Abstract
Transcription factor IIF (TFIIF) cooperates with RNA polymerase II (pol II) during multiple stages of the transcription cycle including preinitiation complex assembly, initiation, elongation, and possibly termination and recycling. Human TFIIF appears to be an α2β2 heterotetramer of RNA polymerase II-associating protein 74- and 30-kDa subunits (RAP74 and RAP30). From inspection of its 517-amino-acid (aa) sequence, the RAP74 subunit appears to comprise separate N- and C-terminal domains connected by a flexible loop. In this study, we present functional data that strongly support this model for RAP74 architecture and further show that the N- and C-terminal domains and the central loop of RAP74 have distinct roles during separate phases of the transcription cycle. The N-terminal domain of RAP74 (minimally aa 1 to 172) is sufficient to deliver pol II into a complex formed on the adenovirus major late promoter with the TATA-binding protein, TFIIB, and RAP30. A more complete N-terminal domain fragment (aa 1 to 217) strongly stimulates both accurate initiation and elongation by pol II. The region of RAP74 between aa 172 and 205 and a subregion between aa 170 and 178 are critical for both accurate initiation and elongation, and mutations in these regions have similar effects on initiation and elongation. Based on these observations, RAP74 appears to have similar functions in initiation and elongation. The central region and the C-terminal domain of RAP74 do not contribute strongly to single-round accurate initiation or elongation stimulation but do stimulate multiple-round transcription in an extract system.
RNA polymerase II (pol II) interacts with a number of general and regulatory factors to initiate transcription accurately from a promoter (reviewed in references 34 and 56). In the pathway toward initiation, promoter DNA is bent, and DNA may be wrapped around pol II (24, 40). General factors TATA-binding protein (TBP) (or transcription factor IID [TFIID]), TFIIB, TFIIF, and TFIIE cooperate with pol II to strain the DNA helix around the transcriptional start site before ATP-driven helix opening by TFIIH (34, 56). After initiation, pol II releases from the promoter (promoter clearance or promoter escape), elongates the RNA chain, terminates transcription, and recycles. TFIIF, made up of RAP30 (RNA polymerase II-associating protein of 30 kDa) and RAP74 (58 kDa) subunits, may participate in each of these stages of the transcription cycle.
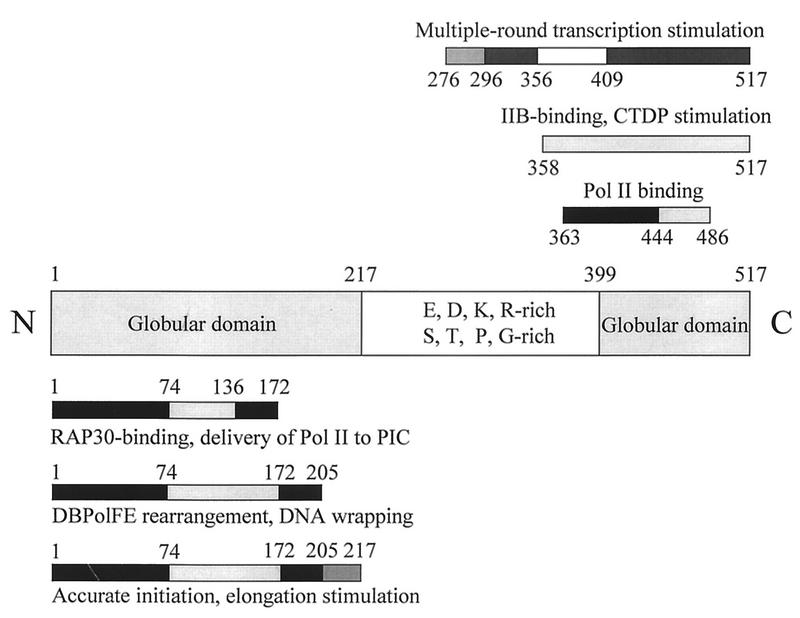
Inspection of its 517-amino-acid (aa) sequence indicates that human RAP74 can be divided into three regions: (i) a highly basic N-terminal domain with significant globular structure (aa 1 to 217); (ii) an overall acidic, highly charged central region lacking in hydrophobic amino acids but rich in E, D, K, R, S, T, G, and P (aa 218 to 398); and (iii) a very basic C-terminal domain (CTD) with globular structure (aa 399 to 517) (2, 15). The N-terminal domain is important for RAP30 binding (54, 55), preinitiation complex assembly (this report), and elongation stimulation (reference 21 and this report). The CTD of RAP74 makes contact with TFIIB (13) and pol II (54) and stimulates the activity of a pol II CTD phosphatase that may have roles in initiation, elongation, termination, and recycling (8).
A pathway for assembly of preinitiation complexes on TATA box-containing promoters has been defined (34, 56). The TBP subunit of TFIID binds to the TATA sequence. Insertion of TBP into the DNA minor groove at TATA induces a 95° bend (23, 25). TFIIB can then enter to form the DB complex, made up of TBP (or TFIID), TFIIB, and promoter DNA. The C-terminal repeats of TFIIB bind DNA upstream and downstream of TATA, stabilizing the DNA bend (26, 33). The N-terminal domain of TFIIB may extend toward the transcriptional start site as a scaffold on which to assemble pol II and TFIIF (34).
To bind efficiently to the promoter, pol II must first bind TFIIF (10, 16, 22). In some cases, the RAP30 subunit has been sufficient to deliver pol II to the promoter (16, 22, 51), but the RAP74 subunit contributes to proper assembly, complex stability, and initiation. For promoters with weak TATA boxes, both RAP30 and RAP74 contribute to template commitment of TFIID, TFIIB, and pol II (49). Furthermore, RAP74 strongly stimulates initiation from supercoiled and premelted templates that are dependent only on TBP, TFIIB, pol II, and TFIIF for accurate transcription (35, 36). In most contexts, therefore, both the RAP30 and RAP74 subunits are important for TFIIF function in complex assembly and initiation.
After fulfilling its role in initiation, TFIIF stimulates the elongation rate of pol II (4, 20, 21, 38, 49). On nonchromatin DNA templates and in the absence of other general factors, TFIIF can accelerate polymerization to up to 1,500 nucleotides per min, close to the estimated physiological rate (20). TFIIF suppresses pausing by pol II (4, 20, 38), but whether this is a cause or effect of rate stimulation is not known. Both the RAP30 and RAP74 subunits of TFIIF are required for elongation stimulation (21, 49), and preliminary mapping studies indicate that the N-terminal domain of RAP74 is most important for elongation (21). Tan et al. (50) identified a class of RAP30 mutants that are impaired for both elongation stimulation and accurate initiation, and these mutants are also defective for binding RAP74, consistent with the requirement of both subunits for elongation. They have also identified classes of RAP30 mutants that are defective only for elongation stimulation or for initiation, not for both. In contrast to their results with RAP30, however, we find that a region within the N-terminal domain of RAP74, that is not essential for RAP30 binding, is nonetheless strongly stimulatory for both initiation and elongation (this report).
An intriguing feature of the RPB1 subunit of pol II is the CTD which has the consensus sequence YSPTSPS tandemly repeated 52 times in human pol II (reviewed in reference 12). Phosphorylation and dephosphorylation of the CTD by the regulated activities of CTD kinases (14, 29, 31, 45, 46) and phosphatases (7) appear to control progression through the transcription cycle. Pol II enters the preinitiation complex with its CTD in a largely unmodified form designated pol IIa (27, 28). During elongation, pol II is converted to the pol IIo form (3, 27), which is heavily phosphorylated on the SP serines of the YSPTSPS consensus sequence, and so hyperphosphorylation of the CTD is thought to be important to establish and maintain the elongation complex. Removal of phosphates from the CTD may be a signal to terminate transcription and recycle pol II to a promoter.
A recently identified CTD phosphatase that catalyzes the dephosphorylation of pol IIo to pol IIa is stimulated by the C-terminal domain of the RAP74 subunit of TFIIF (8). Interestingly, RAP74-dependent stimulation of CTD phosphatase activity is blocked by addition of TFIIB. The C-terminal domain of RAP74 binds to TFIIB (13) and pol II (54), and so TFIIF, TFIIB, and the CTD phosphatase may be components of a multiprotein complex that binds pol II and regulates pol II recycling. In this report, we demonstrate that the central region and the CTD of RAP74 stimulate multiple-round transcription in an extract system consistent with a role for RAP74 in transcriptional recycling.
MATERIALS AND METHODS
Transcription factors and extracts.
Recombinant Saccharomyces cerevisiae TBP and human recombinant TFIIB were the kind gifts of Steven Triezenberg and Fan Shen. The clone for production of TFIIB was the kind gift of Danny Reinberg. Recombinant human RAP30, RAP74, and RAP74 mutants were prepared and quantitated as described elsewhere (13, 52, 53, 54). Construction of new mutants is described below. Calf thymus pol II used in electrophoretic mobility shift experiments was prepared by the method of Hodo and Blatti (19) and was primarily in the IIb form, lacking the CTD. Gel mobility shift experiments with calf thymus pol IIa (the kind gift of Richard Burgess) gave similar results (data not shown).
Human HeLa cells were purchased from the National Cell Culture Center (Minneapolis, Minn.). Extracts of HeLa cell nuclei were prepared as described previously (47). A TFIIF-depleted extract was prepared by immunoprecipitation of TFIIF with anti-RAP30 and anti-RAP74 antibodies (6, 15). The TFIIF-depleted extract was completely dependent on the readdition of RAP30 for activity and was strongly stimulated by addition of RAP74.
Construction of RAP74 mutants.
RAP74(1-217) was constructed by PCR amplification of a plasmid clone encoding RAP74 with primers 5′-CATATGGCGGCCCTAGGCCCT-3′ and 5′-CTCGAGAGACATTTCCAGGT-3′ and subcloning between the NdeI and XhoI sites (cloning sites are underlined) of pET21a (Novagen). Internal deletion mutants RAP74(Δ306-351), RAP74(Δ276-351), and RAP74(Δ219-351) were constructed by using a Quick Change site-directed mutagenesis kit (Stratagene). All mutants were made by using the same primer for the aa 351 position, 5′-GACATTGACAGCGAGGCCTCCTCAGCCCTCTTCATGGCG-3′. For the second oligonucleotide primers, 5′-GGAGGCCTCGCTGTCAATGTCGCTCTGCTCATCGACACCATTGGG-3′, 5′-GGAGGCCTCGCTGTCAATGTCTGACATGTAGTCCACCTCTTGGCC-3′, and 5′-GGAGGCCTCGCTGTCAATGTCGGAGGACATTTCCAGGTCGTCTTCAAGGTC-3′, respectively, were used. The underlined sequences represent the complementary overlap between the two mutant primers.
