Abstract
αPAK in a constitutively active form can exert morphological effects (E. Manser, H.-Y. Huang, T.-H. Loo, X.-Q. Chen, J.-M. Dong, T. Leung, and L. Lim, Mol. Cell. Biol. 17:1129–1143, 1997) resembling those of Cdc42G12V. PAK family kinases, conserved from yeasts to humans, are directly activated by Cdc42 or Rac1 through interaction with a conserved N-terminal motif (corresponding to residues 71 to 137 in αPAK). αPAK mutants with substitutions in this motif that resulted in severely reduced Cdc42 binding can be recruited normally to Cdc42G12V-driven focal complexes. Mutation of residues in the C-terminal portion of the motif (residues 101 to 137), though not affecting Cdc42 binding, produced a constitutively active kinase, suggesting this to be a negative regulatory region. Indeed, a 67-residue polypeptide encoding αPAK83-149 potently inhibited GTPγS-bound Cdc42-mediated kinase activation of both αPAK and βPAK. Coexpression of this PAK inhibitor with Cdc42G12V prevented the formation of peripheral actin microspikes and associated loss of stress fibers normally induced by the p21. Coexpression of PAK inhibitor with Rac1G12V also prevented loss of stress fibers but not ruffling induced by the p21. Coexpression of αPAK83-149 completely blocked the phenotypic effects of hyperactive αPAKL107F in promoting dissolution of focal adhesions and actin stress fibers. These results, coupled with previous observations with constitutively active PAK, demonstrate that these kinases play an important role downstream of Cdc42 and Rac1 in cytoskeletal reorganization.
The small GTP-binding proteins of the Rho subfamily, in particular the ubiquitous Rho, Rac, and Cdc42 proteins, act through a variety of downstream targets which bind to the GTP forms of these p21s (reviewed in references 27 and 54). RhoA signalling is required for maintenance of actin stress fibers and focal adhesions in cultured mammalian cells (46), these activities being mediated by Rho-associated kinases (ROKs) (13, 24, 25, 37). Rac activation produces lamellipodia or membrane ruffles and associated peripheral focal complexes (FCs) perhaps by binding POR1 (47, 56). Cdc42 promotes formation of peripheral actin microspikes, which are structural components of filopodia and retraction fibers, followed by its activation of Rac (19, 39). Cdc42 can antagonize Rho (20, 34), while Rac can promote leukotriene-mediated activation of Rho (43). In addition to their roles in cell morphology, Cdc42, Rac, and Rho participate in regulating transcription both through JNK/stress-activated protein kinase (SAPK)- and p38 mitogen-activated protein (MAP) kinase-regulated pathways (8, 38). Activated forms of these p21s can also stimulate cell cycle progression to promote DNA synthesis in fibroblasts (41). In the yeast Saccharomyces cerevisiae, Cdc42p is required for the control of cell polarity during budding and pseudohyphal growth (28, 62) but plays a role in transcriptional activation via the mating-response MAP kinase pathway (51, 61). Both budding and pheromone-induced signalling in yeast require participation of the PAK-like kinases Cla4p and Ste20p (22, 23).
Mammalian targets of Cdc42 and Rac1 include a number of recently identified proteins, of which the best characterized are PAK kinases (27, 49). Upon binding to either GTP-bound Cdc42 (GTP-Cdc42) or GTP-Rac, PAK undergoes autophosphorylation on multiple sites and is activated (32, 34, 36). PAK function in vivo probably includes activation of JNK/SAPK and p38 MAP kinase pathways (2, 45). Since Rho-p21s are closely linked to cytoskeletal reorganization, the ubiquitously expressed PAKs (32, 36) have been candidate effectors in mediating certain aspects of morphology regulated by Cdc42 and Rac. Expression of constitutively active forms of αPAK causes drastic loss of actin stress fibers and focal adhesions with retraction of the cell periphery, consistent with PAK acting downstream of Cdc42 and Rac (34). Microinjected PAK1 protein has also been shown to promote FC formation and membrane ruffling through N-terminal (nonkinase) interactions (50). The control of the cellular activities of PAK containing various functional domains appears to be complex; p21 activation of PAK is not the only mode of regulation, as capsase-mediated cleavage of PAK2 can generate a catalytically active fragment which regulates morphological changes associated with apoptosis (48).
Comparison of the primary sequences of divergent PAKs revealed two highly conserved functional domains: the serine/threonine kinase domain at the C terminus and a region that includes the p21-binding domain toward the N terminus of the kinase. In this study, we used site-directed mutagenesis to determine that while the N-terminal half of this region binds p21, the C-terminal half is involved in negatively regulating kinase activity. A PAK inhibitor sequence was derived from the conserved region by deletion analysis and then used to analyze the morphological activities of endogenous PAK. Cytoskeletal reorganization in cells expressing Cdc42G12V or RacG12V are markedly affected when the PAK inhibitor is coexpressed, demonstrating that endogenous PAKs can indeed play defined morphological roles downstream of these p21s.
MATERIALS AND METHODS
PAK subcloning and site-directed mutagenesis.
Mutations to the αPAK N-terminal region were constructed by a PCR-based mutagenesis procedure. The 0.75-kb cDNA fragment encoding αPAK1-250 was amplified in two parts with desired mutations introduced at the junction from one PCR primer. The two primers at the central position (∼300 bp from BamHI) were phosphorylated prior to the PCRs to facilitate ligation. The two PCR products were joined with T4 DNA ligase, and the complete mutant 0.75-kb cDNA fragment was reamplified by PCR using PAK complementary primers at the 5′ end (ATGGATCCCGGGATCCATGTCAAATAACGGC; BamHI site underlined) and at the 3′ end, corresponding to the position of an internal BglII site (CCGCTCGAGCTAAGATCTCCTCATCAGACA; XhoI and BglII sites underlined). PCR products were subcloned into the BamHI and XhoI sites of the Bluescript SK(+) vector and sequenced.
The polypeptides encoding αPAK residues 83 to 131, 83 to 149, 89 to 131, and 89 to 149 were expressed from cDNAs amplified by PCR and introduced into pGEX-4T-1 (BamHI and XhoI sites). αPAK83-149 was introduced into the BamHI and XhoI sites in the mammalian pXJ hemagglutinin (HA)-epitope-tagged vectors (34). The primers used to generate PAK mutants and truncation constructs are listed in Table 1.
TABLE 1.
