Abstract
The eukaryotic mRNA 3′ poly(A) tail and its associated poly(A)-binding protein (Pab1p) are important regulators of gene expression. One role for this complex in the yeast Saccharomyces cerevisiae is in translation initiation through an interaction with a 115-amino-acid region of the translation initiation factor eIF4G. The eIF4G-interacting domain of Pab1p was mapped to its second RNA recognition motif (RRM2) in an in vitro binding assay. Moreover, RRM2 of Pab1p was required for poly(A) tail-dependent translation in yeast extracts. An analysis of a site-directed Pab1p mutation which bound to eIF4G but did not stimulate translation of uncapped, polyadenylated mRNA suggested additional Pab1p-dependent events during translation initiation. These results support the model that the association of RRM2 of yeast Pab1p with eIF4G is a prerequisite for the poly(A) tail to stimulate the translation of mRNA in vitro.
The 3′ poly(A) tail of eukaryotic mRNA serves as an important regulator of gene expression. Both in vitro and in vivo experiments have demonstrated a role for this structure in translation (6, 15, 19, 20, 22). Particularly striking examples of the role of the poly(A) tail in translation are found in studies involving vertebrate oocytes, whose timing of progression through meiosis relies on the regulated addition of poly(A) tails to preexisting cytoplasmic mRNAs and their subsequent translational activation (25). The poly(A) tail may also assist in the nucleocytoplasmic transport of mRNA (9) and appears to confer stability to cytoplasmic mRNA (4). To understand the roles of the poly(A) tail more completely, we have analyzed the structure and function of its major associated protein, the poly(A)-binding protein (Pab1p). Pab1p has been found in all eukaryotes examined thus far and is likely to mediate many of the cytoplasmic and possibly some of the nuclear functions of the poly(A) tail. This study focuses on the role of Pab1p in mediating the translational stimulation of mRNA by the poly(A) tail.
Although the function of the poly(A) tail in translation has been the subject of research for many years (11, 21), a relevant target of Pab1p in the translation initiation pathway in yeast has only recently been identified. This target is eukaryotic translation initiation factor 4G (eIF4G) (23). The eIF4G protein is part of the cytoplasmic 5′ cap-binding complex eIF4F, which also contains the cap-binding protein eIF4E. eIF4G has been shown to bind to Saccharomyces cerevisiae Pab1p (23) and more recently the wheat Pab1p (13) in vitro. The association of yeast Pab1p with eIF4G has also been shown to mediate the poly(A) tail- and Pab1p-dependent translational stimulation of mRNA in vitro (24). Current models hold that Pab1p can stimulate recruitment of the 40S ribosomal subunit to mRNA (22) through its association with eIF4G. The eIF4G protein provides the link to the 40S subunit since it can bind to the 40S-associated initiation factor eIF3 (14). The Pab1p-binding site within yeast eIF4G has been narrowed down to a 115-amino-acid region N terminal to the binding site for eIF4E (23, 24). This study was designed to define the region(s) of Pab1p that interacts with this subdomain of eIF4G and thereby stimulates translation in vitro.
Pab1p is the founding member (1, 18) of a large family of proteins containing an RNA recognition motif (RRM). Pab1p also contains a less well defined carboxy-terminal region. The RRM consists of approximately 90 amino acids, and nuclear magnetic resonance and crystal structures of representative RRMs reveal a highly conserved hydrophobic core with a less well conserved surface (reviewed in reference 16). This latter feature undoubtedly supplies the specificity to each protein. Pab1p is one of few proteins in the RRM family that contains four tandem copies of this domain. While all four RRMs of Pab1p presumably share a common three-dimensional fold, they are quite divergent in sequence and therefore probably in function. We previously identified RRM2 of yeast Pab1p as being primarily responsible for its high affinity and specificity for poly(A) RNA (5). Here the importance of RRM2 and the other Pab1p domains in translation is addressed.
To identify domains of the protein that bind to eIF4G and are required for poly(A) tail-dependent translation in vitro, a deletion analysis of S. cerevisiae Pab1p was performed. Specifically, each Pab1 mutant protein was tested for its ability to bind to S. cerevisiae eIF4G in vitro and to stimulate poly(A) tail-dependent translation in yeast extracts. These experiments revealed that RRM2 of Pab1p is required for both of these functions. Additionally, analysis of a site-directed mutant of Pab1p enabled us to functionally separate the binding of Pab1p to eIF4G from its activation of translation in vitro. The results support the hypothesis that the association of RRM2 of Pab1p with eIF4G is a prerequisite for poly(A) tail-dependent translation and that once associated, other regions of Pab1p and/or eIF4G are needed to stimulate translation.
MATERIALS AND METHODS
Construction of Pab1p deletions.
PAB1-1 (5) was used as the template for all PAB1 plasmids created in this study. Of note, the PAB1-1 gene encodes a His6 tag at the amino terminus of Pab1-1 and contains unique restriction sites in the linkers between each of the domains of PAB1. RRM1 was deleted by joining a PvuII site at nucleotide (nt) 25 to a blunted BamHI site at nt 372, thereby deleting amino acids 15 to 129. RRM2 was deleted by inserting OSK01 (GATCACAGGCCT) into the PAB1-1 plasmid with the following ends: a BamHI site (nt 372) and a ClaI site (nt 632). This results in a deletion of amino acids 132 to 216 and an insertion of an alanine prior to amino acid 217. RRM3 was deleted by inserting OSK03 (CGAAACAGTATGAAG) between the ClaI site (nt 632) and a StuI site (nt 927), thus removing amino acids 218 to 311. RRM4 was deleted by joining a blunted BamHI site at nt 1266 to the StuI site (nt 927), which eliminates amino acids 316 to 427. The carboxy-terminal truncation was constructed by fusing blunted BamHI (nt 1266) and SpeI (nt 1819) sites. The downstream BamHI site (nt 1266) was originally made in plasmid pJD13, which contains PAB1 modified only at this site. The final amino acid is residue 429. pab1-105 (contains only RRM1 and RRM2) was made by fusing a blunted ClaI site (nt 632) to a PvuII site at nt 1579. This protein contains a C-terminal LVKLLV hexapeptide following amino acid 217. Finally, the gene encoding the protein containing just RRM2 was constructed in the same manner as pab1-105, with the exception of the starting plasmid, which contained pab1-ΔRRM1 instead of PAB1-1. All of the above were initially constructed in a yeast TRP1 CEN4 vector (pAS414 [7]). These genes were also subcloned into pET11d (Invitrogen) for bacterial overexpression followed by Ni2+ affinity chromatography for purification (see reference 5 for details). Table 1 summarizes the yeast and bacterial strains containing the various pab1 genes.