Three triple-alanine mutations were constructed in RAP74(1-217), using a Quick Change site-directed mutagenesis kit (Stratagene) and appropriate primers. These mutant proteins are named RAP74(1-217)170A3, -173A3, and -176A3. Each of these mutant proteins has the sequence AAA beginning at the indicated amino acid; for example, 170A3 carries V170A, L171A, and N172A (Fig. 4). Mutated RAP74 genes were confirmed by DNA sequencing.
FIG. 4.
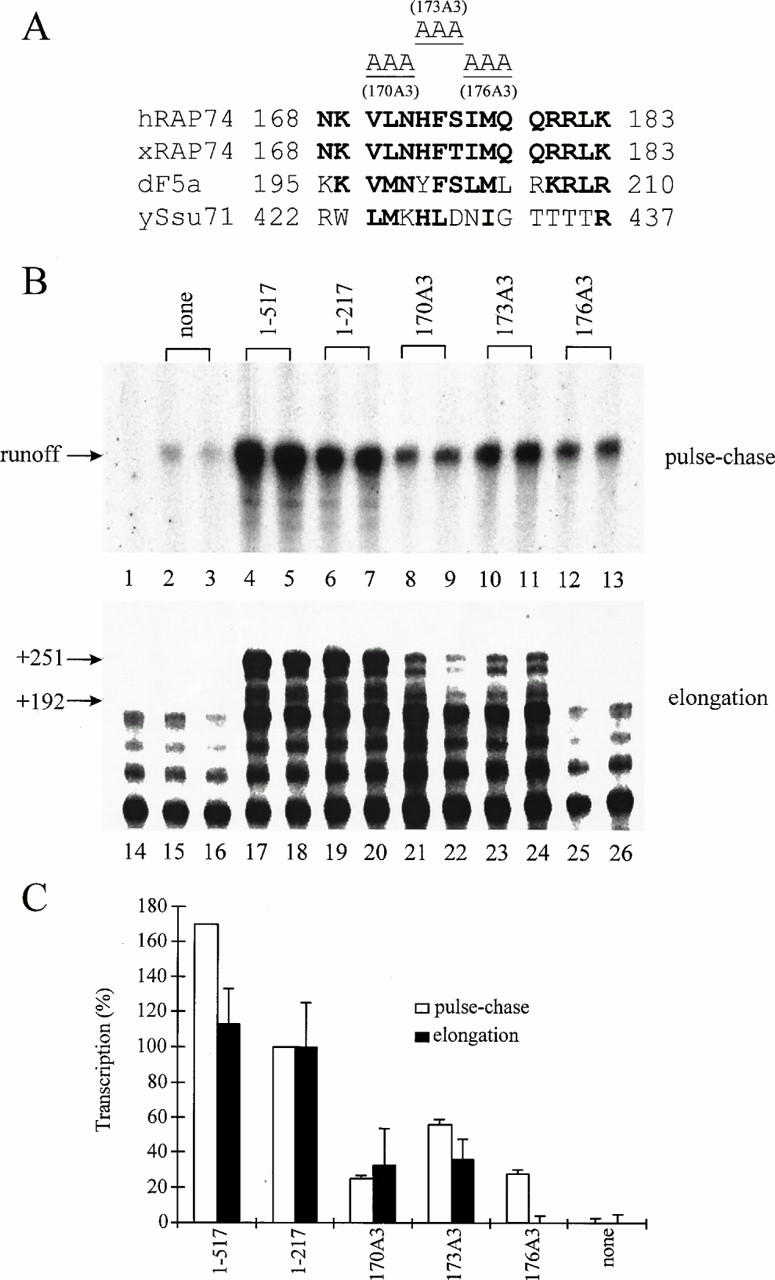
A conserved region of RAP74 between amino acids 170 and 178 is important for both initiation and elongation. (A) Triple-alanine substitutions 170A3, 173A3, and 176A3, constructed in RAP74(1-217), are indicated. Related sequences from human (hRAP74), Xenopus (xRAP74), Drosophila (dF5a), and yeast (ySsu71/Tfgl) cells are shown. (B) RAP74(1-217)170A3, -173A3, and -176A3 were compared with RAP74(1-517) and RAP74(1-217) in pulse-chase initiation (top panel) and in elongation stimulation (lower panel) assays. RAP74 samples were reconstituted with RAP30 in vitro prior to assay. Pulse-chase initiation reactions contained TFIIF-depleted extract with the indicated TFIIF or TFIIF mutant (10 pmol of TFIIF complex) combined with an adenovirus major late promoter template digested with SmaI at position +217. The protocol for the initiation assay was as in Fig. 2 except that the DNA template was not immobilized and the elongation time was 30 min. Elongation stimulation reactions contained salt-washed elongation complexes supplemented with the indicated TFIIF or TFIIF mutant protein (20 pmol) (as in Fig. 3). Lane 1 contains reconstituted TFIIF and α-amanitin 1 μg/ml. Lane 14 contains no added TFIIF. Lanes 2 and 3 contain 10 pmol and lanes 15 and 16 contain 20 pmol of RAP30 but no added RAP74. All other lanes are as indicated above both panels. (C) PhosphorImager quantitation of the data shown in panel B. Elongation stimulation was calculated for the +192 to +251 transcripts.
Electrophoretic mobility shift assay.
The DNA probe for the gel shift assay was the adenovirus major late promoter from positions −53 to +14 relative to the transcriptional start site. The probe was synthesized by PCR, using the primers 5′-32P-CAGGTGTTCCTGAAGG-3′ and 5′-ATGCGGAAGAGAGTGA-3′. The upstream primer was radiolabeled by using [γ-32P]ATP and T4 polynucleotide kinase. After amplification, the DNA probe was gel purified by using a Qiaex kit (Qiagen). Mobility shifts were performed as described by Wang and Burton (54), with some modifications. The reaction mixtures (15 μl) contained 20 mM HEPES (pH 7.9), 20 mM Tris (pH 7.9), 50 mM KCl, 2 mM dithiothreitol, 0.5 mg of BSA (bovine serum albumin) per ml, 10% (vol/vol) glycerol, radiolabeled DNA probe, and proteins, incubated at 30°C. S. cerevisiae TBP (0.3 pmol) was combined with the DNA template (approximately 40 fmol) for 15 min. Recombinant human TFIIB (0.3 pmol) was then added, and the mixture was incubated for 15 min. Calf thymus pol II (0.15 pmol) was incubated with human recombinant TFIIF (0.1 pmol) for at least 5 min prior to addition to the DB complex. For reactions involving separate TFIIF subunits, pol II was incubated with RAP30 for 5 min and then mixed with RAP74 or a RAP74 mutant and preincubated for an additional 5 min before addition to DB and further incubation for 15 min. It appeared that prior addition of RAP30 to pol II aided assembly of RAP74 into DBPolF (defined in Results). Reaction mixtures were loaded onto a 4% polyacrylamide gel containing 0.09% bisacrylamide, 2.5% glycerol, and 0.5× TBE (tris-borate-EDTA). Dried gels were analyzed by autoradiography.
Transcription assays.
Preparation of immobilized templates was adapted from published methods (1, 30). DNA containing the adenovirus major late promoter was synthesized by PCR, using an upstream 5′-biotinylated primer. The template for amplification was a pBluescript II SK(−) vector (Stratagene) containing the adenovirus major late promoter (−258 to +196) subcloned between the XhoI and HindIII sites of the plasmid. The sequence of the upstream primer was 5′-biotin-CCCTCGAGCGGTGTTCCGCGGTCCTCCTCG-3′, and the sequence of the downstream primer was 5′-CGGTGGCGGCCGCTCTAGAACTAGTGGATC-3′. The template extended from positions −263 to +251. Biotinylated DNA was incubated with streptavidin paramagnetic beads (CPG) in 2 M NaCl–1 mM EDTA–10 mM Tris-HCl (pH 7.5) for 15 min at room temperature. Immobilized templates were collected with a magnetic particle separator (CPG), washed four times, and stored at 4°C in phosphate-buffered saline (pH 7.2) containing 1 mg of BSA per ml and 0.03% NaN3.
Pulse-spin and pulse-chase single-round assays.
A 20-μl reaction mixture consisted of 2 μl of paramagnetic beads (about 0.6 μg of DNA) carrying the adenovirus major late promoter and TFIIF-depleted transcription extract (72 μg of total protein) supplemented with recombinant RAP30 and RAP74 or a RAP74 mutant (10 pmol of each), in transcription buffer (12 mM HEPES [pH 7.4], 12% glycerol, 0.12 mM EDTA, 0.12 mM EGTA, 1.2 mM dithiothreitol) containing 60 mM KCl and 12 mM MgCl2. Preinitiation complexes were formed for 60 min at 30°C; 100 μM ATP, CTP, and GTP and 1 μM [α32P]UTP (10 μCi per reaction) were added to initiate transcription for 1 min. For the pulse-spin protocol, template-associated complexes were diluted with 200 μl of transcription buffer containing 60 mM KCl and 1 mg/ml BSA, isolated by centrifugation, and extracted from beads by boiling in 20 μl of 90% (vol/vol) formamide–1% sodium dodecyl sulfate–10 mM Tris (pH 7.9)–1% mM EDTA–0.01% bromophenol blue–0.01% xylene cyanol. For the pulse-chase protocol, instead of dilution and centrifugation of samples, 1 mM ATP, UTP, GTP, and CTP were added 1 min after addition of nucleoside triphosphates (NTPs), and elongation continued for 10 min. Reactions were stopped by addition of 200 μl of 0.1 M sodium acetate (pH 5.4)–0.5% sodium dodecyl sulfate–2 mM EDTA–100 μg of tRNA per ml, followed by phenol-chloroform extraction and ethanol precipitation. Samples were electrophoresed in a 10% polyacrylamide gel containing 1× TBE and 50% (wt/vol) urea. For quantitation of the gel, the signal in the presence of RAP30 and the absence of added RAP74 was used to estimate background (Fig. 2, lanes 21 and 22). The weak signal obtained in the absence of added RAP74 was attributed to residual RAP74 remaining in the TFIIF-depleted extract.
FIG. 2.