Mutagenesis primers
| Oligonucleotide | Sequencea | Mutation |
|---|---|---|
| M1 | 5′AAGAGAAtTCTCGTGCCGCTC3′ | I→N |
| M2 | 5′AAGAGgAATCTCGTGCCGCTC3′ | S→P |
| M3 | 5′gCTTCAGATTTTGAGCATAC3′ | P→A |
| M4 | 5′CCTTCAGATTTTGAGCtTACAATTCtTGTTGGTTTTG3′ | HH→LL |
| M5 | 5′AAGCGGGCCCACTGTTCTGGCATCaCCGTAAAC3′ | G→V |
| M6 | 5′AAGCGGGCCCACTGTTCTGGCtTCCCCGTAAAC3′ | M→K |
| M7 | 5′TTTTTCTTCTtCTtTGACTTGG3′ | EQ→KK |
| M8 | 5′gTTccCTTCTGCTCTGACTTG3′ | KN→GG |
| M9 | 5′CCCACAGGCTGTTCTGcgTGTGT3′ | D→R |
| M10 | 5′CCCACtGGaTGTTCTGGATGTG3′ | QA→LD |
| M11 | 5′CCCACAGGCTGTTCTGGATGTGTTGGAAaaTcATAACTCC3′ | FY→NH |
| Iljz21 | 5′CCTTCAGATTTTGAGCATA3′ | Wild type |
| Iljz24 | 5′AAGAGAAATCTCGTGCCGCTC3′ | Wild type |
| Iljz29 | 5′TTTTTCTTCTGCTCTGACTT3′ | Wild type |
Mutated bases are in lowercase.
Bacterial expression and purification of recombinant PAK proteins.
The plasmid expressing the glutathione S-transferase (GST)/PAK1-250 fusion proteins of each mutant as well as the wild type were constructed by cloning the 750-bp BamHI-XhoI fragments into pGEX-4T-1 (Pharmacia). The full-length PAK mutants were then constructed by adding the BglII-XhoI cDNA fragment encoding αPAK251-544 into the GST/αPAK1-250 mutant plasmids. pGEX plasmids were transformed into Escherichia coli BL21 for protein expression. Recombinant GST fusion proteins from 200-ml cultures were purified by glutathione-Sepharose affinity chromatography (Pharmacia) in a 300-μl column of glutathione-Sepharose. Eluted proteins were stored with 5% glycerol at −70°C.
Expression and purification of GST/αPAK from COS-7 cells.
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. The pXJ-GST mammalian cell expression vector was constructed by replacing the pXJ-HA epitope tag (34) with the coding sequence of GST (amplified by PCR as an EcoRI/BamHI fragment). The 1.6-kb BamHI-XhoI fragment containing the coding region of αPAK (33) was cloned into the BamHI/XhoI sites of pXJ-GST. COS-7 cells were transfected with 5 μg of pXJ-GST vectors by the Lipofectamine (Bethesda Research Laboratories) method. The cells were harvested after 18 h and lysed in buffer containing 40 mM HEPES (pH 7.5), 1% Nonidet P-40, 100 mM NaCl, 1 mM EDTA, 25 mM NaF, 1 mM sodium orthovanadate, and 10 μg each of leupeptin and aprotinin/ml. After centrifugation (12,000 × g) for 10 min at 4°C, supernatant fractions were passed through columns containing 50 μl of glutathione-Sepharose beads. Columns were washed with phosphate-buffered saline containing 50 mM Tris-HCl (pH 8.0) and 0.1% Triton X-100, and GST/PAK fusion proteins were eluted in the same buffer containing 10 mM glutathione and 5% glycerol.
Overlays with [γ-32P]GTP-Cdc42.
Purified GST fusion proteins (0.4 μg) were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 10% polyacrylamide gels and transferred to nitrocellulose membranes. The filter was blocked for 16 h (4°C) in phosphate-buffered saline containing 1% bovine serum albumin, 5 mM dithiothreitol, 0.5 mM MgCl2, and 0.1% Triton X-100 prior to p21 overlay analysis. Cdc42 labeling with [γ-32P]GTP and Western overlays were carried out as described previously (30). The [γ-32P]GTP-Cdc42 signals on the filter were detected at −20°C and quantified by PhosphorImager (Molecular Dynamics) analysis.
Protein kinase assays.
Each 50-μl reaction mix contained 0.5 μg of GST/αPAK, 10 μCi of [γ-33P]ATP (∼6,000 mCi/mmol), and 10 μg of myelin basic protein (MBP) in kinase assay buffer (50 mM HEPES [pH 7.3], 10 mM MgCl2, 2 mM MnCl2, 1 mM dithiothreitol, 0.05% Triton X-100). PAK activation was carried out with 4 μg of GTPγS-Cdc42. The mixture was incubated at 30°C for 30 min. Samples were resolved on 12% polyacrylamide gels and processed for autoradiography.
Microinjection and cell staining.
HeLa cells were cultured in minimal essential medium with 10% fetal bovine serum. Subconfluent cells were microinjected into the nucleus with 50 ng of each expression plasmid DNA/μl, using an Eppendorf microinjector. After 2 or 4 h, cells were fixed in 3% paraformaldehyde for 20 min and stained as described previously (34).
RESULTS
Flanking sequences influence the p21-binding CRIB motif of αPAK.
PAK sequences contain two functional domains, the C-terminal kinase domain and the p21-binding domain near the N terminus whose binding of Cdc42/Rac1 markedly stimulates autophosphorylation (32). In αPAK, a region encompassing residues 67 to 149 (the conserved PAK amino-terminal [PAN] motif) was first found to be functional as the Cdc42/Rac binding domain (32). Burbelo et al. (7) subsequently showed that among a larger family of Cdc42 and Rac1 binders, only a region (the core Cdc42-Rac interaction and binding [CRIB] motif) corresponding to αPAK75-87 shows significant conservation of amino acid residues. In spite of this, a βPAK29-90 construct containing all of these conserved residues (equivalent to αPAK34-95) exhibited extremely weak p21 binding (7).
In light of these observations, we investigated further the requirement for efficient p21 binding by performing [γ-32P]GTP-Cdc42 overlays on six deletion constructs containing portions of the N-terminal region of αPAK (Fig. 1A). For this purpose, cDNA constructs were bordered on the C-terminal sides by BstXI (position 101) and PvuII (position 149) restriction sites present in the αPAK cDNA (32). These E. coli-expressed polypeptides were purified as GST fusion proteins (Fig. 1B, left) and analyzed by using 10 pmol of proteins (Fig. 1B, right), which is within the range for a linear p21-binding response (55). All of these deletion constructs (but not GST) bound Cdc42 with lower affinity than full-length GST/αPAK (whose breakdown fragments also showed robust binding activity [Fig. 1B]). Deletion construct 1 (αPAK1-149) retained 70% of the Cdc42-binding activity of full-length αPAK (Fig. 1C, lane 2). Construct 2 (αPAK1-101), extending the C-terminal limits of the weakly binding construct described by Burbelo et al. (7), still bound GTP-Cdc42 threefold more weakly than αPAK1-149 (lane 3), indicating that residues 102 to 149 enhance binding affinity. Since levels of αPAK61-149 and PAK1-149 binding were similar (Fig. 1C, lane 3), we conclude that the first 60 residues in αPAK do not contribute to p21 binding. Deletion of a further 10 residues N terminal (71 to 149; construct 5) slightly decreased binding further. Surprisingly, αPAK69-149(P73H) exhibited binding eightfold lower than this (Fig. 1C, lanes 4 and 5) (the additional two residues in the mutant construct were added so as not to place the altered residue too close to the GST-PAK junction). Proline 73 is not present in yeast PAK-like kinases or in other CRIB motif-containing proteins (7). From these results, it is apparent that residues 102 to 149, in the region well beyond the CRIB motif (residues 75 to 87), contribute in some way to p21 binding. However, construct αPAK83-149, lacking part of this core binding sequence, is devoid of p21-binding activity (Fig. 4E), indicating that these flanking sequences have no intrinsic binding activity. Comparison of the N-terminal sequences in PAK-like kinases from mammals, invertebrates, and yeast showed that in the region corresponding to αPAK71-131, residues are uniformly conserved (Fig. 2A). This encompasses the CRIB motif and flanking C-terminal sequences; there is no apparent bias of conservation in the N-terminal CRIB motif. The C-terminal sequences are not found in other classes of Cdc42/Rac1 binding proteins, such as ACK, WASP, and MLK3 (1, 7, 31).