TABLE 1.
Strains and plasmids used
| Description | Protein | Expression vectora | Yeast vectorb | Strainc |
|---|---|---|---|---|
| Wild type | Pab1-1p | BAS3059 | BAS3072 | YAS2261 |
| ΔRRM1 | Pab1-100p | BAS3221 | BAS3227 | YAS2235 |
| ΔRRM2 | Pab1-101p | BAS3222 | BAS3228 | YAS2236 |
| ΔRRM3 | Pab1-102p | BAS3223 | BAS3229 | YAS2237 |
| ΔRRM4 | Pab1-103p | BAS3224 | BAS3230 | YAS2238 |
| ΔCterm | Pab1-104p | BAS3225 | BAS3231 | YAS2239 |
| Δ3+4+Cterm | Pab1-105p | BAS3226 | BAS3232 | NA |
| Y83V, F170V + Δ3+4+Cterm | Pab1-106p | BAS3233 | NAd | NA |
| Y83V, F170V | Pab1-16p | BAS3066 | BAS3079 | YAS2025 |
Bacterial expression vectors were derived from pET11-d (Invitrogen) and are contained in the indicated DH5α strain.
The yeast vector contains the indicated pab1 gene in a TRP1 CEN4 vector. These vectors are stored in the indicated DH5α strain.
Yeast strain YAS2031 was transformed with the indicated yeast vector and cured of wild-type PAB1 to yield these strains.
NA, not applicable.
RNA binding analysis.
Gel shifts were performed as described by Deardorff and Sachs (5). Briefly, final buffer and salt conditions were as follows: 10 mM Tris (pH 8.0), 80 mM potassium acetate, 30 mM NaCl, 0.4 mM EDTA, 0.7 mM MgCl2, 0.6 mM dithiothreitol, 2 μg of tRNA per ml, 0.6% glycerol, 15 μg of bovine serum albumin per ml, and 1.5% polyvinyl alcohol. The samples were run on a 4% acrylamide (40:1) native gel in 0.4× Tris-borate-EDTA at 80 V for 50 min. Data were quantitated with a Molecular Dynamics PhosphorImager and the ImageQuant program. The data were then fit to the equation y = 1/[1 + (Kd/x)], where y is the fraction of the oligo(A)20 shifted from the origin and x is the free protein concentration. Binding to poly(A)-Sepharose (Pharmacia) was determined using 25 μl of resin with indicated concentrations of proteins in a final binding volume of 100 μl of PBST (150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, 0.1% Triton X-100 [pH 7.3]). Following incubation for 1 h at 4°C, the resin was washed three times in 750 μl of PBST. The resin was then boiled in 25 μl of sodium dodecyl sulfate (SDS) gel loading buffer, of which 20 μl was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by staining with Coomassie brilliant blue.
eIF4G binding analysis.
Glutathione S-transferase (GST) fusions of the two yeast eIF4G Pab1p-binding sites were overexpressed in Escherichia coli BAS3024 and BAS3035 (23). Bacterial cell lysates (5 μl) containing one of these proteins were diluted 100-fold in PBST containing 1 mM phenylmethylsulfonyl fluoride and incubated with 25 μl of glutathione-Sepharose (Pharmacia) for 1 h at 4°C. The resin was washed three times in 750 μl of PBST. Preincubated Pab1p-poly(A) (100 μl) was then added to the resin, which contained approximately 125 to 250 pmol of the eIF4G fragment, for 1 h at 4°C. The resin was again washed three times in 750 μl of PBST and then boiled in 25 μl of SDS gel loading buffer. Binding was measured by SDS-PAGE with 20 μl of the eluate for Coomassie brilliant blue staining or with 1 μl of a 1:20 dilution of the eluate for Western analysis. Western blots were performed with a polyclonal antibody to Pab1p and are as described by Tarun and Sachs (23).
In vitro translation.
Translation extracts were prepared from yeast cells as described previously (10, 22). For studies using the addition of recombinant Pab1p variants, an extract from YAS1874 (MATa MAK10::URA3 PEP4::HIS3 ade2 his3 leu2 trp1 ura3) was treated with 60 U of micrococcal nuclease per 100 μl of extract for 5 min at 26°C (the reaction was quenched with 2 mM EGTA) and then with a 1:50 dilution of anti-Pab1p monoclonal antibody 1G1 (80 μg/μl) (2). Various concentrations of the recombinant proteins were incubated with 50 ng of polyadenylated luciferase (LUCpA) mRNA in a 7.5-μl mixture containing other translational components prior to the addition of 7.5 μl of extract. Translation was allowed to proceed for 40 min at 26°C before quick freezing in liquid nitrogen. Translation was quantified by adding 10 μl of the above-described mixture to 50 μl of luciferase assay system mixture (Promega) and measuring activity in a TD-20e Luminometer (Turner).
Extracts were also prepared from YAS2261, YAS2235, YAS2236, YAS2237, YAS2238, YAS2239, and YAS2025. These extracts were not treated with nuclease or with the monoclonal antibody but were treated with EGTA to a final concentration of 2 mM; 50 ng of either LUC (luciferase), capLUC, LUCpA, or capLUCpA mRNA was added to the extracts. Translation was measured as described above. All extracts used in this study had optical densities at 260 nm of between 106 and 140.
Yeast methods.
