the lagging-strand assay, plus- and minus-strand RNA transcripts served as substrates on dot blots to avoid background hybridization to the vector. In addition, we have performed the lagging-strand, ELFH, and one or both 2-D gel approaches on cell cultures manipulated in the same way, thereby avoiding as much as possible laboratory-to-laboratory variations in cell culture, synchronization methods, or other factors that could affect experimental outcomes.
By using the modified lagging-strand assay, we detected approximately the same hybridization bias (∼80%) to the minus-strand template in the DHFR gene as was observed in two previous studies on CHOC 400 cells synchronized in early S phase by release from aphidicolin (5, 16). However, unlike the study of Burhans et al. (5), we did not detect a prominent switch in template bias in the region of ori-β (Fig. 5); rather, the hybridization bias increased gradually toward the positive strand from one end of the intergenic region to the other (with minor undulations), approximating 50% near the MAR. In addition, when we examined hybridization bias to probes C and D, which defined the origin of bidirectional replication in a previous study (5), a strand switch between the two fragments was not detected, and it was shown experimentally that the strand assignments of the two immobilized transcripts were correct.
In this regard, our data are more compatible with those of Gilbert et al. (16), who also observed a relatively gradual change in hybridization bias throughout the 5′ half of the intergenic region in CHOC 400 cells (the region to the right of probe DGK was not examined in their study). Importantly, the position at which hybridization to the two templates was equal in that study did not coincide with the strand switch site at ori-β defined by Burhans et al.; instead, equal hybridization to the two templates lay 5 to 6 kb to the right, which is 5 to 6 kb left of the 50:50 position detected in the present study (i.e., the MAR [Fig. 5]). It is possible to imagine that the position at which Okazaki fragments switch templates differs in experiments performed in different laboratories if cell culture conditions are capable of affecting the relative initiation frequencies in different portions of a broad initiation zone. However, such a change in the position of the switch is difficult to fit to a model in which a single, narrowly localized origin is dictated by the position of a genetic replicator.
In Fig. 5, we have purposely presented the lagging-strand assay data as a percentage of total counts hybridizing to each strand of a probe pair (rather than as a ratio of hybridization to the two separate strands [5]) to emphasize that significant hybridization occurs to both strands in the intergenic region. Given that there is no background hybridization to the cloned inserts used in this experiment, this result must mean that replication forks are moving in both directions at any of the positions examined in this study. Therefore, in this case, the position at which hybridization to the two templates is equal (i.e., 50%) corresponds not to a single origin of bidirectional replication but, rather, to the position at which the numbers of forks moving in the two directions are equal (in our study, approximately at the MAR). An important corollary is that a switch in hybridization bias from one strand to another would suggest the presence of a fixed origin of bidirectional replication only if the switch were very abrupt at that position and if the hybridization biases to the right and left of the switch site were equal and opposite throughout the length of the replicon under study.
In theory, in a domain containing a broad zone of initiation flanked by two passively replicated regions (the DHFR and 2BE2121 genes in this case), one should observe nearly infinite hybridization bias to the lagging-strand template in one flank, which should change gradually throughout the zone to approach infinite bias to the lagging-strand template in the other flank. Our data approximate this model but deviate from it somewhat. As in two other studies (5, 16), hybridization to the lagging-strand template in the DHFR gene averaged only ∼80%, with values ranging from 66 to 90%. Therefore, in only some experiments did strand bias achieve the apparent 10- to 20-fold biases measured in the same material by the neutral-alkaline 2-D gel method in this and several previous studies (Fig. 10) (11, 13, 39). The source of this discrepancy is unclear, although the low levels of radioactivity incorporated into the DHFR and 2BE2121 genes in the very early S phase in the lagging-strand assay can introduce more variability into measurements (and, thus, the signal-to-noise ratio) than in the intergenic region itself, where the values are higher. It is also possible that there is some contamination of the Okazaki fraction with fragmented leading strands, which would reduce biases somewhat and which could vary from experiment to experiment. In the 2BE2121 gene, Okazaki fragment hybridization to the two templates is roughly equal, in agreement with the results for 2-D gels, which also detected a low level of initiation in the early S phase leading to forks moving in both directions (2).
Results from the ELFH assay on the same starting material as used in the lagging-strand assay show that radioactivity is distributed throughout the entire intergenic zone after a pulse time of only 1.5 min delivered ∼4 min after removal of aphidicolin (Fig. 8). These data are similar to those obtained by Gilbert et al. (16) (Fig. 1), except that the region around ori-γ was not examined in that study. Interpolation of their data in this region therefore led them to suggest a single, somewhat asymmetric peak of early labelling centered at ori-β. With the inclusion of clones for the ori-γ region in the present study, we suggest that the data are more compatible with a broad zone of initiation, with the regions around ori-β and ori-γ being somewhat preferred, as originally suggested by in-gel renaturation experiments on early-labelled DNA (26).