FIG. 1.
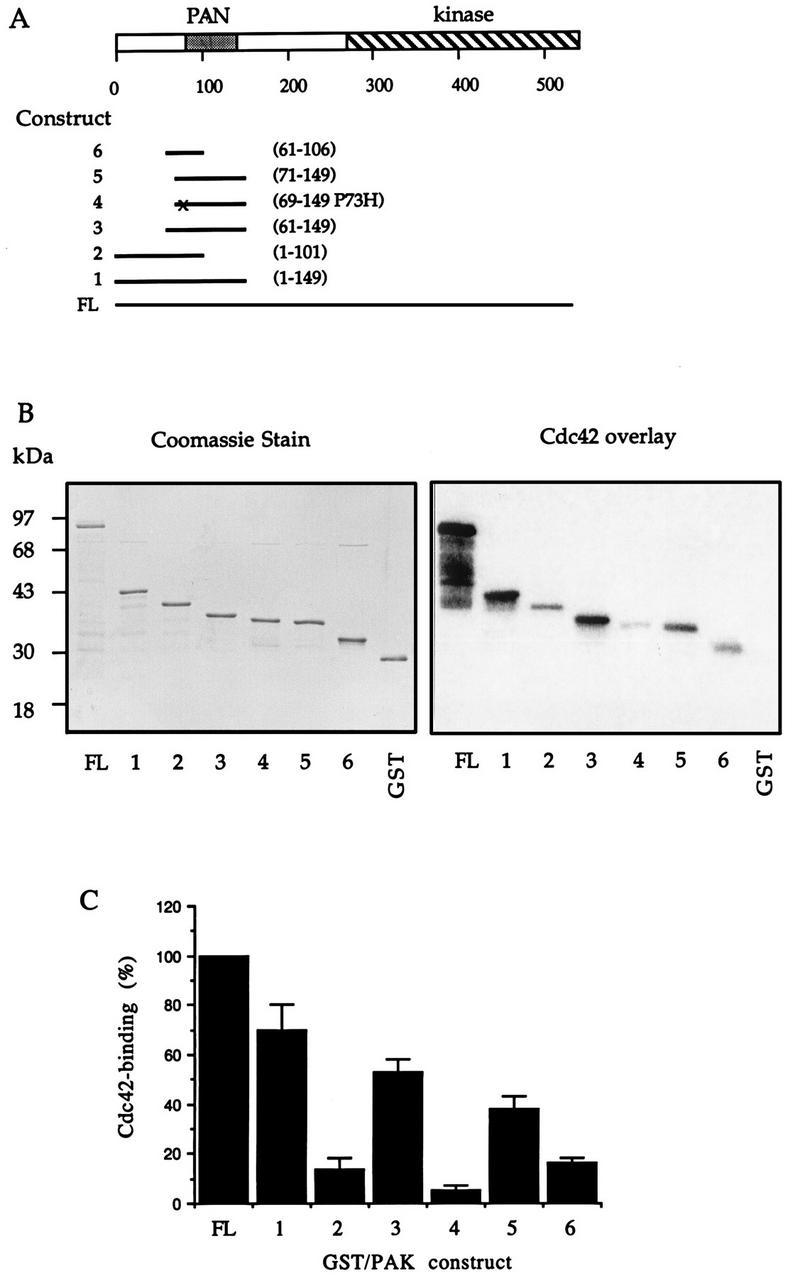
Sequences affecting efficiency of p21 binding to αPAK. (A) Schematic diagram representing regions of αPAK expressed and purified from E. coli. Amino acid numbers at the beginning and the end of each fragment are indicated on the right. The point mutation present in construct 4 is marked by “x.” FL, full length. (B) GST/αPAK constructs were expressed and purified as GST fusion proteins, and 1 μg of each protein was resolved by SDS-PAGE (12% gel) and stained with Coomassie brilliant blue (left); 10 pmol of each of these proteins was analyzed by an overlay binding assay using [γ-32P]GTP-Cdc42 (right). (C) The Cdc42-binding signals shown in panel B (right) were quantified on a PhosphorImager (Molecular Dynamics). The means of data from two independent experiments are shown.
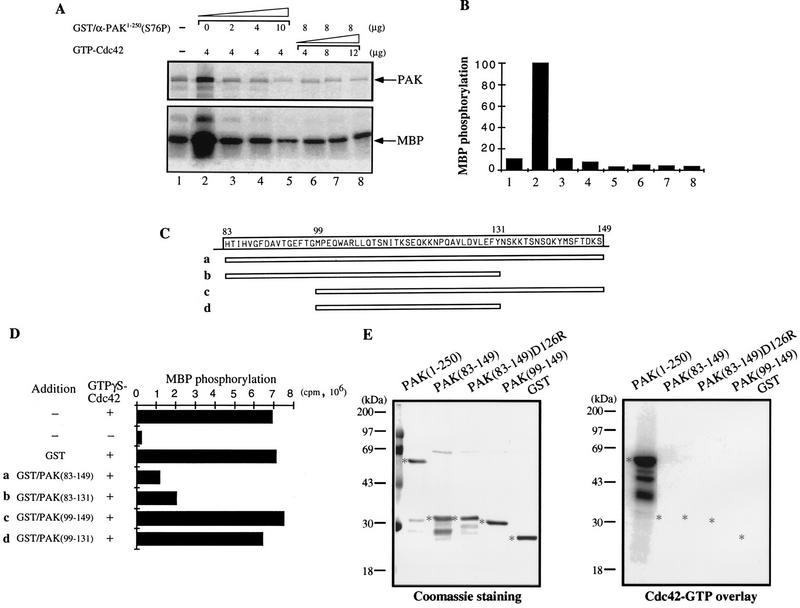
FIG. 4.