The TRP1 CEN4 plasmids containing the PAB1 variants were transformed into YAS2031 (MATa pab1::HIS3 ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100) containing plasmid pPAB1URA3CEN. Trp+ transformants were selected on solid YMD-Trp medium. These transformants were restruck onto YMD-Trp medium containing 1 mg of 5-fluoro-orotic acid (8) per ml to cure cells of pPAB1URA3CEN. Growth rates were determined by growing these strains in liquid YMD-Trp medium at 30°C.
RESULTS
Purification of yeast Pab1p deletion variants.
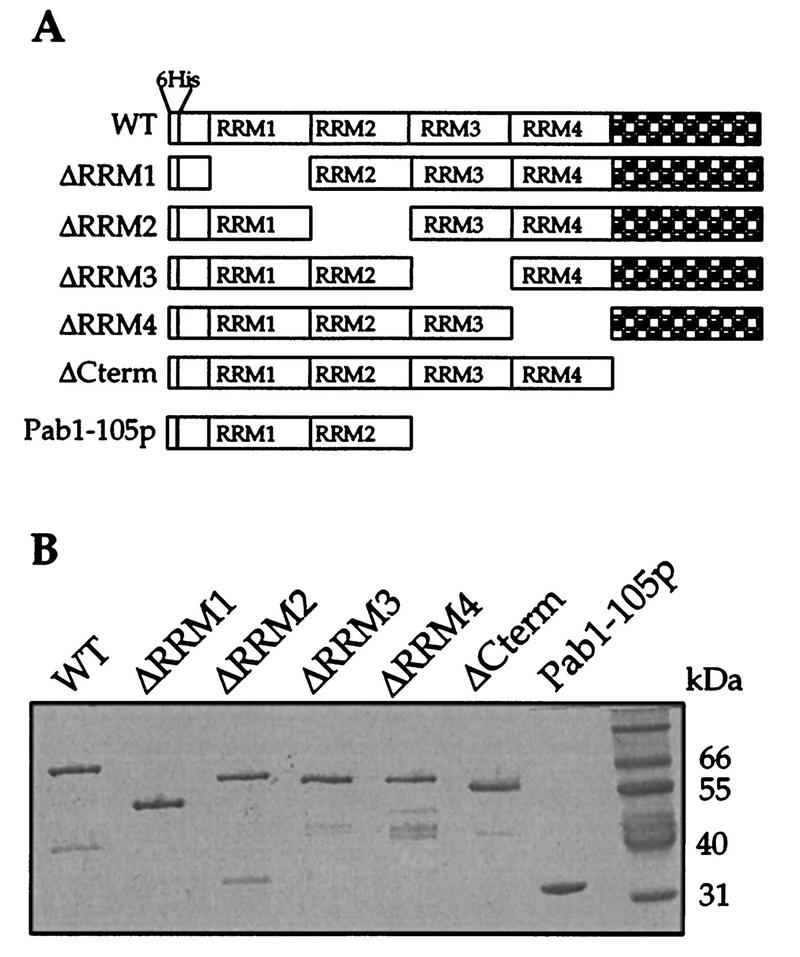
We have previously reported the construction of a modified yeast PAB1 gene which contains convenient restriction enzyme sites between each of the four RRMs and encodes a hexahistidine tag at the protein’s N terminus (5). The presence of the restriction sites allowed for the exact deletions of each RRM, while the N-terminal histidine tag allowed for the rapid purification of the Pab1 protein from bacterial extracts by Ni2+-agarose affinity chromatography. Figure 1A shows the pab1 genes that were created for the experiments described in this report. Both single- and multiple-domain dropouts were made, and each was transformed into yeast cells and expressed as a recombinant protein in E. coli. Figure 1B displays a Coomassie blue-stained gel containing each of the purified recombinant Pab1p variants isolated from the E. coli extracts. These proteins were sufficiently pure to allow for further analysis.
FIG. 1.
Construction and purification of Pab1p variants. (A) Schematic diagram of the Pab1p deletion constructs used in this study. The following amino acids were omitted from each of the constructs: Pab1-ΔRRM1p, 15 to 129; Pab1-ΔRRM2p, 132 to 216; Pab1-ΔRRM3p, 218 to 311; Pab1-ΔRRM4p, 316 to 427; Pab1-ΔCtermp, 430 to 584; and Pab1-105p, 218 to 584. WT, wild type. (B) Purified Pab1p variants. An SDS–10% polyacrylamide gel stained with Coomassie brilliant blue is shown. Positions of molecular weight standards are indicated to the right.
Poly(A)-binding properties of the Pab1p variants.
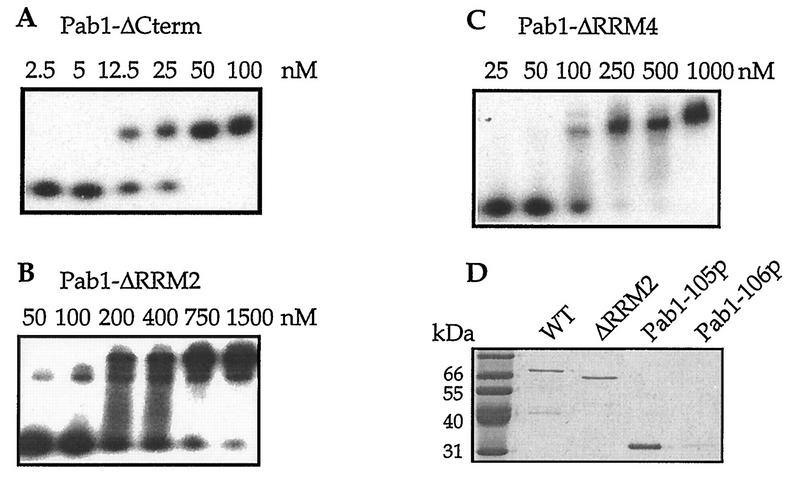
The interaction between yeast Pab1p and eIF4G depends on the presence of poly(A) RNA and on the ability of Pab1p to bind to the RNA (23). To rule out the possibility that a Pab1p variant would be unable to bind eIF4G as a result of its inability to associate with poly(A), we assessed the poly(A)-binding activity of each variant. To do so, a gel mobility shift assay was used to measure the equilibrium dissociation constant (Kd) of each Pab1 protein for oligo(A)20 as previously described (Fig. 2A; Table 2) (5). All of the single-deletion Pab1p variants were able to bind to the RNA in these assays with submicromolar Kds. Of these proteins, the Pab1p missing RRM2 had the lowest affinity for oligo(A)20 (approximately 10-fold lower than that of wild-type Pab1p). This finding is consistent with results from previous work that examined the effects of deletions and of point mutations within each of Pab1p’s RRMs on oligo(A) binding (3, 5, 12). Each of the other single-RRM deletion proteins exhibited oligo(A)20 affinities that were intermediate in value to the full-length Pab1 and the Pab1-ΔRRM2 proteins (Table 2).