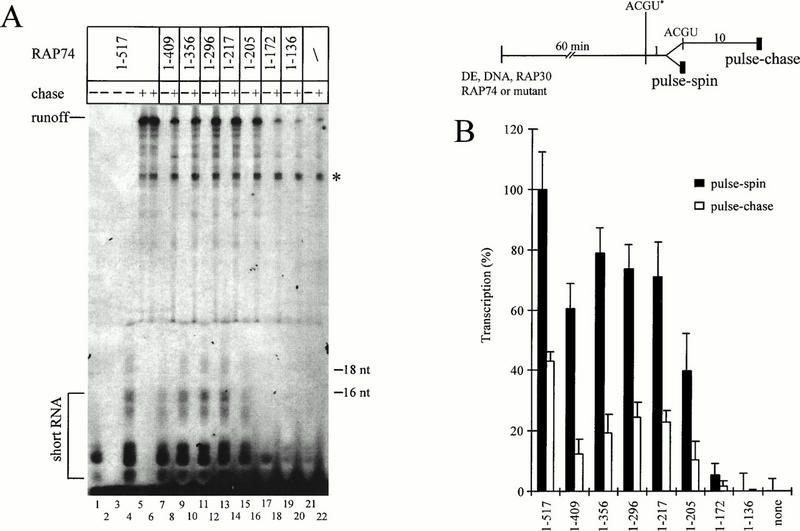
Regions of RAP74 required for accurate initiation. (A) Short and runoff RNAs accurately initiated from the adenovirus major late promoter. All lanes contained TFIIF-depleted extract (DE; 72 μg of total protein), recombinant human RAP30 (10 pmol), and RAP74 or a RAP74 mutant (10 pmol), preincubated with immobilized template for 60 min. Transcripts were initiated with all four NTPs and radiolabeled with [α-32P]UTP (ACGU*) for 1 min. For the pulse-chase protocol, samples were chased by addition of 1 mM each NTP for 10 min (+). For the pulse-spin protocol, initiated complexes were diluted with transcription buffer and centrifuged briefly to isolate short, template-associated RNAs (−). The approximate sizes of short RNAs can be estimated by comparison to 5′-phosphorylated 16- and 18-nucleotide (nt) DNA markers. In lane 1, AMPPCP (a β-γ nonhydrolyzable ATP analog) and 2′,3′-dideoxy-ATP (ddA) were substituted for ATP. In lane 2, AMPPCP was substituted for ATP. In lane 3, 1 μg of α-amanitin per ml was included in the reaction. The gel band indicated with an asterisk is not a pol II transcript because it is synthesized in the presence of 1 μg of α-amanitin per ml (data not shown). (B) PhosphorImager quantitation of the data shown in panel A combined with data from two other experiments, reported as average ± standard deviation. Short transcripts generated in the presence of RAP74 are expressed as 100%.
Elongation stimulation assay.
Stimulation of pol II elongation was determined by adding TFIIF subunits to transcriptionally engaged pol II molecules that were washed free of associated elongation factors. A 20-μl reaction mixture consisted of 2 μl of paramagnetic beads carrying the adenovirus major late promoter, transcription extract derived from HeLa cell nuclei (108 μg of total protein), and transcription buffer containing 60 mM KCl and 12 mM MgCl2. Samples were incubated for 60 min at 30°C to form preinitiation complexes. Transcription was initiated by addition of 100 μM ATP, GTP, CTP, and 1 μM (10 μCi) [α-32P]UTP (2-μl volume). After 1 min, elongation complexes were diluted in 200 μl of transcription buffer containing 500 mM KCl and 1 mg of BSA per ml and isolated by centrifugation. The purpose of this treatment was to remove accessory factors from the elongation complex. The 500 mM KCl wash appeared to be effective because elongation rate stimulation was highly dependent on addition of both RAP30 and RAP74 (Fig. 3). Complexes were diluted with transcription buffer containing 60 mM KCl and 1 mg of BSA per ml and isolated by centrifugation two times. Complexes were then resuspended in 20 μl of transcription buffer containing 60 mM KCl and 12 mM MgCl2; recombinant RAP30 and RAP74 or a RAP74 mutant was added (10 pmol each), and the mixture was incubated for 5 min; 100 μM ATP, GTP, CTP, and UTP were added in 2 μl, and transcripts were elongated for 2 min. By using this protocol but allowing 5 min to elongate RNA chains, most of the observed pol II transcripts were extended to close to the +251 runoff position (data not shown). Transcripts were isolated by phenol extraction and ethanol precipitation and electrophoresed in a 6% polyacrylamide gel, as described above. Accurate transcription was quantitated for all of the transcripts from +122 to +251. Elongation stimulation was determined by subtracting the average value for samples containing RAP30 but no RAP74 as background (Fig. 3A, lanes 14 and 15) and expressed as a percentage of the highest signal obtained for RAP74 (lane 3).
FIG. 3.
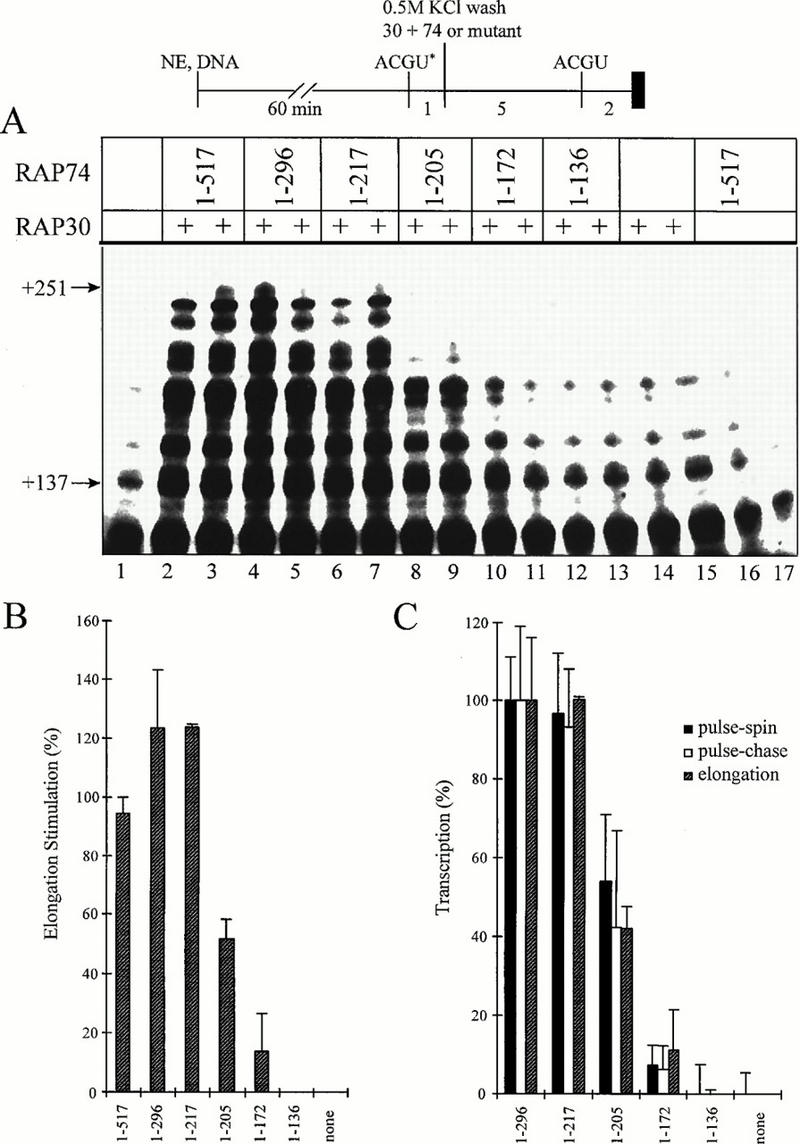
The region of RAP74 between aa 172 and 217 is critical for both accurate initiation and elongation stimulation. (A) Elongation stimulation assay. 32P-labeled short RNAs were accurately initiated from the adenovirus major late promoter immobilized on beads. Complexes were washed with 0.5 M KCl to remove associated elongation factors from pol II. RAP30 (10 pmol) and RAP74 or a RAP74 mutant (10 pmol) were added and incubated for 5 min. All four NTPs (100 μM each) were added, and RNA chains were allowed to elongate for 2 min. NE, nuclear extract. (B) PhosphorImager quantitation of the data shown in panel A for transcripts between +137 and +251 in length. (C) Comparison of elongation stimulation (B), pulse-spin, and pulse-chase data (from Fig. 2). Values for RAP74(1-296) are reported as 100% of signal.
Multiple-round and Sarkosyl block assays.
Transcription was initiated from an adenovirus major late promoter template digested with SmaI to produce a 217-base runoff transcript. The source of transcription factors was TFIIF-depleted extract (72 μg of total protein) supplemented with recombinant RAP30 and RAP74 (10 pmol of each, except as noted). For all reactions, preinitiation complexes were formed for 60 min at 30°C. For the multiple-round assay, 600 μM ATP, CTP, and GTP and 25 μM [α32P]UTP (10 μCi per reaction) were added, and transcription continued for the indicated times (Fig. 5 and 6). For the 10-min cold–multiple-round assay, 600 μM ATP, CTP, and GTP and 25 μM UTP were added and incubated for 10 min, and then [α-32P]UTP (10 μCi per reaction) was added and transcription continued for 60 min (Fig. 5). For the single-round Sarkosyl block assay (18), 600 μM ATP and CTP and 25 μM [α32P]UTP (10 μCi per reaction) were added and incubated for 1 min; 600 μM GTP and 0.25% (wt/vol) Sarkosyl were added and transcription continued for 59 min (Fig. 5), 79 min (Fig. 6B and C), or 99 min (Fig. 6D). Since 0.05% Sarkosyl was previously shown to be sufficient to block new initiation by pol II (18), Sarkosyl was added in fivefold excess over the amount necessary to constrain transcription to a single round. As a control, Sarkosyl was added to reactions before NTPs, causing initiation to be completely blocked, indicating that the level of detergent added in experiments was sufficient to block reinitiation (data not shown).
FIG. 5.