The C terminus of the PAN motif inhibits PAK activation by GTP-Cdc42. (A) The PAK N-terminal fusion protein GST/αPAK1-250(S76P) inhibits PAK activation by GTP-Cdc42. The kinase activity of bacterially expressed GST/αPAKL404S was assayed in the absence (lane 1) or presence (lane 2 to 8) of the indicated amounts of GTP-Cdc42. The autoradiograph shows inhibition due to the indicated amounts of GST/αPAK1-250(S76P) added during the kinase reactions. Signals of αPAK autophosphorylation and MBP phosphorylation are indicated by arrows. (B) Quantification of the MBP phosphorylation shown in panel A. (C) Schematic diagram of four peptides in the PAN motif region. The corresponding amino acid sequence is shown at the top. These peptides were expressed as GST fusion proteins. SDS-PAGE analysis of 1 μg of each purified protein showed single appropriately sized bands (not shown). (D) One microgram of bacterially expressed GST/αPAKL404S was assayed for kinase activity to MBP in the presence of excess GTP-Cdc42 (4 μg) and 4 μg of each inhibitory peptide. The MBP phosphorylation signals were quantified on a PhosphorImager. (E) One microgram each of GST/αPAK and GST proteins was resolved by SDS-PAGE (11% gel) and blotted onto a polyvinylidene difluoride membrane. The proteins were stained with Coomassie blue (left), and their p21 binding was analyzed by overlay with [γ-32P]GTP-Cdc42 (right). Asterisks indicate positions of protein bands.
FIG. 2.
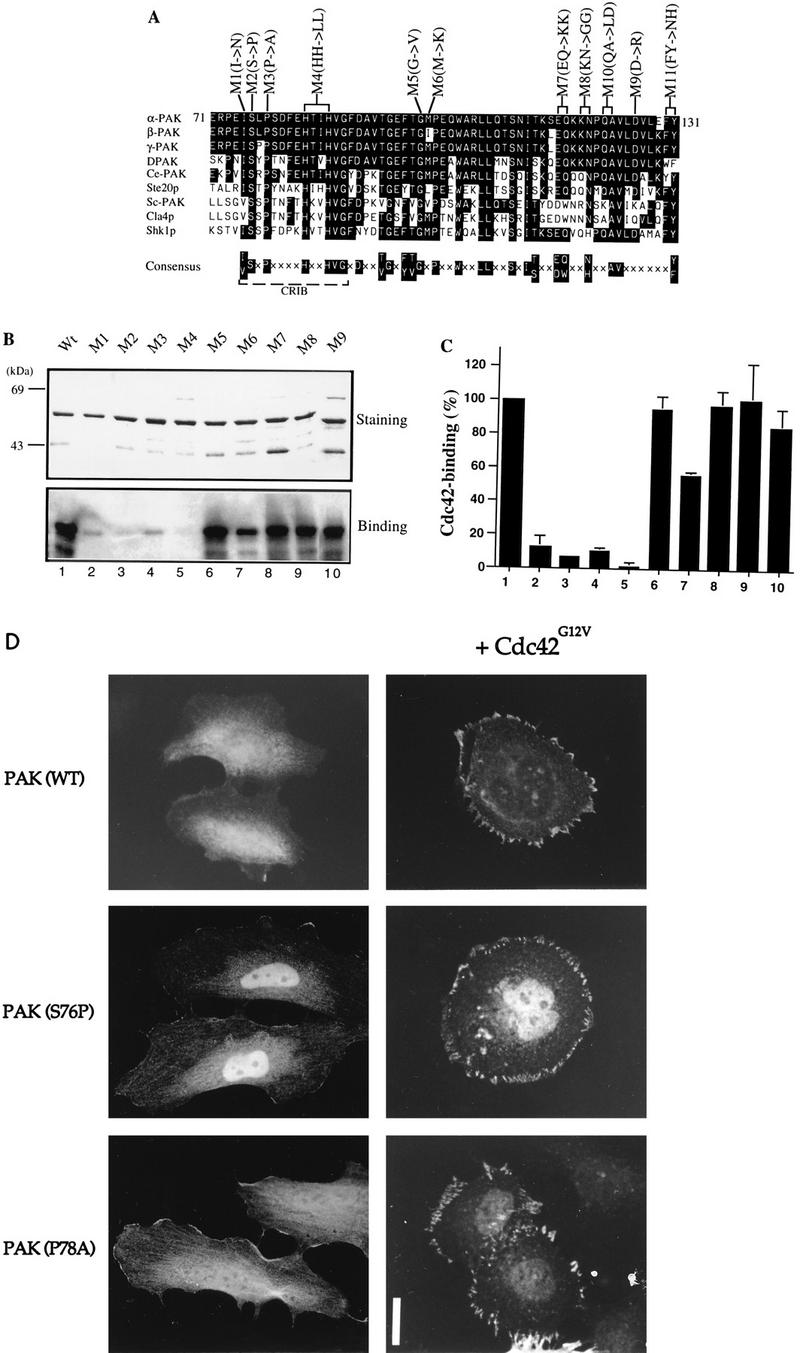
Mutations in the N terminus of the PAN motif abolish p21 binding. (A) Sequence alignment of PAN motifs of PAK-related proteins from rat (α-, β-, and γ-PAK), Drosophila (DPAK), Caenorhabditis elegans (Ce-PAK), S. cerevisiae (Ste20p, Cla4p, and Sc-PAK), and Schizosaccharomyces pombe (Shk1p). Accession numbers are given elsewhere (27). The conserved residues are boxed in black, and a consensus of these is shown below. Amino acid substitutions corresponding to each of the mutant constructs are shown. (B) The first 250 amino acids of each αPAK mutant and wild-type (Wt) construct were purified as GST fusion proteins, and 1 μg of each protein was resolved by SDS-PAGE (11% gel) and stained with Coomassie brilliant blue (top); p21 binding to bands containing 0.4 μg of each protein was determined by overlays with [γ-32P]GTP-Cdc42 (bottom). (C) The Cdc42 binding signals in panel B (bottom) were quantified on a PhosphorImager; the means of two independent experiments are shown. (D) Expression constructs encoding HA-αPAK mutants as indicated were transfected into HeLa cells alone or with FLAG-Cdc42G12V. Typical cells stained for αPAK are shown; in all cases, the cells were also stained with antipaxillin to confirm that peripherally located PAK was in FCs. Bar, 10 μm.
Amino acid substitutions that affect p21 binding do not prevent PAK recruitment to Cdc42-driven FCs.
To investigate the function of the conserved residues in the PAN motif, we introduced amino acid substitutions at beginning, middle, and end positions indicated in Fig. 2A. To analyze their p21-binding activities, residues 1 to 250 of each mutant were purified and subjected to Western overlay with [γ-32P]GTP-labeled Cdc42 (Fig. 2B); mutants M10 and M11 could not be recovered as stable GST fusion proteins. As shown in Fig. 2C, substitutions of conserved residues in the N-terminal portion of the PAN motif, M1 (I75N), M2 (S76P), M3 (P78A), and M4 (H83/86L), caused drastic loss of p21 binding; M6 (M100K) also showed reduced binding, but the C-terminal mutants had normal p21 binding. These amino acid substitution results concur with previously published data (7) showing that the N-terminal portion of the PAN motif functions as the core p21-binding region.