FIG. 2.
Poly(A) binding by the Pab1p variants. Determination of equilibrium dissociation constants of Pab1p variants for oligo(A)20 by gel mobility shift analysis. Shown are representative autoradiograms for Pab1-ΔCterm (A), Pab1-ΔRRM2 (B), and Pab1-ΔRRM4 (C). The analysis was also performed for each of the Pab1p variants discussed in this study. 32P-labeled oligo(A)20 was incubated with increasing amounts of the indicated Pab1p variants. The percentage of radiolabeled RNA being shifted was used to calculated the Kd values (see Materials and Methods). These values are reported in Table 2. (D) Binding of Pab1p variants to poly(A)-Sepharose. Eluates from poly(A)-Sepharose resin incubated with the indicated Pab1p variants were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining. Initial binding concentrations of the Pab1 proteins were 3 μM for the wild type (WT), ΔRRM2, and ΔRRM4 and 6 μM for Pab1-105p and Pab1-106p.
TABLE 2.
Summary of data
| Construct | Kd (nM) for oligo(A)20a | eIF4G binding | Translationb | Generation time (h)c | Tail/capd | Synergisme |
|---|---|---|---|---|---|---|
| Wild type | 30 ± 5 | Yes | 100 ± 0.0 | 2.15 ± 0.05 | 1.68 ± 0.28 | 6.37 ± 1.38 |
| ΔRRM1 | 104 ± 31 | Yes | 71.6 ± 19.0 | 2.45 ± 0.15 | 0.21 ± 0.06 | 5.61 ± 1.51 |
| ΔRRM2 | 200 ± 25 | No | 0.2 ± 0.1 | 3.05 ± 0.05 | 0.008 ± 0.002 | 1.48 ± 0.21 |
| ΔRRM3 | 22.5 ± 2.5 | Yes | 246 ± 150 | 2.25 ± 0.05 | 1.65 ± 0.23 | 5.69 ± 0.93 |
| ΔRRM4 | 65 ± 0 | Yes | 28.4 ± 4.80 | 2.65 ± 0.05 | 1.12 ± 0.13 | 4.22 ± 0.22 |
| ΔCterm | 14.5 ± 3.5 | Yes | 55.3 ± 10.2 | 2.65 ± 0.05 | 0.15 ± 0.04 | 6.34 ± 0.96 |
| Pab1-105p | 1,250 ± 50 | Yes | 41.6 ± 16.7 | Dead | NDf | ND |
| Pab1-16p | 1,450g | Yes | 4.9 ± 2.7 | 3.83 ± 0.33 | 0.043 ± 0.005 | 3.05 ± 0.31 |
Average of at least two experiments.
Measured at 30°C in YMD medium lacking Trp.
ND, not determined.
Reported by Deardorff and Sachs (5).
The Pab1 protein truncated immediately C terminal to RRM2 (Pab1-105p) exhibited a 40-fold decrease in affinity for oligo(A)20 (Table 2). This relatively poor affinity was unexpected since it was previously shown that RRM2 is responsible for the high affinity of Pab1p for poly(A) (3, 5, 12). Additionally, experiments using a similar derivative of Pab1p from Xenopus laevis found it to exhibit near-wild-type affinity for oligo(A)23 (12). Future work will be needed to clarify the differences between these studies.
Both the in vitro association assays of Pab1p with eIF4G and the stimulation of in vitro translation by the poly(A) tail use poly(A) RNAs greater than 70 residues in length. Therefore, we also tested the binding of some of the variants to poly(A)-Sepharose resin. This resin has poly(A) RNAs of approximately 100 residues, and as a result assays using it should more clearly reflect the ability of Pab1p to bind poly(A) in the experiments described below. The Pab1p concentrations used in this binding assay are also similar to those used in the eIF4G binding assay and to the Pab1p concentrations in crude yeast extracts (22). The Pab1p that was retained on the poly(A)-Sepharose resin was eluted, resolved by SDS-PAGE, and then visualized by Coomassie brilliant blue staining. As shown in Fig. 2D, all of the Pab1p variants that were tested under these conditions bound to the poly(A)-Sepharose resin with similar efficiencies. Specificity of binding was shown by the inability of the Pab1-105 variant containing RNA-binding inactivating mutations in each of its RRMs (Pab1-106p) to bind to the resin (Fig. 2D). These experiments demonstrate that the Pab1p variants are indeed able to bind to poly(A) RNA, and they therefore eliminate the possibility that any failure to bind to eIF4G would be due to a failure to bind poly(A).
RRM2 of Pab1p is required for association with yeast eIF4G.