However, it is important to point out that in the absence of perfectly synchronous entry of the cell population into the S phase, the ELFH assay cannot distinguish between fixed initiation sites in the ori-β and ori-γ regions as opposed to two somewhat preferred regions within a broad zone of potential sites. For example, labelling near the MAR could result either from initiations occurring in that region or from early forks that have already reached that region from fixed sites at ori-β and ori-γ in the 4-min interval after removal of aphidicolin.
Therefore, it was important to show directly that the labelling patterns we observed in the lagging-strand and ELFH assays on cells sampled ∼4 min after entry into the S phase do, indeed, reflect a delocalized initiation mode. Accordingly, 2-D gel analyses were performed on cell populations that were either synchronized and sampled identically (Fig. 9) or performed on the very same preparations used to examine Okazaki template strand bias (Fig. 10). These studies show that initiation sites are scattered widely in the intergenic zone even in the first few minutes of the S phase, as indicated by the presence of both bubbles and single forks in the same fragments on neutral-neutral 2-D gels (Fig. 9) and by the presence of forks moving into intergenic fragments from both ends, as detected in neutral-alkaline gels (Fig. 10). Importantly, these findings argue that kinetic in vitro and steady-state in vivo approaches see the initiation reaction in essentially the same way.
Finally, when the lagging-strand assay was applied to asynchronous CHOC 400 cell cultures in the present study, only a very small bias to either of the template strands could be detected at any position in the intergenic zone or the two flanking genes. This is the predicted result if 85 to 90% of amplicons are read through passively by forks from a small number of active amplicons, which was suggested by the results of neutral-neutral and neutral-alkaline 2-D gel assays performed on log-phase cultures (10, 13), as well as earlier intrinsic labelling studies (20). We do not understand the difference between the results of our experiments on log-phase cells and those of Burhans et al. (5). However, it has been suggested that the CHOC 400 cultures that were thought to be asynchronous in those studies may have been inadvertently synchronized (16).
In summary, the lagging-strand, ELFH, and neutral-neutral and neutral-alkaline 2-D gel studies reported here, as well as all previous experimental approaches that have been applied to the DHFR locus in CHOC 400 cells, all paint a very similar picture of the initiation reaction in the DHFR locus. These findings are summarized in the model in Fig. 11, where several amplicons are arrayed side by side to suggest that (i) initiation probably occurs at different locations within the intergenic zone in different copies of the amplicon, with the regions of ori-β and ori-γ being somewhat preferred, and (ii) only some amplicons sustain active initiation events in any one S period, with the remainder being replicated passively at later times in the S phase. Whether an initiation reaction with these characteristics might be regulated by genetic replicators, which could presumably be located at or near ori-β and ori-γ, remains to be determined.
FIG. 11.
Model for initiation in the DHFR locus. Several DHFR amplicons are arrayed side by side instead of end to end as they are in the genome, to emphasize that (i) initiation occurs at different sites within the intergenic region in each copy of the DHFR domain in the early S phase but probably somewhat more frequently near the ori-β and ori-γ regions, and (ii) only ∼15% of the amplicons actually sustain active initiation events, with the remainder being replicated passively by forks from distant amplicons.
An important caveat is that our studies here have focused on the initiation reaction in the amplified DHFR domain of CHOC 400 cells, which could differ somewhat from the initiation reaction in the single-copy locus in CHO cells, which contributed to the data set in the previous lagging-strand analysis on this locus (5). Furthermore, a PCR-based nascent-strand abundance assay that detected a rather sharp peak of initiation at ori-β examined CHO cells exclusively (34; the remainder of the intergenic zone was not examined in this study). It is conceivable, for example, that the initiation reaction has become delocalized as a consequence of the amplification process. However, the spectra of replication intermediates in the intergenic zone in the single-copy and amplified DHFR domains are nearly identical when analyzed by neutral-neutral or neutral-alkaline 2-D gel approaches (11). Thus, we believe that the differences must lie elsewhere. Studies are under way in our laboratory to determine whether the PCR-based nascent-strand abundance assay detects a well-defined initiation spike at ori-β in the same starting material that suggests a zone on 2-D gels.
ACKNOWLEDGMENTS
We thank John Kolman and Geoff Wahl (Salk Institute) and Mike Leffak (Wright State University) for very helpful advice concerning the lagging-strand assay. We also thank Howard Cedar (Hebrew University) and Dave Gilbert (Syracuse University) for sharing genomic clones. We are indebted to Carlton White and Kevin Cox for excellent technical assistance.
This work was supported by NIH grant GM26108 to J.L.H.