Recent reports that Cdc42-binding-deficient Ste20p mutants are functionally competent (23, 44) led us to consider whether the cellular activities of PAK could occur without its directly binding Cdc42. The normally cytoplasmic wild-type kinase is translocated to FCs by activated Cdc42 or Rac1 (34). We therefore tested whether Cdc42G12V could relocalize the p21-binding-deficient PAK mutants described above. αPAK mutant M4 was not investigated due to its partially activated state (see Fig. 4 and references 48 and 50). As shown in Fig. 2D, wild-type αPAK and the αPAK mutants M2 (S76P) and M3 (P78A), which were distributed throughout the cytoplasm and nucleus, became associated with peripheral FCs in a manner indistinguishable from that of wild-type αPAK when coexpressed with Cdc42G12V. These kinases were not activated by Cdc42G12V when coexpressed in COS-7 cells (data not shown) or by GTPγS-Cdc42 when tested in vitro (Fig. 3). Thus, PAK recruitment to FCs can be independent of direct association with Cdc42.
FIG. 3.
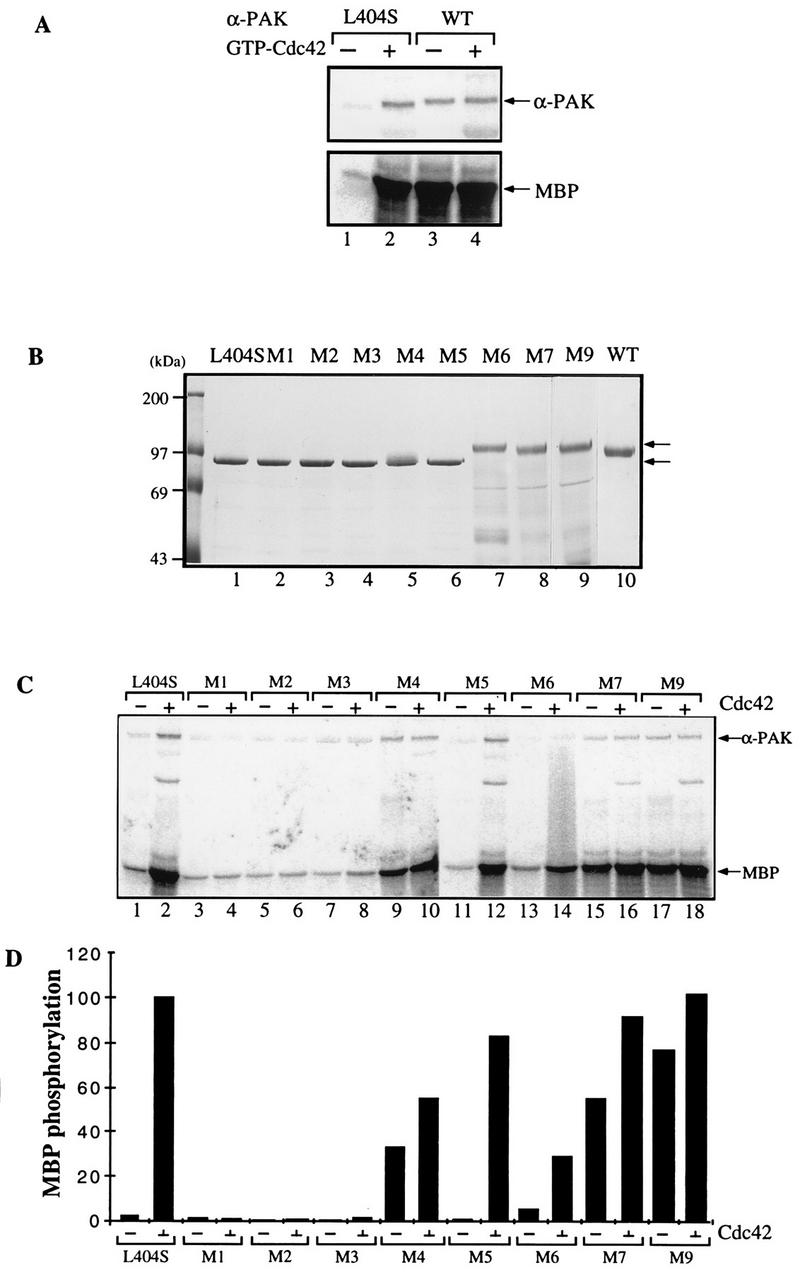
Mutations in the C-terminal portion of the PAN motif activate αPAK. (A) Activity of bacterially expressed GST/αPAK and GST/αPAKL404S in the presence or absence of GTP-Cdc42. Purified GST/αPAK or GST/αPAKL404S was assayed for kinase activity with MBP at 20 μM [γ-33P]ATP in the presence or absence of GTP-Cdc42. The kinase reaction was carried out at 30°C for 30 min. WT, wild type. (B) The GST/αPAKL404S mutants containing additional mutations in the PAN motif were expressed and purified from E. coli. The GST/PAK bands (arrows) correspond to 1 μg of the purified proteins stained with Coomassie brilliant blue. (C) Proteins shown in panel B were assayed for kinase activity to MBP in the presence or absence of GTP-Cdc42. Arrows indicate positions of autophosphorylated PAK and phosphorylated MBP bands. (D) Quantification of the MBP phosphorylation shown in panel C.
The C-terminal portion of the PAN motif may negatively regulate the kinase.
The wild-type αPAK is toxic in E. coli, probably due to its constitutive kinase activity in this bacterium (34). We were able to obtain, by serendipitous in vivo selection, an attenuated form of αPAK where leucine 404 is replaced by serine; this mutant exhibited low intrinsic kinase activity when purified as a GST fusion protein and was activated in vitro by GTP-bound Cdc42 or Rac1, albeit with slower than wild-type activation kinetics (33). A comparison of purified recombinant wild-type αPAK with αPAKL404S is shown in Fig. 3A. This αPAKL404S enabled us to assess the effects of additional mutations on kinase activation.