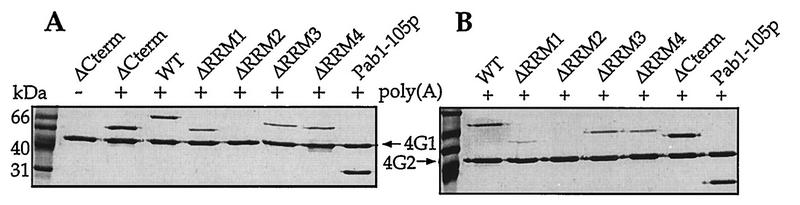
Having established that all of the Pab1p variants bound to poly(A) with reasonable affinity, we next investigated their ability to associate with the Pab1p-binding regions of yeast eIF4G1 and eIF4G2. Each of the two yeast homologs of eIF4G contains a 115-amino-acid region that binds to Pab1p (23, 24). These 115-amino-acid fragments were individually fused to GST, expressed in E. coli, and immobilized on a glutathione-Sepharose resin. To define the region of Pab1p that is responsible for the interaction, each Pab1p variant, at a concentration of between 1.5 and 6 μM, was tested for its association with the immobilized eIF4G. As shown in Fig. 3 and Table 2, all Pab1p variants containing RRM2 were capable of associating with both eIF4G fragments, while the Pab1p variant lacking RRM2 failed to bind at all concentrations tested. None of the Pab1p variants bound to GST (data not shown). Another aspect of the interaction between Pab1p and eIF4G is the requirement for poly(A) binding by Pab1p. While the data shown in Fig. 3 and elsewhere (23, 24) are consistent with this requirement, a recent study (13) has reported that wheat Pab1p does not require poly(A) binding in order to interact with wheat eIF4F (the complex which contains eIF4G). Whether this discrepancy is due to actual differences between Pab1p and eIF4G from yeast and wheat has not yet been determined.
FIG. 3.
RRM2 is required for Pab1p binding to eIF4G. Glutathione resins containing the Pab1p-binding region of eIF4G1 (GST-eIF4G1/187-300p) (A) or eIF4G2 (GST-eIF4G2/200-315p) (B) were incubated with poly(A) and the indicated Pab1p variant. Eluates from these resins were then resolved by SDS-PAGE (12% gel) and visualized by Coomassie brilliant blue staining. Initial concentrations for the Pab1p variants in the binding reaction were 1.5 μM except for Pab1-ΔRRM1p and Pab1-105p, which were at 3 μM. Poly(A) was used at a concentration of 58 μM AMP. WT, wild type.
Of particular note, Pab1-105p, which contains only the two N-terminal RRMs, also interacts with eIF4G (Fig. 3). This observation partially confirms the assignment of RRM2 of Pab1p as directly interacting with eIF4G. However, a recombinant fragment of Pab1p containing only RRM2 was unable to bind to the eIF4G fragments (data not shown). This inability may be due to a very weak poly(A)-binding activity of the RRM2 protein [Kd for oligo(A)20 of >10 μM (data not shown)]. In summary, these eIF4G binding data indicate that the eIF4G-binding site of Pab1p includes RRM2. They also indicate that this region of Pab1p interacts with both eIF4G1 and eIF4G2. Finally, the inefficient binding of Pab1p lacking RRM1 to eIF4G suggests that this region may also contribute to eIF4G binding.
RRM2 of Pab1p is also required for the high-affinity, specific binding of Pab1p to poly(A) (3, 5, 12, 17). This could suggest that the RRM2 requirement for Pab1p binding to eIF4G stems solely from the poly(A) recognition activity of this domain and that this binding enables another domain of Pab1p to bind to eIF4G. However, no other domain of Pab1p is indispensable for eIF4G binding in our assays, indicating that some feature of RRM2 of Pab1p is required for contacting eIF4G. Another argument holds that binding to eIF4G merely requires high affinity (i.e., Kd of <150 nM) for oligo(A)20. Thus, all single-domain deletions other than Pab1-ΔRRM2p can bind to eIF4G. However, this seems unlikely because the Pab1-105p variant, which displays even lower affinity for oligo(A)20 than Pab1-ΔRRM2p (Table 2), still efficiently associates with eIF4G.
Pab1p domain requirements for poly(A) tail-dependent translation in vitro.
With the identification of RRM2 of Pab1p as being required for binding to eIF4G, it was important to determine whether this domain, and perhaps others, was necessary for the stimulation of translation in vitro. To test the Pab1p deletion variants for their ability to mediate poly(A) tail-dependent translation, a varied approach was taken. The three assays reported here are all based on a method that uses yeast extracts and a luciferase reporter system (10). In these extracts, it was shown that the poly(A) tail has a stimulatory activity independent of the 5′ cap structure and that these two structures can give rise to synergistic effects (10, 22). Moreover, these two effects are dependent on Pab1p (22, 24) and the Pab1p-binding site on eIF4G (24). Initially, we tested for the ability of the Pab1p deletion variants to rescue translation of uncapped, polyadenylated (LUCpA) mRNA in an immunoneutralized wild-type extract. Then, both poly(A) tail-dependent translation and the cap-poly(A) tail synergism were analyzed in extracts prepared from strains harboring each of the Pab1p variants. While these three tests are all based on published procedures, slight variations were used and will be discussed when relevant.
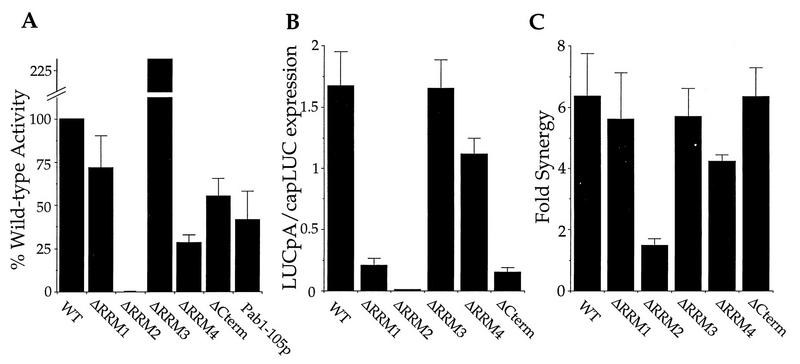
The initial experiments with immunoneutralized extracts proved to be a useful screening procedure for the activity of each of the Pab1p variants. These tests enabled the use of a single wild-type extract that was treated identically for all trials. Following a brief nuclease treatment to rid the extract of endogenous mRNA (see Materials and Methods), the endogenous Pab1p was neutralized with a monoclonal antibody (2, 22) whose epitope is within RRM2 (data not shown). Upon addition of recombinant full-length Pab1p, translation of LUCpA mRNA was stimulated to levels approximately 40% of the level in a nonneutralized extract (reference 22 and data not shown). The Pab1p deletion variants that bound to eIF4G all displayed activation to levels within 30 to 70% of that for the full-length recombinant version (Fig. 4A; Table 2). In contrast, Pab1-ΔRRM2p, which does not bind to eIF4G, was inactive in this assay. This result is consistent with the requirement for the interaction of Pab1p with eIF4G for poly(A) tail-dependent translation. The observed activities in these tests were due to the recombinant proteins (and not titration of the antibody off of the endogenous Pab1p), as one Pab1p variant, Pab1-16p, was recognized by the antibody (data not shown) but exhibited no activity in this assay (see below). In addition, it has been previously shown that immunodepleted extracts are also rescued by recombinant Pab1p (22).