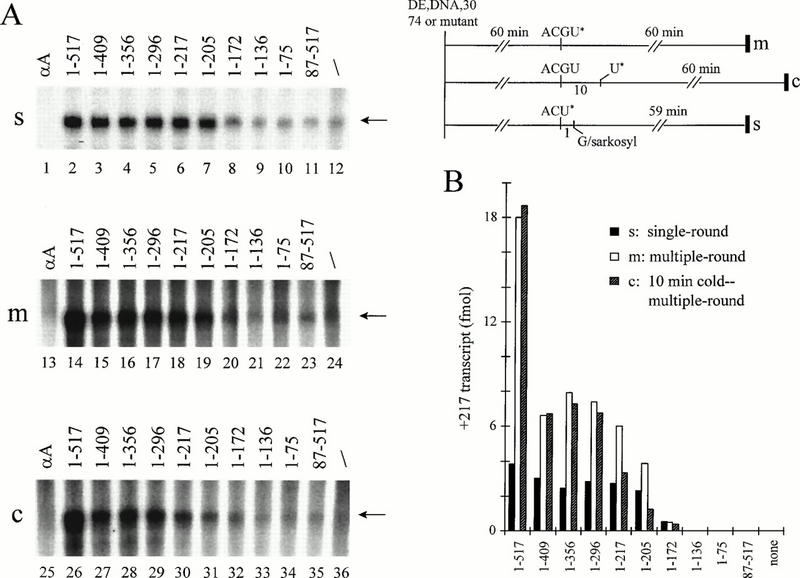
The CTD of RAP74 stimulates multiple-round transcription. (A) Single-round and multiple-round transcription assays. All samples contained TFIIF-depleted extract (DE; 72 μg of protein), recombinant human RAP30 (10 pmol), and RAP74 or a RAP74 mutant (10 pmol). In the single-round protocol (s), reinitiation was blocked by addition of the anionic detergent sarkosyl. In the multiple-round protocol (m), transcription was allowed to proceed for 60 min. In the 10-min cold–multiple-round protocol, (c), transcription with all four NTPs was allowed to proceed for 10 min before addition of radiolabel and incubation for 60 min. Reactions labeled αA contained RAP74 and 1 μg of α-amanitin per ml. (B) PhosphorImager quantitation of the data in panel A.
FIG. 6.
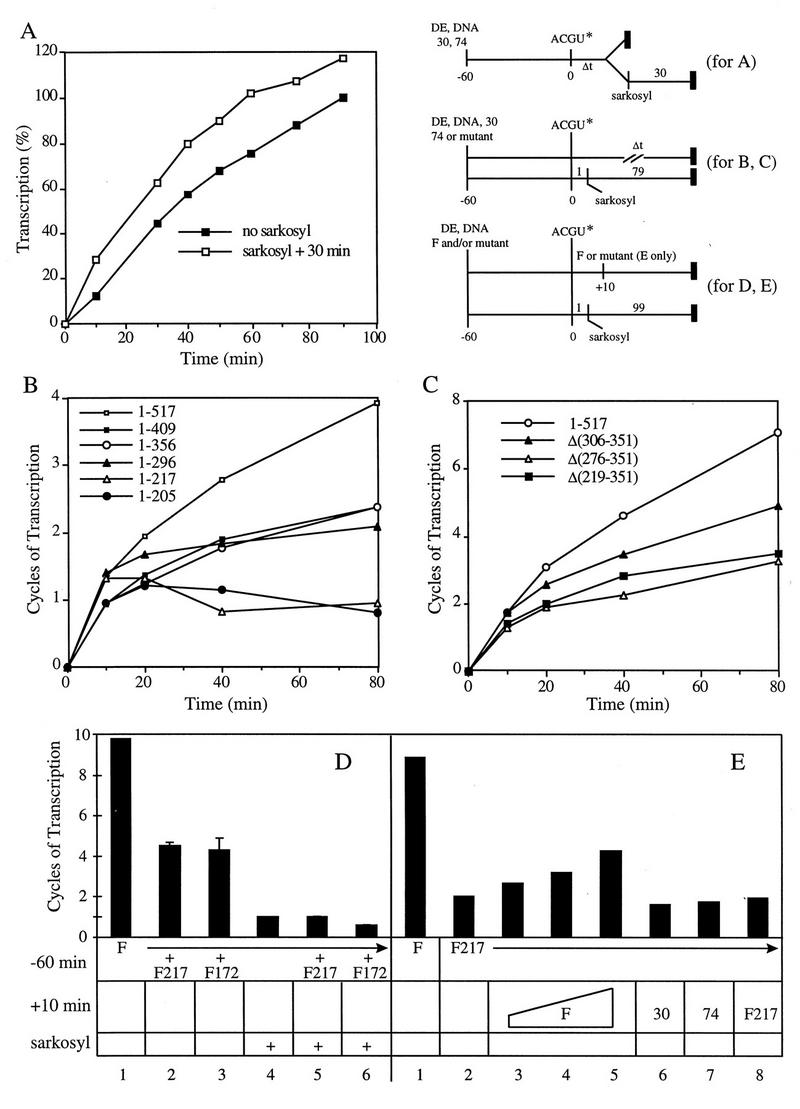
The central region and the CTD of RAP74 cooperate to stimulate multiple-round transcription. (A) New initiation occurred throughout the 90-min course of the reaction. Assays were done by the multiple-round protocol (Fig. 5) and stopped at the indicated times (filled squares), or instead of stopping the reactions, sarkosyl was added to block new initiation and transcription continued for an additional 30 min to complete any previously initiated chains (open squares). (B and C) Both the central region and the CTD of RAP74 contributed to multiple-round transcription. Multiple-round transcription was determined by the protocol used for Fig. 5 except that reactions were stopped at the indicated times. Single-round transcription was estimated by using a Sarkosyl block procedure with a 79-min elongation. Cycles of transcription were estimated as transcription in the absence (time indicated) or presence (79-min elongation) of Sarkosyl. (D and E) TFIIF containing RAP74(1-217) (abbreviated F217) and F172 competed with TFIIF (F) for transcription complex formation and inhibited multiple-round transcription. The reaction protocol was the same as used for panels B and C except that the reaction time after NTP addition was 100 min. TFIIF or mutants were added to the reaction at −60 min or +10 min, as indicated. (D) Reactions in columns 1 to 6 contained 2 pmol of TFIIF; reactions in columns 2 and 5 also contained 2, 4, 10, or 20 pmol F217 (data points were combined within the error bar because the effect was essentially maximal with 2 pmol of F217); reactions in columns 3 and 6 contained 2 or 20 pmol of F172. (E) The reaction in column 1 contained 2 pmol of TFIIF; reactions in columns 2 to 8 contained 2 pmol of F217; reactions in columns 3, 4, and 5 contained 2, 4, or 10 pmol of TFIIF added at +10 min; reactions in columns 6, 7, and 8 contained 10 pmol of RAP30, RAP74, or F217 added at +10 min.
For the experiment shown in Fig. 6A, using the multiple-round transcription protocol, 0.25% Sarkosyl was added at the indicated times to block new initiation, and transcription was continued for an additional 30 min to complete all previously initiated chains (18). This was a control experiment to demonstrate that new initiation occurs throughout the course of the reaction. Transcription was quantitated and background was selected as described above.
G-less cassette pol II trap.
To characterize multiple-round transcription in the extract system, a G-less cassette trap for pol II was used (48). The template was plasmid pML(C2AT)19, the kind gift of Michele Sawadogo, containing the adenovirus major late promoter fused to a G-less cassette at position +11 (42, 43). Preinitiation complexes were formed for 1 h. Reactions contained TFIIF-depleted extract supplemented with 10 pmol of recombinant RAP30 and RAP74; 600 μM ATP, GTP, and CTP, 1 mM 3′-O-methyl-GTP, and 25 μM [α32P]UTP (10 μCi per reaction) were added to reactions as indicated in Fig. 7. At t = +1 min, 0.05% Sarkosyl was added to some reactions to estimate single-round transcription. In the presence of ATP, CTP, UTP, and 3′-O-methyl-GTP, a transcript of 390 bases was synthesized. Under this condition, pol II stalled after insertion of 3′-O-methyl-GMP into the RNA chain at position +390. The template was digested with PvuII to allow simultaneous detection of stalled transcription at +390 and runoff transcription at position +602. In control experiments, 0.05% Sarkosyl was found to be sufficient to constrain transcription to a single round (data not shown); 0.05% sarkosyl was used in this experiment because, unexpectedly, the early elongation complex formed from the G-less cassette template was much more sensitive to disruption with Sarkosyl than that initiated from the wild-type promoter (data not shown). The promoters in these two plasmids are identical from positions −256 to +10, and we do not know the explanation for the observed difference in Sarkosyl sensitivity. For chase reactions, 1 mM GTP and UTP were added and elongation continued for 10 or 60 min, as indicated in Fig. 7.
FIG. 7.
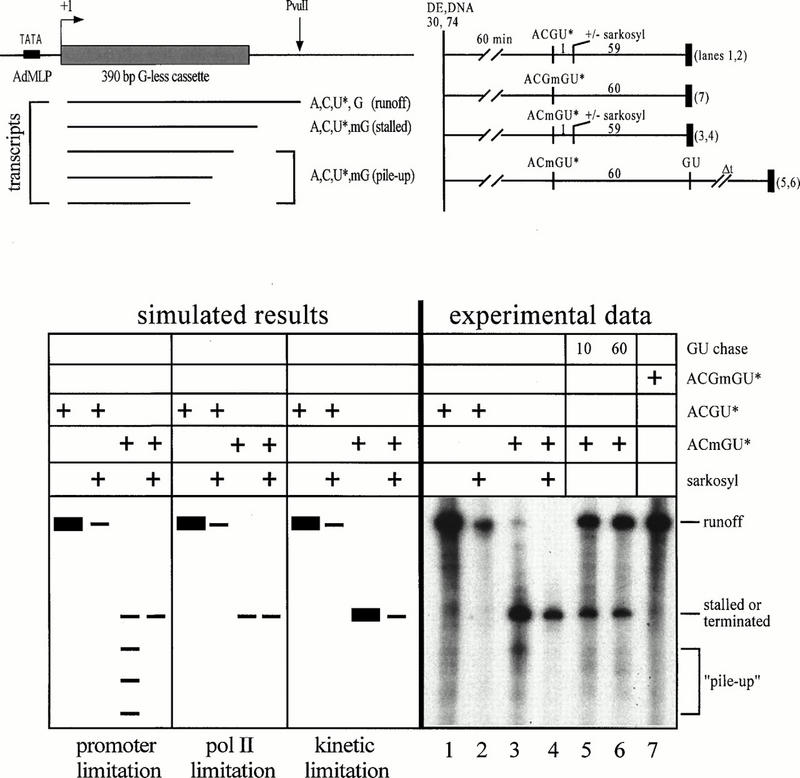
Multiple-round transcription in an extract system can be described by a kinetic limitation model. The template contains the adenovirus major late promoter (AdMLP) fused to a G-less cassette that extends to position +389. The template was digested with PvuII at position +602. Transcripts formed in the presence of the chain terminator 3′-O-methyl-GTP (mG) and in the absence of GTP stalled at the end of the G-less cassette. When GTP was included in the reaction, pol II continued transcription to the +602 runoff position. The template and protocols are summarized at the top. Expected results for the promoter limitation, pol II limitation, and kinetic limitation models are shown on the left and discussed in the text. Experimental data are shown on the right.