Those mutants that could be expressed as full-length GST/αPAKL404S fusion proteins were purified and analyzed on SDS–8% polyacrylamide gels (Fig. 3B). Mutants M1, M2, M3, and M5 showed the same electrophoretic migration as the inactive αPAKL404S (lane 1); part of the M4 protein appeared as a higher band (lane 5). By contrast, M6, M7, and M9, with mutations in the C-terminal portion of the PAN motif, exhibited a retarded electrophoretic mobility indistinguishable from that of constitutively active wild-type (autophosphorylated) αPAK protein (lane 10). Since equivalent mutations in PAK1-250 did not affect its electrophoretic mobility (Fig. 2B), the retarded mobility of these mutant full-length kinases probably reflects kinase autophosphorylation. Indeed, all of the shifted mutants had elevated activity toward MBP in the absence of p21 (Fig. 3C and D). Mutants M1, M2, and M3, with amino acid substitutions which abolished p21-binding activity, showed only a basal level of kinase activity, which was not stimulated by GTPγS-Cdc42 (Fig. 3C and D). PAK mutants M4, M7, and M9, which were apparently constitutively phosphorylated, showed robust kinase activity toward MBP even without GTP-Cdc42 (Fig. 3C and D). Mutant M6 was an exception; despite being fully shifted, it exhibited a rather low constitutive activity which was increased by GTPγS-Cdc42. Taken together, these results indicated an autoinhibitory role for the residues in the C-terminal portion of the PAN motif.
Constructing a minimal PAK-inhibitory polypeptide.
We then determined whether the N-terminal half of the kinase harbored inhibitory sequences by testing GST/αPAK1-250 (S76P), deficient in Cdc42 binding (Fig. 2C), for its ability to inhibit PAK activation in vitro. Increasing amounts of GST/αPAK1-250(S76P) were added into the kinase assay reactions in the presence of excess GTP-Cdc42 (Fig. 4A, lanes 2 to 5). Both autophosphorylation and kinase activities were inhibited (Fig. 4A and B). Additional GTP-Cdc42 did not overcome this inhibition (Fig. 4B, lanes 6 to 8). Thus, αPAK activation can be directly inhibited by an excess of N-terminal regulatory sequences. To localize the autoinhibitory region, four smaller fusion proteins covering different lengths of the C-terminal region of the PAN motif were similarly tested (Fig. 4C and D). αPAKL404S activation by GTPγS-Cdc42 was clearly prevented by GST/αPAK83-149 and GST/αPAK83-131 but not by the smaller GST/αPAK99-149 and GST/αPAK99-131. GST/αPAK83-149 did not bind p21 (Fig. 4E) since it lacks critical residues 71 to 82.
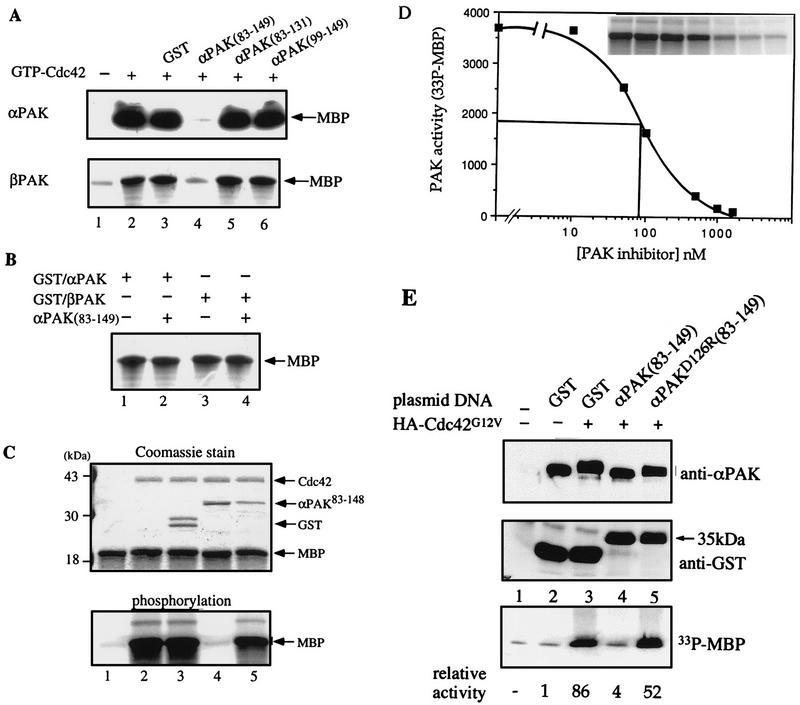
To test whether this inhibitory domain could suppress wild-type αPAK activation, we used purified wild-type αPAK and βPAK expressed in COS-7 cells, which are recovered in forms exhibiting basal kinase activity (34). Excess GST/αPAK83-149 was highly effective in inhibiting αPAK and βPAK activation in vitro by GTP-Cdc42, whereas GST/αPAK83-131 was not (Fig. 5A). However, GST/αPAK83-149 did not inhibit kinase activity of E. coli-expressed active wild-type (autophosphorylated) GST/αPAK or GST/βPAK (Fig. 5B). This finding suggests that the inhibitory domain functions primarily by preventing autophosphorylation (Fig. 4A) and consequential activation of the kinase. Replacing D126 with R resulted in substantial loss of the inhibitory function of αPAK83-148 (Fig. 5C; compare lanes 4 and 5). The inhibitor was highly potent in vitro, with an apparent Ki of 90 nM (Fig. 5D). When a cDNA encoding GST/αPAK83-149 was cotransfected with αPAK and Cdc42G12V into COS-7 cells, it completely blocked PAK activation (Fig. 5E; compare lanes 3 and 4), while the equivalent D126R mutant was ineffective (lane 5). This was evident in terms of both αPAK mobility and its activity toward MBP (as quantified at the bottom of Fig. 5E). The GST/inhibitor construct was detected as a single 35-kDa band, indicating its stability under in vivo expression conditions.
FIG. 5.
Specific inhibition of αPAK and βPAK activation by the GST/αPAK83-149 polypeptide. (A) In each assay, 0.2 μg of αPAK or βPAK purified from COS-7 cells was assayed for kinase activity toward MBP in the absence (lane 1) or presence of 2 μg of GTPγS-Cdc42 (lanes 2 to 6) and 2 μg of indicated PAK-derived polypeptides (lanes 4 to 6). GST was used as a control (lane 3). Kinase reactions were carried out with 10 μM [γ-33P]ATP and 10 μg of MBP in a final volume of 30 μl; half of the reaction was analyzed on 12% polyacrylamide gels. The MBP region of the autoradiograph is shown. (B) Bacterially expressed (active) GST/αPAK (1 μg) and GST/βPAK (1 μg) were assayed for MBP kinase activity (as described above) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 5 μg of GST/αPAK83-149. (C) A mutation (D126R) in GST/αPAK83-148 interferes with its inhibitory activity. Top, Coomassie blue-stained gel (12%); bottom, corresponding autoradiograph of the MBP region. Each reaction was carried out with 0.2 μg of αPAK which was activated in the presence of 2 μg of Cdc42-GTPγS. Lanes 4 and 5 contained 1.5 μg of GST/αPAK83-148 or GST/αPAK83-148(D126R), a level ∼15-fold higher than the calculated Ki of the wild-type inhibitor (D). GST alone has no effect on activation (the larger GST form contains attached polylinker-derived sequence). Kinase reactions were as for panel A. (D) Concentration dependence of αPAK inhibition was determined by using 50 ng of kinase (=18 nM) under activation conditions as for panel A while varying the GST/αPAK83-148 concentration. MBP phosphorylation was quantified on a PhosphorImager. For reference, the data of MBP phosphorylation is shown in the inset. (E) Inhibition of αPAK activation by coexpression of GST/αPAK83-149 in COS-7 cells. Cells (in 100-mm-diameter dishes) were transfected with 3 μg of FLAG-αPAK (lanes 2 to 5) with or without 3 μg of HA-Cdc42G12V expression plasmid as indicated. Expression of GST, GST/αPAK83-149, or the mutant (D126R) version (6 μg of plasmid per dish) was detected in the total extract by using anti-GST antibodies. The activity of the anti-FLAG immunoprecipitates, recovering equivalent amounts of the kinase (top), was determined by phosphorylation of MBP (counts quantified at the bottom).