FIG. 4.
RRM2 is required for poly(A) tail-dependent translation in vitro. (A) Reconstitution of translation in Pab1p immunoneutralized extracts with the recombinant Pab1p variants and LUCpA mRNA. The percentage of reconstitution, relative to the wild type (WT), achieved by the addition of the indicated Pab1p variants to the immunoneutralized extract is plotted on the y axis. Values given are the averages of multiple experiments with three different extracts. Each protein was tested over a range of concentrations, and the maximal activation value was used to represent the percent reconstitution. On average, a nonneutralized extract gave 150 U of luminescence in the absence of added protein, and a neutralized extract gave 2.2 U of luminescence. (B) Poly(A) tail-dependent translation in extracts containing different Pab1p variants. Extracts from yeast strains harboring the indicated Pab1p were prepared and assayed for the indicated LUC mRNA translation. Values on the y axis represent the ratio of LUCpA translation to capLUC translation. The translation of capLUC mRNA serves as an internal standard to control for variation in the overall translational activity of each extract. (C) Synergistic activation of translation in extracts containing different Pab1p variants. The ratio of the amount of translation of capLUCpA mRNA to the sum of capLUC and LUCpA mRNA translation [capLUCpA/(capLUC + LUCpA)] within the indicated extract is plotted on the y axis. For panels B and C, the plotted ratios represent the average of at least two experiments with each of two independently prepared extracts. Representative luminescence values for capLUC mRNA translation in each extract were as follows: WT, 39.8; ΔRRM1, 20.3; ΔRRM2, 23.4; ΔRRM3, 79.8; ΔRRM4, 29.4; and ΔCterm, 5.8.
While the results presented above are consistent with our hypothesis that binding to eIF4G by Pab1p is a prerequisite for poly(A) tail-dependent translation, we wanted to be sure that no artifacts resulted from the use of the anti-Pab1p antibody and recombinant Pab1 protein. Therefore, we prepared extracts from cells containing each of the Pab1p deletion variants in place of the wild-type Pab1p. These extracts allowed for the analysis of the translational activity of these proteins in their own native-like environments. Furthermore, since strains harboring Pab1p deleted for RRM2 are viable (see Table 2 for growth rates), it was important to determine whether extracts from such strains were capable of mediating poly(A) tail-dependent translation in vitro.
These new extracts afforded us the ability to test for both stimulation of translation of LUCpA mRNA as well as capLUCpA (capped and polyadenylated) mRNA. These two tests required neither the antibody nor recombinant protein. Furthermore, we found that nuclease treatment was not beneficial. In fact, while qualitatively similar results were achieved with nuclease treatment (data not shown), enhanced synergistic effects were observed without such treatment. The nuclease treatment, however, has proved to be required for the efficient reconstitution by Pab1p of Pab1p-immunoneutralized extracts.
As shown in Fig. 4B and summarized in Table 2, translation was stimulated by the poly(A) tail only in extracts prepared from strains producing Pab1p containing RRM2. This effect is most clearly exemplified by comparing the ratios of translational activity of capped, unadenylated (capLUC) mRNA to that of LUCpA mRNA. The use of capped transcripts provided an internal standard for each extract by which to judge the tail-dependent effects. To address the possibility that the extract harboring Pab1-ΔRRM2 is lacking some factor that responds to the Pab1p-poly(A) signal, recombinant Pab1p was added to these extracts. This addition was found to specifically rescue translation of LUCpA and not unadenylated mRNA (data not shown), suggesting that this extract is not deficient in something other than the Pab1p activity. We did observe differences in the ability of individual extracts to translate LUCpA versus capLUC mRNA. In particular, extracts containing the Pab1-ΔRRM1 and Pab1-ΔCterm proteins had 7- and 10-fold reductions in their ability to translate LUCpA relative to wild-type extracts (Fig. 4B; Table 2). The basis for these differences in the translational activities among the extracts awaits further investigation. Nonetheless, the present data strongly support the notion that RRM2 of Pab1p is required for its translational activity.
Another measure of Pab1p-dependent translation is the measure of synergy between the 5′ cap and poly(A) tail. Previously, it has been shown that the presence of both a cap and a tail leads to synergistic effects on translation in vitro (10, 22). Furthermore, this synergy is dependent on Pab1p (22) and on its ability to interact with eIF4G (24). Synergy is measured as the ratio of translation of capLUCpA mRNA to the sum of the translations of capLUC and LUCpA mRNAs. A ratio of 1.0 indicates that there is no synergy between these two terminal mRNA structures. Again, we observed a role for RRM2 of Pab1p in translational synergy since only the extract containing Pab1-ΔRRM2p failed to exhibit significant synergy (Fig. 4C; Table 2). The value for synergy in the Pab1-ΔRRM2p extract slightly exceeds 1.0, perhaps due to a slight increase in the functional half-life of capLUCpA over LUCpA and capLUC mRNAs (21a). As with the poly(A) tail-dependent effects, it is likely that the ability of Pab1p to interact with eIF4G via RRM2 is a prerequisite for the cooperative effects of the cap and tail.