RESULTS
The N-terminal domain of RAP74 supports preinitiation complex assembly.
A primary function of TFIIF in accurate initiation is to deliver pol II to the promoter; therefore, we used an electrophoresis mobility shift assay to determine which regions of RAP74 were required to bring pol II into a stable complex with adenovirus major late promoter DNA, TBP, TFIIB, and RAP30 (DBPolF complex) (Fig. 1). By itself, TBP did not efficiently induce a shift of the promoter fragment (lane 2). Upon addition of TFIIB, however, a DB complex consisting of TBP, TFIIB, and promoter DNA was observed (lane 3). When pol II was added to DB, a weak DBPol shift was seen (lane 4). RAP30 alone did not stimulate pol II binding to DB (lane 6), nor did RAP74 (data not shown). The TFIIF complex and separately added RAP30 and RAP74 subunits, however, supported assembly of DBPolF (lanes 5 and 7). RAP74(1-172) was minimally required to support assembly (lane 10). A number of RAP74 mutants that have been shown to be defective for RAP30 binding (54, 55) failed to support formation of DBPolF (lanes 11 to 15). The different mobilities of complexes containing RAP74 deletion mutants may be attributable to differences in the charge or the degree of DNA bending or flexibility in the complex. Because RAP74(1-517), RAP74(1-296), RAP74(1-205), and RAP74(1-172) are predicted to carry charges of 0, −4, +6, and +7; however, it is difficult to account for all observed mobility differences solely on the basis of charge. For instance, RAP74(1-172) is predicted to be more basic than RAP74(1-205) and yet supported a DBPolF complex with a higher mobility. Furthermore, RAP74(1-205) and (1-172) have different transcriptional activities (see below).
FIG. 1.
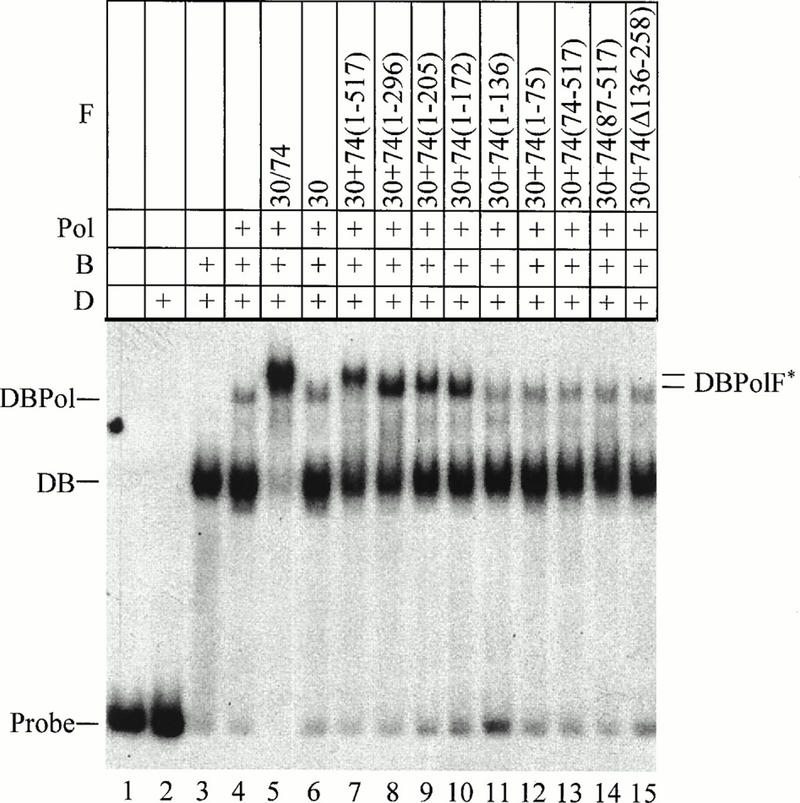
Electrophoretic mobility shift assay to analyze the requirement for RAP74 to form DBPolF. The probe was the adenovirus major late promoter between positions −53 and +14. D, recombinant yeast TBP (0.3 pmol); B, recombinant human TFIIB (0.3 pmol); Pol, calf thymus pol II (0.15 pmol); F, recombinant human TFIIF complex or RAP30 and RAP74 or a RAP74 mutant, added separately (0.1 pmol). DBPolF* indicates the different mobilities of complexes containing different RAP74 mutants.
Tyree et al. (51) reported DBPolF30 and DBPolF74 complexes, which formed with only the RAP30 or the RAP74 subunit of TFIIF, but these investigators used a different promoter and Drosophila TBP, TFIIB, pol II, and human TFIIF rather than yeast TBP, human TFIIB, human TFIIF, and bovine pol II, as used in this study. Killeen et al. (22) and Flores et al. (16) also found DBPol30 and DABPol30 complexes under buffer conditions very different from those used here. Although RAP74 was not essential for assembly in those studies, it was strongly stimulatory. This paper is the first report that shows the minimal region of RAP74 that stimulates incorporation of pol II into DBPolF.
The N-terminal domain of RAP74 supports accurate initiation.
To determine which regions of RAP74 are most important for initiation, RAP74 mutants were tested for accurate initiation and runoff transcription from the adenovirus major late promoter (Fig. 2). RNA within the early elongation complex was visualized directly on gels in the pulse-spin protocol or extended to the runoff position in the pulse-chase protocol. Both protocols limited transcription to a single round. In the pulse-spin protocol, pol II remained close to the promoter, blocking a second round of transcription. In the pulse-chase protocol, a 1,000-fold excess of unlabeled UTP was added during the chase; so, although reinitiation could occur, it was not detected.
By several criteria, short transcripts produced in the pulse-spin protocol were inferred to be accurately initiated from the adenovirus major late promoter. Short RNAs were template associated because they could be isolated by centrifugation with beads (lanes designated “−” for no chase). Short RNAs were chased to the predicted runoff position of +251 with addition of NTPs (lanes designated “+”). As expected for pol II transcripts, synthesis of short RNAs was completely dependent on ATP with a hydrolyzable β-γ bond (compare lanes 1 and 2). Synthesis of short RNAs was sensitive to 1 μg of α-amanitin per ml (compare lanes 3 and 4) and was RAP30 and RAP74 dependent (lane 21 and data not shown).
RAP74 supported synthesis of the highest yield of short RNAs (lane 4), but RAP74(1-409), RAP74(1-356), RAP74(1-296), and RAP74(1-217) also supported transcription (lanes 7, 9, 11, and 13). RAP74(1-205) supported a lower yield of short RNA than RAP74(1-217) (compare lanes 13 and 15). RAP74(1-172) was barely active, and RAP74(1-136) was inactive (lanes 17 and 19). RAP74(74-517) and RAP74(87-517) were also inactive (reference 54 and data not shown). For accurate initiation, therefore, the region of RAP74 between aa 1 and 205 was necessary, and the region between aa 205 and 217 was stimulatory. Comparison of the activities of deletion mutants showed that the regions between aa 136 and 217 and aa 1 and 74 were very important for initiation.
A very similar conclusion was reached from inspection of runoff transcripts (Fig. 2A, lanes designated “+”; Fig. 2B, white bars). Because RNA was labeled by comparable procedures in the pulse-chase and pulse-spin reactions, yields of transcript in the pulse-chase reactions are reported as percentage of the highest signal observed using the pulse-spin protocol. RAP74 had the highest activity in runoff transcription (lanes 5 and 6). RAP74(1-296) and RAP74(1-217) were about 75% as active as RAP74 (lanes 12 and 14). RAP74(1-205) had reduced activity (lane 16). RAP74(1-172) was almost inactive (lane 18), and RAP74(1-136) was inactive (lane 20). These data confirmed that the most important region of RAP74 for supporting accurate initiation was located within aa 1 to 217 and that the region between aa 172 and 217 was critical for activity.
RAP74(1-409) and RAP74(1-356) had surprisingly low activities in runoff transcription (lanes 8 and 10), a result that has been reproduced in several experiments. RAP74(1-409) also has a partial defect in elongation stimulation (data not shown). Deletion of sequence between aa 296 and 356 relieves these defects, because RAP74(1-296) and RAP74(1-217) have higher activities than RAP74(1-409) and RAP74(1-356) in runoff transcription (compare lanes 8, 10, 12, and 14). Because RAP74(1-409) and RAP74(1-356) appeared to initiate more efficiently than to form runoff transcripts, these mutants may form complexes with a tendency to release abortive transcripts.
Background transcription in this and other experiments appeared to be due to residual RAP74 in the TFIIF-depleted extract (lanes 19 to 22). Accurate transcription was not detected when RAP30 was omitted from the reaction (data not shown), indicating that RAP30 was more efficiently depleted than RAP74. Another observation was that a significant proportion of short RNAs were released as abortive transcripts. Comparing the ratio of accurately initiated transcripts in the pulse-spin protocol (Fig. 2B, black bars) to runoff products observed in the pulse-chase protocol (white bars), we found that less than half of the short transcripts were recovered at the runoff position.
The N-terminal domain of RAP74 stimulates elongation by pol II.
Because preinitiation complex assembly (Fig. 1) and single-round initiation (Fig. 2) were supported by the N-terminal domain of RAP74, we wondered whether the same or distinct regions might stimulate elongation by pol II (Fig. 3). Consistent with previous reports (21, 49), both RAP30 and RAP74 were required to stimulate elongation (Fig. 3A, lanes 1 and 14 to 17). RAP74(1-217) stimulated elongation to about the same extent as RAP74(1-296) and RAP74. RAP74(1-205), however, stimulated elongation at a reduced level, and RAP74(1-172) was inactive or nearly so. The region between aa 1 and 205, therefore, appeared to be essential for elongation stimulation, and the region between aa 205 and 217 was strongly stimulatory. RAP74(74-517) and RAP74(87-517), from which N-terminal sequences were deleted, did not stimulate pol II elongation (reference 21 and data not shown).