PAK inhibitor prevents certain cytoskeletal rearrangements induced by Cdc42 and Rac1.
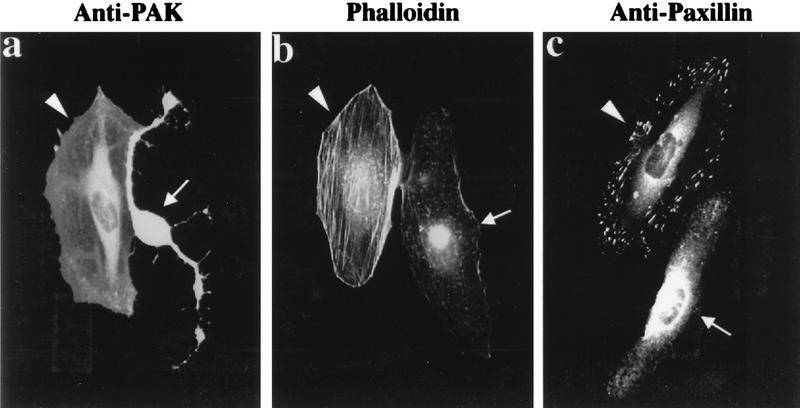
In HeLa cells, expression of Cdc42G12V induces formation of filopodia and cell retraction (34); stress fibers and Rho-dependent focal adhesions disappear with production of new peripheral FCs. Rac1G12V-expressing cells, which are characterized by cell flattening, membrane ruffling, and formation of bead-like FCs at the cell periphery, also lose stress fibers (39). In (human) HeLa cells, γPAK represents the predominant form corresponding to hPAK65 (36). Injection of a plasmid expressing the PAK83-149 inhibitor sequence had no effect on the morphology of HeLa cells; the inhibitor was cytosolic, and its disposition was apparently unaffected by Cdc42 or Rac (data not shown). However, its expression dramatically affects cytoskeletal reorganization elicited by Cdc42 and Rac1 (Fig. 6). In each field, we show a typical p21-injected cell, a cell doubly injected with p21 plus PAK inhibitor, and uninjected control cells. Compared with HA-Cdc42G12V injection alone (Fig. 6a and b), cells coinjected with plasmid encoding αPAK83-149 did not exhibit a clear reorganization of actin to the periphery 2 h after injection. Peripheral actin microspikes were always absent in the doubly injected cells. Typically both Cdc42G12V- and Rac1G12V-injected cells coexpressing αPAK83-149 (Fig. 6c and d) retained internal Rho-type FCs while forming peripherally located FCs. With Cdc42G12V plus αPAK83-149, these peripheral complexes were clearly larger than with Cdc42G12V alone (Fig. 6c). The formation of these new FCs indicates that the p21s are functional in the presence of the kinase inhibitor and that PAK kinase activity is not required for FC formation. We monitored the behavior of cells to 4 h postinjection (Fig. 6e to h); over this time period, coexpression of αPAK83-149 still prevented the production of peripheral actin microspikes in Cdc42G12V-expressing cells (Fig. 6e and f) but did not block membrane ruffles due to RacG12V (Fig. 6g and h). This lack of effect on ruffles, seen here as a stained and wavy outline, was also observed on phase-contrast microscopy (data not shown). PAK inhibition also prevented the loss of actin stress fibers in both Cdc42G12V- and Rac1G12V-expressing cells (Fig. 6f and h).
FIG. 6.
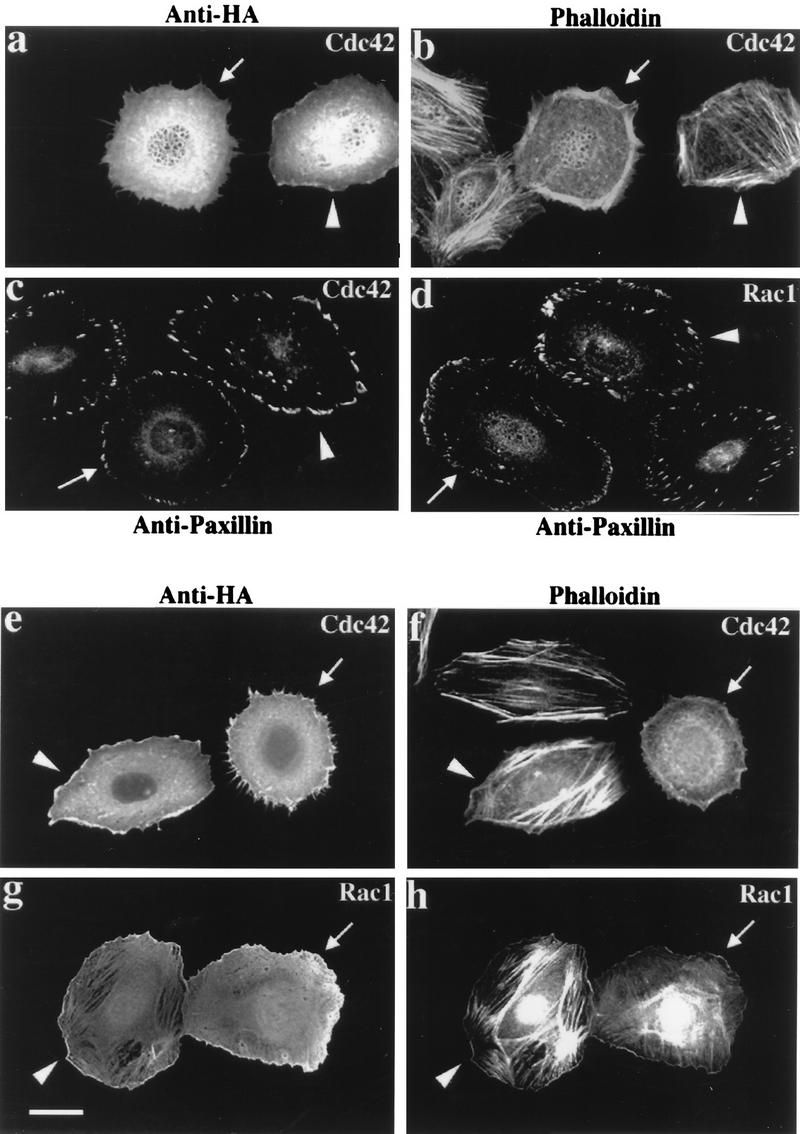
Morphological effects of Cdc42 and Rac are blocked by inhibitory PAK83-149. HeLa cells were examined 2 h (a to d) or 4 h (e to h) after plasmid microinjection. In each case, a constitutively active mutant (G12V) form of Cdc42 or Rac1 was used. The top panels show that coinjection of the PAK inhibitor (arrowheads) blocks the reorganization of actin stress fibers normally seen with Cdc42 (arrows). Analysis of FCs by paxillin staining (c and d) shows production of peripherally located FCs to occur in the presence of the PAK inhibitor (arrowheads), but the resultant structures are thicker and there is no apparent loss of the existing internal focal adhesions. Even after 4 h, Cdc42G12V-expressing cells (e) do not form filopodia or retraction fibers when injected with the PAK inhibitor: with both Cdc42G12V and RacG12V, PAK inhibition leads to an overall increase in the intensity of stress fiber staining (f and h).