REFERENCES
- 1.Anachkova B, Hamlin J L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989;9:532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T A, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 3.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 4.Burhans W C, Selegue J E, Heintz N H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci USA. 1986;83:7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burhans W C, Vassilev L T, Caddle M S, Heintz N H, DePamphilis M L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- 6.Caddle M S, Lussier R H, Heintz N H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990;211:19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- 7.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 9.Dijkwel P A, Hamlin J L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988;8:5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkwel P A, Hamlin J L. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol Cell Biol. 1992;12:3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkwel P A, Hamlin J L. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol Cell Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkwel P A, Vaughn J P, Hamlin J L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkwel P A, Vaughn J P, Hamlin J L. Replication initiation sites are distributed widely in the amplified CHO dihydrofolate reductase domain. Nucleic Acids Res. 1994;22:4989–4996. doi: 10.1093/nar/22.23.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrova D S, Giacca M, Demarchi F, Biamonti G, Riva S, Falaschi A. In vivo protein-DNA interactions at human DNA replication origin. Proc Natl Acad Sci USA. 1996;93:1498–1503. doi: 10.1073/pnas.93.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert D M, Miyazawa H, DePamphilis M L. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamlin J L, Dijkwel P A. On the nature of replication origins in higher eukaryotes. Curr Opin Genet Dev. 1995;5:153–161. doi: 10.1016/0959-437x(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 18.Hamlin J L, Dijkwel P A, Vaughn J P. Initiation of replication in the Chinese hamster dihydrofolate reductase domain. Chromosoma. 1992;102:17–23. doi: 10.1007/BF02451781. [DOI] [PubMed] [Google Scholar]
- 19.Handeli S, Klar A, Meuth M, Cedar H. Mapping replication units in animal cells. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- 20.Heintz N H, Hamlin J L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci USA. 1982;79:4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huberman J A, Riggs A D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 22.Kalejta R F, Hamlin J L. Composite patterns in neutral/neutral two-dimensional gels demonstrate inefficient replication origin usage. Mol Cell Biol. 1996;16:4915–4922. doi: 10.1128/mcb.16.9.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalejta R F, Lin H B, Dijkwel P A, Hamlin J L. Characterizing replication intermediates in the amplified CHO dihydrofolate reductase domain by two novel gel electrophoretic techniques. Mol Cell Biol. 1996;16:4923–4931. doi: 10.1128/mcb.16.9.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly R E, DeRose M L, Draper B W, Wahl G M. Identification of an origin of bidirectional DNA replication in the ubiquitously expressed mammalian CAD gene. Mol Cell Biol. 1995;15:4136–4148. doi: 10.1128/mcb.15.8.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leu T H, Anachkova B, Hamlin J L. Repetitive sequence elements in an initiation locus of the amplified dihydrofolate reductase domain in CHO cells. Genomics. 1990;7:428–433. doi: 10.1016/0888-7543(90)90178-w. [DOI] [PubMed] [Google Scholar]
- 26.Leu T H, Hamlin J L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989;9:523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levenson V, Hamlin J L. A general protocol for evaluating the specific effects of DNA replication inhibitors. Nucleic Acids Res. 1993;21:3997–4004. doi: 10.1093/nar/21.17.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine A J, Kang H S, Billheimer F E. DNA replication in SV40-infected cells. I. Analysis of replicating SV40 DNA. J Mol Biol. 1970;50:549–568. doi: 10.1016/0022-2836(70)90211-1. [DOI] [PubMed] [Google Scholar]
- 29.Linskens M H, Huberman J A. Ambiguities in results obtained with 2D gel replicon mapping techniques. Nucleic Acids Res. 1990;18:647–652. doi: 10.1093/nar/18.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milbrandt J D, Heintz N H, White W C, Rothman S M, Hamlin J L. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc Natl Acad Sci USA. 1981;78:6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirkovitch J, Mirault M E, Laemmli U K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 32.Mosca P J, Dijkwel P A, Hamlin J L. The plant amino acid mimosine may inhibit initiation at origins of replication in Chinese hamster cells. Mol Cell Biol. 1992;12:4375–4383. doi: 10.1128/mcb.12.10.4375. . (Erratum, 13:1981, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nawotka K A, Huberman J A. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol Cell Biol. 1988;8:1408–1413. doi: 10.1128/mcb.8.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelizon C, Diviacco S, Falaschi A, Giacca M. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol Cell Biol. 1996;16:5358–5364. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Tasheva E S, Roufa D J. Densely methylated DNA islands in mammalian chromosomal replication origins. Mol Cell Biol. 1994;14:5636–5644. doi: 10.1128/mcb.14.9.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassilev L, Johnson E M. An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol Cell Biol. 1990;10:4899–4904. doi: 10.1128/mcb.10.9.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vassilev L T, Burhans W C, DePamphilis M L. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol. 1990;10:4685–4689. doi: 10.1128/mcb.10.9.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaughn J P, Dijkwel P A, Hamlin J L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]