The activities of the RAP74 mutants for elongation stimulation (Fig. 3B) appeared to be very similar to the activities obtained for accurate initiation and runoff transcription (Fig. 2B), as if very similar RAP74 N-terminal domain sequences were important for both processes. To challenge this idea, pulse-spin, pulse-chase, and elongation stimulation data were plotted on the same graph, using the sample containing RAP30 but no RAP74 as background and the sample containing RAP74(1-296) as 100% signal. The RAP74(1-296) sample was selected as the highest value in order to eliminate the influence of the CTD on initiation from the comparison. As can be seen in Fig. 3C, the region of RAP74 required to support accurate initiation, runoff transcription, and elongation stimulation was the same.
To investigate this issue in more detail, three triple-alanine substitution mutants were constructed within this critical region (Fig. 4). The target for mutation, between aa 170 and 178, was selected because this region is evolutionarily conserved (54), and interestingly, this sequence is strongly predicted to be α-helical for diverse eukaryotic species, including S. cerevisae, Drosophila melanogaster, Xenopus laevis, and humans. RAP74(1-217)170A3, -173A3, and -176A3 were constructed in the RAP74(1-217) deletion mutant. The triple-alanine mutants showed significant defects in both accurate initiation and elongation stimulation (Fig. 4), as expected from the results with deletion mutants (Fig. 3C). The region of RAP74 between aa 170 and 178, therefore, was very important for both accurate initiation and elongation.
The central region and CTD of RAP74 stimulate multiple-round transcription.
RAP74 mutants were next tested for the ability to support multiple-round transcription in vitro (Fig. 5). In one protocol, NTPs were added with [α32P]UTP radiolabel, and transcription was allowed to continue for 60 min. In a modified protocol, unlabeled NTPs were added for 10 min before addition of radiolabel (10-min cold-multiple round), and transcription continued for 60 min. For comparison, we performed a single-round protocol in which Sarkosyl (0.25%) was added 1 min after addition of NTPs to block new initiation (18). The final specific activity of radiolabel was the same in all three procedures, and so the intensities of transcription signals can be compared directly. For the Sarkosyl block assay (Fig. 5A), the observed transcriptional activities of RAP74 mutants were very similar to those determined in the pulse-spin initiation assay (Fig. 2).
Cycles of transcription were estimated by dividing the yield of transcripts in the absence of Sarkosyl (multiple round) by the yield of transcripts in the presence of sarkosyl (single round). By this estimation, full-length RAP74 supported approximately four cycles of transcription in 60 min (Fig. 5B). However, RAP74(1-409), RAP74(1-356), RAP74(1-296), and RAP74(1-217) supported only about two rounds of accurate transcription. The region between aa 409 and 517, therefore, was important for multiple-round transcription. For RAP74(1-517), RAP74(1-409), and RAP74(1-356), cycles of transcription were not diminished in the 10-min cold–multiple-round protocol. The transcription system, therefore, did not become limiting for factors or substrates within 70 min. In contrast, RAP74(1-217) and RAP74(1-205) had a reduced ability to support multiple rounds of transcription after a 10-min incubation with unlabeled NTPs. In the presence of RAP74(1-217) and RAP74(1-205), therefore, some transcription factor(s) appeared to become limiting for initiation at later reaction times.
To characterize new initiation in the extract, the yield of transcripts was determined at different times after addition of NTPs, and transcripts continued to accumulate for more than 90 min (Fig. 6A). The increase in transcription was due to new initiation rather than slow elongation of chains initiated at earlier times, because when Sarkosyl was added to rescue all previously initiated chains, transcripts nonetheless continued to accumulate for the entire 90 min. If chains were initiated at early times and slowly elongated, the Sarkosyl rescue curve would achieve its maximal value at an early time, instead of tracking the curve without Sarkosyl, as observed.
To better understand the activities of RAP74 mutants in multiple-round transcription, cycles of transcription were determined as a function of time for RAP74 mutants (Fig. 6B and C). Mutants for which the graph has a positive slope at the +40- and +80-min time points were judged to be capable of supporting multiple-round transcription. By this criterion, RAP74 was most active, followed by RAP74(1-409) and RAP74(1-356). RAP74(1-296) had a very weak capacity to support multiple-round transcription, and RAP74(1-217) and RAP74(1-205) were inactive. Although defective for multiple-round transcription, RAP74(1-296) and RAP74(1-217) were highly active in single-round transcription (Fig. 2). The region of RAP74 between aa 409 to 517 was important for multiple-round transcription, but the region between aa 217 and 356 stimulated multiple-round transcription, even in the absence of the CTD. To further test the importance of the central region, three internal deletion mutants were constructed and tested (Fig. 6C). RAP74(Δ306-351), RAP74(Δ276-351), and RAP74(Δ219-351) all were shown to support multiple-round transcription at a reduced level compared to RAP74. Each of these mutants, however, supported single-round transcription almost as well as RAP74 [about 75% wild-type activity (data not shown), similarly to RAP74(1-217)]. RAP74(Δ306-351) was most active for multiple-round transcription, and RAP74(Δ276-351) and RAP74(Δ219-351) had lower activity. Although the central region stimulated multiple-round transcription, the CTD supported this activity in the absence of aa 219 to 351, as indicated by the positive slope of the graph for RAP74(Δ276-351) and RAP74(Δ219-351) at late time points (Fig. 6C). Therefore, both the CTD and the central region of RAP74 contributed to multiple-round transcription.
An alternative explanation for the observed defects of RAP74 deletion mutants in multiple-round transcription might be that mutants were less stable than RAP74 during the reaction time course. We did not believe that this would be the case because the central loop of RAP74 appears to be the region that is most sensitive to proteolysis (data not shown), and RAP74(1-296) and RAP74(1-217), which show the most dramatic defects in multiple-round transcription, have most or all of the central region removed. Also, a spectrum of defects in multiple-round transcription was noted (Fig. 6B and C); thus, different mutants would have to have different stabilities in approximate proportion to their length, which seemed unlikely. Furthermore, a 5- to 10-fold functional excess of each mutant protein was added to reactions during a 1-h preincubation; thus, RAP74 mutants would have to be stable in the extract under reaction conditions during the preincubation but not after addition of NTPs, which seemed unlikely. In a Western blot analysis, RAP74(1-517), RAP74(1-217), RAP74(1-172), and RAP30 remained intact throughout the reaction time course (data not shown). Most convincingly, experiments shown in Fig. 6D and E demonstrated the stability of RAP74 mutants during the reaction and the selective defect of RAP74(1-217) in multiple-round transcription. The experiment shown in Fig. 6D demonstrated that F217 [TFIIF complexes containing RAP74(1-217)] and F172 appeared to compete with TFIIF for formation of transcription complexes. Addition of F217 to a reaction containing TFIIF inhibited multiple-round transcription (compare columns 1 and 2) but not single-round transcription (compare columns 4 and 5). This was expected because RAP74(1-217) is active for single-round but not multiple-round transcription (Fig. 6B). Since RAP74(1-172) was nearly inactive for transcription (Fig. 2), this mutant inhibited both multiple-round (compare columns 1 and 3) and single-round (compare columns 4 and 6) transcription. The inhibitory effect of RAP74(1-172) was consistent with our observation that this mutant assembled into the DBPolF complex (Fig. 1). The competitive effects of RAP74(1-217) and (1-172) persisted during the 100-min time course, indicating that these proteins remained active for complex assembly. The experiment shown in Fig. 6E shows that late addition of TFIIF rescued a reaction containing F217 for multiple-round transcription (compare column 1 to columns 3 to 5) but that readdition of F217 did not (compare columns 2 and 8). Rescue of the reaction with TFIIF was not complete because of inhibition by F217 (Fig. 6D). If the defect of F217 at late reaction times were due to degradation or inactivation, readdition of F217 would be expected to rescue transcription, but this was not observed (column 8). Furthermore, because readdition of TFIIF did rescue multiple-round transcription, the presence of F217 did not cause the irreversible inactivation of another general factor, such as TFIIB or pol II. Therefore, these data demonstrated that RAP74(1-217) had a specific defect in multiple- but not single-round transcription and that this defect was not attributable to degradation or inactivation of the mutant protein. These experiments strengthen our argument that the CTD and central region of RAP74 have a specific role in transcriptional recycling.
New initiation resulted from previously unused pol II molecules initiating from previously unused promoters.
In the extract system, new initiation events could involve reuse of promoters, reuse of pol II, or initiation from many promoters from which pol II is enabled to initiate at various times. We used a G-less cassette template as a pol II trap to discriminate between these possibilities (48). With the G-less cassette, an adenovirus major late promoter transcript can be synthesized with only ATP, CTP, and UTP, omitting GTP (42). Pol II stalls at the first position at which GMP must be incorporated. If promoters are reused, multiple pol II molecules pile up at the end of the G-less cassette, resulting in a ladder of transcripts each one shorter by about 30 nucleotides (48). The number of rungs on the ladder corresponds to the number of pol II molecules that have stalled on the template. If pol II must be reused for new initiation at later reaction times, trapping pol II at the end of the G-less cassette is expected to inhibit new initiation.
We have therefore considered three models to characterize multiple-round transcription in the extract system: (i) promoter limitation (promoter reuse), (ii) pol II limitation (pol II reuse), and (iii) kinetic limitation (new initiation from different promoters using different pol II molecules at various times). Promoter limitation results if only a small number of promoters are bound by the necessary set of DNA-binding transcription factors (TFIID, major late transcription factor, etc.), and these factors remain committed to the same promoter through multiple transcription rounds. Sawadogo’s laboratory has set up a transcription system with purified and recombinant components in which functional promoter limitation was demonstrated (48). Alternatively, in the pol II limitation model, only a small fraction of pol II molecules have the capacity to accurately initiate and reinitiate. If pol II is limiting in concentration and if pol II molecules are trapped at the end of the G-less cassette, multiple-round transcription will be inhibited. Pol II limitation can arise by functional limitation of any transcription factor that remains tightly associated with pol II through the transcription cycle. A third possibility is the kinetic limitation model. If a transcription factor has to be in a particular form (i.e., phosphorylation state) to be active, then the availability of active transcription complexes at any one time might be controlled.
To discriminate between these models, we did the experiment shown in Fig. 7. The template contained the adenovirus major late promoter fused to a G-less cassette (43). We anticipated that a low level of GTP in the extract might allow elongation past the end of the cassette, and in fact, this is what we observed (data not shown). This problem was overcome by addition of the chain terminator 3′-O-methyl-GTP. Because the plasmid template was digested with PvuII at position +602, stalling at the end of the cassette could be compared to runoff transcription.