Binding to eIF4G is not sufficient for Pab1p-dependent translation.
Having established that both the binding of Pab1p to eIF4G and the activation of poly(A) tail-dependent translation by Pab1p required RRM2, we evaluated the Pab1-16 protein. This protein contains the following substitutions within the highly conserved RNP1 motif of RRM1 and RRM2: Y83V and F170V, respectively (5). These substitutions result in an approximately 100-fold decrease in the affinity of the recombinant Pab1-16 protein for oligo(A)20 (5). Cells harboring Pab1-16p grow more slowly than a wild-type strain and become inviable when the eIF4E gene CDC33 is also mutated (24).
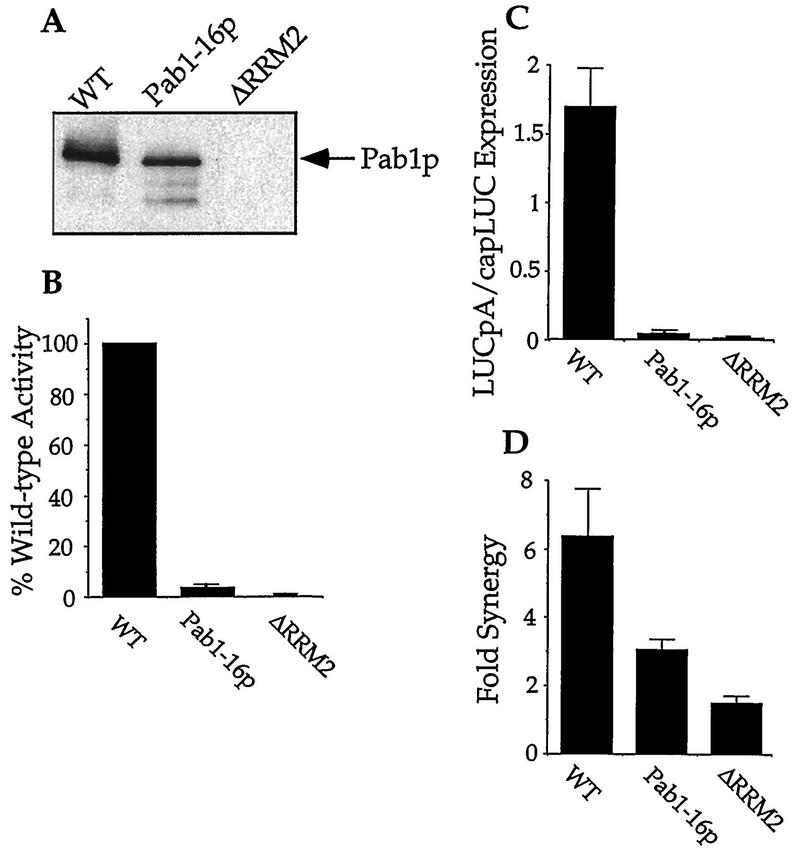
Recombinant Pab1-16p did associate with the Pab1p-binding site of eIF4G2 (Fig. 5A) and eIF4G1 in an RNA-dependent manner (data not shown). However, this protein was unable to restore poly(A) tail-dependent translation in immunoneutralized extracts (Fig. 5B). Furthermore, extracts from cells containing Pab1-16p exhibited nearly undetectable levels of LUCpA mRNA translation (Fig. 5C) and only intermediate levels of synergy with the cap structure (Fig. 5D). This intermediate value for synergy probably indicates that these point mutations did not completely inactivate the translational activity of the protein, as Pab1-16p still associated with eIF4G. Similarly, partial loss-of-function mutations in the Pab1p-binding site on eIF4G result in the loss of translation of LUCpA mRNA without destroying the synergism between the cap and the poly(A) tail (24). Each of these observations on the properties of Pab1-16p suggests that while the ability of Pab1p to associate with eIF4G is a prerequisite for translational stimulation, there are probably other regions of Pab1p and/or eIF4G that respond to this binding event and then stimulate translation. We cannot, however, rule out at this time that Pab1-16p has a decreased affinity for eIF4G in the extracts. Therefore, its deficiencies could result from inefficient eIF4G binding.
FIG. 5.
The Pab1-16 protein associates with eIF4G but does not activate poly(A) tail-dependent translation. (A) Pab1-16p associates with eIF4G. Glutathione resin containing the GST-eIF4G2/200-315p fusion protein was incubated with poly(A) and the indicated Pab1 protein. Eluates from these resins were then resolved by SDS-PAGE (10% gel). The Pab1 proteins were visualized by Western analysis with a Pab1p polyclonal antibody. The initial concentration for Pab1-16p in the binding reaction was 3 μM; Pab1-1p and Pab1-ΔRRM2p were at 1.5 μM. Poly(A) was used at a concentration of 58 μM AMP. WT, wild type. (B) Pab1-16p fails to reconstitute Pab1p-dependent activation of LUCpA translation. Immunoneutralized extracts were supplemented with the indicated Pab1p and then assayed for their translation of LUCpA mRNA. (C) Extracts containing Pab1-16p do not support LUCpA mRNA translation. Translation extracts from strains harboring the indicated Pab1 protein were programmed with either LUCpA or capLUC mRNA. The ratio of the translation of these two mRNAs is plotted. (D) Extracts containing Pab1-16p exhibit decreased levels of translational synergy. The ratio of the amount of translation of capLUCpA to the sum of capLUC and LUCpA mRNA translation within the indicated extract is plotted on the y axis. A representative of the values of capLUC mRNA translation in the Pab1-16p extracts used for panels C and D was 15.6. (See the legend to Fig. 4 for details of panels B to D.)
DISCUSSION
It has previously been shown that Pab1p from yeast can interact with the 5′ cap-binding complex eIF4F, which contains eIF4G (23, 24). Here, we extend this finding by showing that this function requires RRM2 of Pab1p. The functional consequences of the interaction between RRM2 and eIF4G are demonstrated by the need for RRM2 in poly(A) tail-dependent translation in yeast extracts. Our analysis of the Pab1-16p mutation also reveals that while association of Pab1p with eIF4G is a prerequisite for translational stimulation, other features of this interaction may also be required.