Simulated results based on the three models are indicated with experimental data that are most consistent with a kinetic limitation model (Fig. 7). RAP74 supported approximately 4 to 10 rounds of initiation in 80 min (Fig. 5 and 6B to E), and this result was confirmed for the G-less cassette template by comparing multiple-round (without Sarkosyl) and single-round (with Sarkosyl) runoff transcription (Fig. 7; compare lanes 1 and 2). When 3′-O-methyl-GTP was included in the reaction, multiple-round transcription was not noticeably inhibited (compare lanes 3 and 4 with lanes 1 and 2). These data most closely resemble the results expected for the kinetic limitation model. Stalling of pol II at the end of the G-less cassette was efficient, because the runoff transcript was barely detectable in the presence of the chain terminator (lane 3). A single pile-up product corresponding to approximately 10% of stalled transcripts was observed above background. This gel band appears to be a pile-up product because it was chased to the runoff position with addition of GTP (compare lane 3 with lanes 5 and 6), and it was not observed in lanes 1, 5, and 6, as expected for a terminated transcript. We have not detected a second pile-up product (which would be expected at a level of only 1% of the stalled transcript), and so few promoters appear to be used as many as three times (lane 3). Because about four cycles of transcription were observed and only 10% of promoters were used even twice, the extract system was generally in active promoter excess. Because new initiation was not inhibited by trapping pol II, the data were not consistent with a pol II limitation model (compare lanes 3 and 4 with lanes 1 and 2). A very slow rate of true pol II recycling may occur in this system, but most new initiation required the recruitment of unused pol II molecules.
About 20% of the transcripts stalled at the end of the G-less cassette were terminated or arrested, because they were not chased with addition of GTP (compare lane 3 with lanes 5 and 6). This low frequency of escape from the pol II trap was not sufficient to complicate interpretation of the experimental results because escape of 20% of pol II molecules cannot account for four rounds of new initiation. Also, the sample in lane 7 showed that inclusion of 3′-O-methyl-GTP in the reaction did not affect multiple-round transcription.
DISCUSSION
From primary sequence (2, 15), RAP74 is proposed to have highly basic N- and C-terminal domains separated by a highly charged, overall acidic, and flexible central region that is rich in charged amino acids, E, D, K, and R, and also S, T, P, and G. In this report, we show that these primary sequence features correspond to distinct N- and C-terminal functional domains (Fig. 8). The N-terminal domain is sufficient to support preinitiation complex assembly, single-round initiation, and elongation by pol II. The central region and CTD of RAP74 have a small stimulatory effect on initiation, but their most important function is in multiple-round transcription. Deletion of just the central region or just the CTD of RAP74 creates a protein with partial function in multiple-round transcription.
FIG. 8.
N- and C-terminal domains of RAP74, which were originally proposed from sequence analysis (2, 15), correspond to distinct functional domains. The N-terminal domain has most of the functions required for single-round initiation and elongation. The central region and CTD of RAP74 function in multiple-round transcription. Regions shaded dark are very important for activity. IIB, TFIIB; CTDP, CTD phosphatase; PIC, preinitiation complex.
The N-terminal domain of RAP74 stimulates initiation and elongation.
The RAP74 N-terminal domain extending from aa 1 to 217 supports most RAP74 functions for preinitiation complex assembly (Fig. 1), accurate initiation (Fig. 2), and elongation stimulation (Fig. 3 and 4). In this work, we have mapped a critical region for these functions between aa 136 and 217. RAP74(1-172) binds tightly to the RAP30 subunit, but RAP74(1-136) does not (54, 55). Consistent with its capacity to bind RAP30, RAP74(1-172) is minimally sufficient to support pol II assembly into DBPolF (Fig. 1). Fulfilling this role in assembly, however, is not sufficient to support TFIIF function in initiation or elongation, which minimally requires RAP74(1-205) (Fig. 2). RAP74(1-217) is significantly more active than RAP74(1-205) in both accurate initiation and elongation stimulation (Fig. 2 and 3).
Recent site-specific DNA photo-cross-linking studies show that RAP74 induces a significant conformational change within the DBPolFE preinitiation complex (17, 39, 40). This conformational change is referred to as isomerization to compare it with isomerization of the Escherichia coli preinitiation complex, which involves similar changes in protein conformation (41) and wrapping of promoter DNA around RNA polymerase (11, 37). Interestingly, RAP74(1-205), which is minimally required for accurate initiation, also minimally supports isomerization of DBPolFE, as indicated by development of a number of specific DNA photo-cross-links with the RPB1 and RPB2 subunits of pol II, with RAP30, and with TFIIE34 (17, 39, 40). Because RAP74(1-172) is sufficient for tight RAP30 binding and DBPolF assembly but insufficient for isomerization of DBPolFE, accurate initiation or elongation stimulation, RAP74(1-217) and RAP74(1-205) appear to have functions that are not communicated to pol II directly through the RAP30 subunit. Photo-cross-linking studies have indicated that RAP74 approaches the adenovirus major late promoter at numerous positions extending all the way from −56 to −61 upstream of TATA to +26 downstream of +1 (about 280 Å of B-form DNA) (17, 40). This extensive cross-linking footprint is the basis for one argument in favor of DNA wrapping around pol II in DBPolFE and also for an α2β2 heterotetrameric form of TFIIF in the complex (40). Because RAP74 interacts with promoter DNA and induces isomerization of DBPolFE, RAP74 appears to support DNA wrapping both by contacting DNA directly and by modifying the contacts of RAP30, pol II, and TFIIE34 with DNA (17, 39, 40). The region of RAP74 between aa 172 and 205, therefore, appears to stimulate transcription by helping DNA to wrap around pol II. Wrapping may result from direct interactions between DNA and aa 172 to 205, or this region may be involved in protein-protein interactions that facilitate wrapping. Recent work indicates that the region from aa 172 to 205 is involved in dimerization of RAP74 (40). It is not clear whether the adjacent region of RAP74 from aa 205 to 217, which also stimulated initiation and elongation, contributed to DNA wrapping or another function. It is interesting that DBPolF complexes containing RAP74(1-517), RAP74(1-296), RAP74(1-205), and RAP74(1-172) had different electrophoretic mobilities that could have been caused by differences in the degree of DNA bending or flexibility (Fig. 1). Differences in mobility might relate to the degree of DNA bending caused by partial or complete isomerization of DBPolFE in the presence of TFIIF mutants.
RAP74(1-217), RAP74(1-205), and RAP74(1-172) showed a spectrum of decreasing activities in both accurate initiation and elongation stimulation (Fig. 3C). Triple alanine mutants in this region, RAP74(1-217)170A3, -173A3, and -176A3, were also significantly affected for both initiation and elongation (Fig. 4). The 170A3 mutant was more defective in initiation than 173A3, but they had very similar defects in elongation. Perhaps more interestingly, the 176A3 mutant was partially active in initiation but inactive for elongation stimulation. The initiation assay is more complex than the elongation assay, because initiation is influenced by all of the general transcription factors and some regulatory factors in the extract system, and the elongation assay may involve only elongating pol II and TFIIF. In the initiation assay, therefore, general or regulatory factors could partially complement TFIIF activity. 176A3, therefore, might be partially complemented for its function in initiation by interaction with a general or regulatory transcription factor, which is present in the cell extract but absent in salt-washed elongation complexes. Conceivably, readdition of such a factor to the elongation complex might relieve the 176A3 defect in elongation.
Because the same region of RAP74 contributed strongly to both initiation and elongation, RAP74 may perform similar roles in the two processes. In preinitiation complex assembly, the role of RAP74 appears to be to wrap DNA around pol II and to isomerize the complex (40). If RAP74 has a similar role in elongation, it is also likely to involve DNA wrapping around pol II. In initiation, DNA wrapping is induced around eukaryotic RNA polymerase I (44), RNA polymerase II (24, 40), and prokaryotic RNA polymerase (11, 37).
It was somewhat surprising that the sequence requirements for RAP74 were so similar for initiation and elongation, because a very different conclusion was reached for the RAP30 subunit of TFIIF (50). Although RAP30 mutations that fail to bind RAP74 were found to be severely defective for both initiation and elongation, mutations in other regions of RAP30 affected initiation and elongation in different ways. RAP30 mutations within a presumed pol II binding region were defective in elongation stimulation but not in initiation. RAP30 mutations within a DNA-interacting region were defective for accurate initiation but not elongation stimulation (50). These results may indicate that RAP30 has distinct roles in initiation and elongation, although RAP74 appears to fulfill a common role in both processes. Because DNA-binding regions of RAP30 appear to be critical for initiation but dispensable for elongation, RAP30 might interact specifically with DNA in the preinitiation complex. During elongation, the DNA sequence encountered by pol II is in flux, and RAP30 may make less extensive template contacts. RAP30 is also likely to make reduced protein-protein contacts as initiation factors dissociate from the elongation complex.
The central region and CTD of RAP74 stimulate multiple-round transcription.
When cycles of transcription were plotted against time for multiple-round transcription reactions, the resulting curve showed a high initial rate of RNA synthesis followed by a lower rate (Fig. 6B and C). The initial burst of synthesis persisted for 10 to 15 min, and then the lower rate dominated the reaction. This lower rate represents multiple-round transcription and is likely to represent a recycling mechanism for pol II or an essential pol II transcription factor. RAP74 mutants from which C-terminal and/or central regions were removed were found to be defective for this low rate of transcription at later reaction times.
In Fig. 7, we showed that multiple-round transcription in the extract conformed to what we call a kinetic limitation model. If this system had fit the pol II limitation model, this would have demonstrated true pol II recycling, because release of pol II from the template would then have been required to support new initiation. Establishment of a pol II limited system that can support multiple-round transcription will require that recycling be much more efficient than in the extract system. According to the kinetic limitation model, new initiation events at later reaction times were largely dependent on unused pol II molecules that were recruited to (or activated at) previously unused promoters. The extract system was found to have an excess of active promoters, and pol II and general factors were estimated to be in significant excess over the level of adenovirus major late promoter transcripts that were produced (references 5, 9, and 29 and our unpublished data). In a system that contains an excess of active promoters and a presumed excess of transcription factors, it is somewhat difficult to understand why all the active promoters are not occupied and utilized at once. One way to view this kind of regulation is that a particular factor may require modification (i.e., phosphorylation or dephosphorylation) to be activated for initiation, such that although the factor is not limiting in concentration, it is limiting in activity.