The most obvious finding of this study is the requirement for RRM2 of Pab1p in mediating binding to eIF4G and in stimulating poly(A) tail-dependent translation in vitro. What is less clear is the role of the other domains of Pab1p, which include three RRMs and a 17-kDa C-terminal region. Our in vitro translation data could suggest that RRM1 and RRM4 and the C-terminal region of Pab1p are also involved in mediating the Pab1p-poly(A) tail translation function. Specifically, we found that deletion of RRM4 had an effect in the reconstitution assay, while deletion of RRM1 had a significant effect in extracts prepared from strains containing this Pab1p mutant (compare Fig. 4A and B). Yeast RRM4 has been previously shown to have significant RNA-binding activity (5), and this activity may contribute to the translation function of the protein. As a result of its proximity to RRM2, RRM1 may provide structural support to RRM2 and thereby enhance the function of this latter domain. The C-terminal region of Xenopus Pab1p has been suggested to mediate Pab1p dimerization (12), and this could also serve to heighten the translation function of Pab1p. Further experiments will be needed in order to understand how other regions of Pab1p serve to enhance the translational function of RRM2.
We are unable to conclude from our data that RRM2 directly contacts eIF4G because of our inability to demonstrate an interaction between the isolated RRM2 and eIF4G. One possible explanation for this observation is that an isolated RRM2 lacks structural integrity. Consistent with this proposal, we have been unable to observe significant poly(A) binding by this domain in isolation (data not shown), which presumably prevents its RNA-dependent interaction with eIF4G. The minimal Pab1p which we could show has both eIF4G-binding activity and translational activity (Pab1-105p) contains both RRM1 and RRM2. Thus, it is possible that additional essential stabilizing contacts with RRM2 are made through RRM1. Because RRM1 can be singly deleted from Pab1p without destroying the interaction with eIF4G and Pab1p-dependent translation, we assume that these proposed stabilizing contacts can also be supplied by the other Pab1p RRMs.
Why does Pab1p require poly(A) in order for it to interact with eIF4G? RNA binding may place Pab1p in an appropriate conformation to bind to eIF4G. Alternatively, Pab1p may place the poly(A) into a conformation suitable for contacting a latent eIF4G RNA-binding site. The eIF4G protein may also contact both Pab1p and poly(A). The requirement for a Pab1p-poly(A) complex for the interaction with eIF4G may exist so as to prevent association of either non-mRNA-associated Pab1p or naked poly(A) with eIF4G. This would create a more stringent and specific requirement for Pab1p-dependent activation of translation.
An exciting yet unexpected result was provided by the analysis of Pab1-16p. This protein was originally constructed with the goal of disrupting its RNA-binding activity (5), but our investigation has now suggested an additional role for its altered residues in Pab1p-dependent translation. This protein exhibits a reduction in the affinity for poly(A) RNA (5) yet still interacts with eIF4G (Fig. 5A). However, this protein is incapable of stimulating the translation of uncapped, polyadenylated (LUCpA) mRNA in vitro (Fig. 5B and C). This result indicates that a simple binding event is insufficient for mediating poly(A) tail-dependent translation. The nature of the defect of Pab1-16p will require further investigation. As mentioned above, the possibility that Pab1-16p has a decreased affinity for eIF4G will be pursued. The deficiency is unlikely to be due to the reduction in affinity for poly(A), as Pab1-105p has a similar affinity yet still activates translation in vitro. This finding is also interesting in light of genetic analysis of pab1-16. This allele exhibits synthetic lethality with cdc33-1 (24), which is a mutant allele of the gene encoding eIF4E. In the latter mutant, cap-dependent translation is compromised. Thus, a possible reason for the observed synthetic lethality is the deleterious effects of losing or reducing both cap-dependent and poly(A) tail-dependent translation in vivo.
Future analysis of the interaction between Pab1p and eIF4G will involve site-directed mutagenesis of Pab1p. This approach will allow for the identification of the amino acid side chains that contact eIF4G, thus more precisely defining the interaction surface. It should also identify other residues within Pab1p that are dispensable for association with eIF4G but are required for translational stimulation. Site-directed mutagenesis, which has been used to examine the RNA binding of Pab1p (5), should alleviate any potential structural perturbations caused by the deletion of the ∼90-amino-acid RRMs and thus create fewer ambiguities in the data interpretation. Nevertheless, the deletion analysis of Pab1p presented here has given us the ability to define a region of the protein that is involved in translation initiation in vitro and has therefore provided us with the region of the protein which will be subject to more intensive investigation.
ACKNOWLEDGMENTS
We thank members of the Sachs laboratory for thoughtful comments on the manuscript.
This work was supported by grants to A.B.S. from the American Cancer Society (82666) and the Hellman Family Fund.
REFERENCES
- 1.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J, Paddy M, Swanson M. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1993;136:102–112. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burd C G, Matunis E L, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deardorff J A, Sachs A B. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 6.Gallie D R. The cap and the poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 7.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 8.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:302–318. [PubMed] [Google Scholar]
- 9.Huang Y, Carmichael G C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 12.Kuhn U, Pieler T. Xenopus poly(A) binding protein:functional domains in RNA binding and protein-protein interactions. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 13.Le H, Tanguay R L, Balasta M L, Wei C C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factor eIF-iso4G and eIF4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 14.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M, Sonenberg N, editors. Translational control. Vol. 30. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 15.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translation initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai K, Oubridge C, Ito N, Avis J, Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 17.Nietfeld W, Mentzel H, Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachs A B, Bond M W, Kornberg R D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 19.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 20.Sachs A B, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 21.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eucaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 21a.Tarun, S., and A. B. Sachs. Unpublished data.
- 22.Tarun S, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 23.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 24.Tarun S Z, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF-4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]