In the extract system, the rate at which CTD phosphatases convert pol IIo to pol IIa may become limiting for initiation. When ATP and GTP are added, CTD kinases such as TFIIH (14, 29, 45, 46) and P-TEFb (31) can phosphorylate pol IIa to pol IIo. If this conversion is efficient, most pol II will be in the pol IIo form, but only the small fraction that remains in the initiating pol IIa form is expected to assemble into preinitiation complexes (7, 27). Pol IIo requires a CTD phosphatase to convert it to the pol IIa form for new initiation. The initial burst of RNA synthesis observed in Fig. 6B and C may reflect utilization of the pool of pol IIa in the extract. The low rate of RNA accumulation at later reaction times may reflect the rate of conversion from pol IIo to pol IIa. In our system, true recycling of pol II after template runoff was not efficient, because trapping pol II at the end of a G-less cassette did not inhibit multiple-round transcription (Fig. 7). On the other hand, multiple-round transcription in the extract appeared to reflect a physiological recycling system, because this process involved activation of transcription complexes as a function of time. Additionally, multiple-round transcription was completely dependent on the presence of either the central region or CTD of RAP74 (Fig. 6B and C). As an example, RAP74(1-217) was completely inactive for multiple-round transcription because this mutant failed to support new initiation at late reaction times, but RAP74(1-217) was highly active for the initial burst of transcription (Fig. 6B).
RAP74, TFIIB, and the CTD phosphatase may be components of a pol II recycling mechanism. The CTD of RAP74 stimulates a CTD phosphatase (8), and this region of RAP74 binds TFIIB (13) and pol II (54). TFIIB blocks stimulation of CTD phosphatase activity by RAP74 (8). Dephosphorylated pol IIa preferentially enters the preinitiation complex, and CTD kinases phosphorylate pol II within the preinitiation complex or shortly after initiation (7, 27). During elongation, pol II is hyperphosphorylated on the CTD (7, 28). Because RAP74 stimulates and TFIIB blocks stimulation of CTD phosphatase activity (8), we suggest that TFIIB may be present in elongation complexes to block CTD dephosphorylation in order to prevent premature termination. RNA processing beyond the 3′ mRNA cleavage and polyadenylation sequence (AAUAAA) may provide a signal to relieve the TFIIB block to CTD phosphatase activity, allowing RAP74 to stimulate conversion of pol IIo to pol IIa. Interestingly, the 3′-end cleavage factor complex (CPSF-CstF) interacts with the CTD and associates with elongating pol II (32). We suggest that conversion of transcriptionally engaged pol IIo molecules to the pol IIa form may induce termination, with pol IIa released in the appropriate form to recycle to a promoter.
RAP74 has distinct functions in bringing pol II to the promoter, in isomerization of the preinitiation complex, in elongation stimulation, perhaps in termination, and in pol II recycling. These specialized functions must be regulated for progression through the transcription cycle. For instance, RAP74 is unlikely to stimulate elongation rate and the activity of the CTD phosphatase during elongation, because this would simultaneously stimulate elongation and induce premature termination. It will be of great interest to identify all of the components of the initiation, elongation, termination, and recycling complexes to begin to unravel how pol II monitors its progress through the transcription cycle.
ACKNOWLEDGMENTS
We thank Steven Triezenberg, Fan Shen, Richard Burgess, and Stephan Reimers for proteins and Danny Reinberg and Michelle Sawadogo for clones. We gratefully acknowledge David Arnosti, Benoit Coulombe, James Geiger, and Steven Triezenberg for providing valuable criticisms of the manuscript. Augen Pioszak, Victoria Sutton, Jessica Metcalf-Burton, Hiroe Taki, Julia Clay, and Nadine Kobty helped with production of RAP74 mutants as part of undergraduate internship programs. Julia Xiaozhu Pan helped with purification of proteins.
This work was supported by a grant from the American Cancer Society, by Michigan State University, and by the Michigan State University Agricultural Experiment Station. Verna C. Finkelstein also contributed funds to support this work.
REFERENCES
- 1.Arias J, Dynan W S. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J Biol Chem. 1989;264:3223–3229. [PubMed] [Google Scholar]
- 2.Aso T, Vasavada H A, Kawaguchi T, Germino F J, Ganguly S, Kitajima S, Weissman S M, Yasukochi Y. Characterization of cDNA for the large subunit of the transcription initiation factor TFIIF. Nature. 1992;355:461–464. doi: 10.1038/355461a0. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomew B, Dahmus M E, Meares C F. RNA contacts subunits IIo and IIc in HeLa RNA polymerase II transcription complexes. J Biol Chem. 1986;261:14226–14231. [PubMed] [Google Scholar]
- 4.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton Z F, Ortolan L G, Greenblatt J. Proteins that bind to RNA polymerase II are required for accurate initiation of transcription from the adenovirus 2 major late promoter. EMBO J. 1986;5:2923–2930. doi: 10.1002/j.1460-2075.1986.tb04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton Z F, Killeen M, Sopta M, Ortolan L G, Greenblatt J. RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol. 1988;8:1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers R S, Dahmus M E. Purification and characterization of a phosphatase from HeLa cells which dephosphorylates the C-terminal domain of RNA polymerase II. J Biol Chem. 1994;69:26243–26248. [PubMed] [Google Scholar]
- 8.Chambers R S, Wang B Q, Burton Z F, Dahmus M E. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J Biol Chem. 1995;270:14962–14969. doi: 10.1074/jbc.270.25.14962. [DOI] [PubMed] [Google Scholar]
- 9.Cochet-Meilhac M, Nuret P, Courvalin J C, Chambon P. Animal DNA-dependent RNA polymerases. 12. Determination of the cellular number of RNA polymerase B molecules. Biochim Biophys Acta. 1974;353:185–192. doi: 10.1016/0005-2787(74)90183-x. [DOI] [PubMed] [Google Scholar]
- 10.Conaway R C, Garrett P, Hanley J P, Conaway J W. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors α and βγ promote entry of polymerase into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig M L, Suh W-C, Record M T. HO · and DNase I probing of Eς70 RNA polymerase-λPR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 12.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 13.Fang S M, Burton Z F. RNA polymerase II-associated protein (RAP) 74 binds transcription factor (TF)IIB and blocks TFIIB-RAP30 binding. J Biol Chem. 1996;271:11703–11709. doi: 10.1074/jbc.271.20.11703. [DOI] [PubMed] [Google Scholar]
- 14.Feaver W J, Gileadi O, Li Y, Kornberg R D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 15.Finkelstein A, Kostrub C F, Li J, Chavez D P, Wang B Q, Fang S M, Greenblatt J, Burton Z F. A cDNA encoding RAP74, a general initiation factor for transcription by RNA polymerase II. Nature. 1992;355:464–467. doi: 10.1038/355464a0. [DOI] [PubMed] [Google Scholar]
- 16.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forget D, Robert F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley D K, Roeder R G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- 19.Hodo H G, III, Blatti S P. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1977;16:2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- 20.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 21.Kephart D D, Wang B Q, Burton Z F, Price D H. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 22.Killeen M, Coulombe B, Greenblatt J. Recombinant TBP, transcription factor IIB, and RAP30 are sufficient for promoter recognition by mammalian RNA polymerase II. J Biol Chem. 1992;267:9463–9466. [PubMed] [Google Scholar]
- 23.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 24.Kim T-K, Lagrange T, Wang Y-H, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y-J, Geiger J H, Hahn S, Sigler P B. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:512–520. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 26.LaGrange T, Kim T-K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laybourn P J, Dahmus M E. Transcription-dependent changes in the C-terminal domain of mammalian RNA polymerase subunit IIa/o. J Biol Chem. 1989;264:6693–6698. [PubMed] [Google Scholar]
- 28.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 30.Marshall N F, Price D H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxy-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 32.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 33.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 34.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 35.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 36.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 37.Polyakov A, Severinova E, Darst S A. Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 38.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 40.Robert, F., M. Douziech, D. Forget, J. Greenblatt, Z. F. Burton, and B. Coulombe. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 41.Roe J H, Burgess R R, Record M T. Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-λ PR promoter interaction: assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 42.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawadogo M, Roeder R G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 44.Schultz P, Celia H, Riva M, Sentenac A, Oudet P. Three-dimensional model of yeast RNA polymerase I determined by electron microscopy of two-dimensional crystals. EMBO J. 1993;12:2601–2607. doi: 10.1002/j.1460-2075.1993.tb05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serizawa H, Conaway R C, Conaway J W. A carboxy-terminal domain kinase associated with RNA polymerase II transcription factor δ from rat liver. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serizawa H, Conaway R C, Conaway J W. Multifunctional RNA polymerase II initiation factor δ from rat liver: relationship between carboxy-terminal kinase, ATPase, and DNA helicase activities. J Biol Chem. 1993;268:17300–17308. [PubMed] [Google Scholar]
- 47.Shapiro K J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 48.Szentirmay M N, Sawadogo M. Sarkosyl block of transcription reinitiation by RNA polymerase II as visualized by the colliding polymerases reinitiation assay. Nucleic Acids Res. 1994;22:5341–5346. doi: 10.1093/nar/22.24.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan S, Aso T, Conaway R C, Conaway J W. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 50.Tan S, Conaway R C, Conaway J W. Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc Natl Acad Sci USA. 1995;92:6042–6046. doi: 10.1073/pnas.92.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 52.Wang B Q, Kostrub C F, Finkelstein A, Burton Z F. Production of human RAP30 and RAP74 in bacterial cells. Protein Expression Purif. 1993;4:207–214. doi: 10.1006/prep.1993.1027. [DOI] [PubMed] [Google Scholar]
- 53.Wang B Q, Lei L, Burton Z F. Importance of codon preference for production of human RAP74 and reconstitution of the RAP30/74 complex. Protein Expression Purif. 1994;5:476–485. doi: 10.1006/prep.1994.1067. [DOI] [PubMed] [Google Scholar]
- 54.Wang B Q, Burton Z F. Functional domains of human RAP74 including a masked RNA polymerase II-binding domain. J Biol Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 55.Yonaha M, Aso T, Kobayashi Y, Vasavada H A, Yasukochi Y, Weissman S M, Kitajima S. Domain structure of a human general transcription initiation factor, TFIIF. Nucleic Acids Res. 1993;21:273–279. doi: 10.1093/nar/21.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]