Expression of αPAK83-149 blocks PAK function in vivo.
We then tested whether αPAK83-149 could block the morphological effects of αPAK itself. Certain mutant forms of αPAK require no binding of Cdc42 to be fully activated in cultured cells, and their expression results in loss of actin stress fibers and focal adhesions and in the concomitant retraction of peripheral membranes (34). We found that αPAKL107F (6) has greater activity when recovered from COS-7 cells than HAαPAKEQ116/117KK (M7) and HA-αPAKD126R (M9) proteins (data not shown) or the chimeric PAK-CC that we reported previously (34). Injection of a plasmid encoding αPAKL107F elicited rapid changes in cell morphology, in line with our previous observations. HeLa cells injected with plasmid encoding αPAKL107F (Fig. 7) behaved very differently from those coinjected with an αPAKL107F plasmid and with a plasmid encoding the inhibitory αPAK83-149. Notably, the coinjected cells did not undergo the characteristic retraction (Fig. 7a), nor was there any evident loss of stress fibers or focal adhesions (Fig. 7b and c), although both cells clearly expressed αPAKL107F (Fig. 7a). Injection with plasmid encoding HA-αPAK83-149 showed that expression of the inhibitor by itself led to no distinct changes in actin or vinculin staining (data not shown). These results show that inhibitory αPAK83-149 is able to block PAK activity in vivo. The inhibitor presumably interferes with intracellular activation of kinase, since αPAKL107F recovered from COS-7 cells displayed the retardation in mobility characteristic of hyperphosphorylated and activated PAK only when the inhibitor was not coexpressed (data not shown).
FIG. 7.
The PAK inhibitor can block effects of constitutively active PAK in vivo. The effects of microinjected αPAKL107F on HeLa cells (arrows) include cell retraction (a) seen in cells stained for expressed PAK, loss of actin stress fibers (b), and dissolution of paxillin-stained focal adhesions (c). All three effects could be blocked by coinjection of an expression construct encoding αPAK83-149 (arrowheads).
DISCUSSION
Activation of PAK.
PAK kinases can be directly and potently activated by the GTP-bound forms of Cdc42 and Rac1, unlike Rho-binding kinases such as ROK or PKN, which are activated only weakly on binding to Rho-GTP (13, 25, 37, 59). Our results indicate that the PAK N-terminal PAN motif integrates two functions, p21 binding and kinase inhibition. Binding of Cdc42/Rac initiates release of a negative constraint embodied in sequences immediately C terminal of the binding site to allow autophosphorylation and kinase activation. The autoinhibitory sequence, like the kinase domain, is highly conserved among PAKs from yeasts to humans but is absent from other Cdc42 effectors, including ACK (31) and WASP (1, 18, 53). Native purified PAKs are regulatable kinases with low intrinsic activity (4, 32); however, for reasons that are not apparent, wild-type PAKs are constitutively active when expressed in E. coli (34). This precludes use of recombinant protein for regulatory studies unless a suitable derivative such as αPAKL404S is employed. Use of this attenuated kinase, which undergoes autophosphorylation identical to that of the wild type upon Cdc42 activation in vitro (34), revealed that amino acid substitutions in the autoinhibitory sequence produce activated kinases (Fig. 3). PAK1L107F (6) and mPAK3, containing mutations equivalent to positions F96, G98, and P100 in αPAK (2), are similarly active. The effects of mutations dispersed throughout αPAK83-131 suggest that they act by altering the conformation of the inhibitory region. The behavior of the H83/86L mutant may reflect overlapping of p21-binding and inhibitory domains. The data suggest a model in which the inhibitory region forms a complex with the kinase domain, rendering it inactive, but does not do so when the domain is already activated (with phosphorylated T422).
Other studies have pointed to a role for N-terminal regions of PAK-related proteins in negatively regulating kinase activity. Expression of PAK1 (αPAK) lacking the first 231 amino acids, but not of full-length PAK1, yielded active kinase (45). S6/H4 (γPAK-like) kinase can be proteolytically cleaved in vitro to yield an activated catalytic fragment (∼40 kDa) (4). Caspase-mediated cleavage of PAK2 (γPAK) at position 212 during apoptosis activates the kinase (49) and may play an important part in the morphological changes seen during cell death. By specifically inhibiting PAK, it should be possible to dissect the role of PAK in apoptosis.
Many protein kinases are regulated by autoinhibition in which substrate-like sequences complex with the catalytic site to block substrate binding and in some cases autophosphorylation. Binding of an allosteric activator can disrupt conformation in the autoinhibitory domain (16, 52). Residues 580 to 595 in myosin light-chain kinase resemble a consensus substrate site and constitute an autoinhibitory domain (10, 14); a corresponding synthetic peptide inactivates the proteolytically activated kinase (17). The N-terminally located autoinhibitory domain of protein kinase C (PKC) is well documented (9, 12, 42). In these cases, the pseudosubstrate inhibitory domain primarily blocks phosphorylation of exogenous substrates. In contrast, the αPAK inhibitor sequence (83 to 149) cannot inhibit active (autophosphorylated) forms of αPAK and βPAK (Fig. 5C). Its mechanism of action therefore probably involves blocking autophosphorylation events. If a pseudosubstrate-like peptide exists within PAK83-149, its conformation must be stabilized by flanking sequences since we cannot derive a smaller inhibitory peptide (Fig. 3). The PAK autoinhibitory domain may partially overlap and be stabilized by the p21-binding domain (Fig. 4), resembling yeast PKC1, where the Rho1-binding domain lies within a pseudosubstrate sequence (40).
Morphological roles for PAK downstream of Cdc42 and Rac.
The finding that PAK83-149 can potently inhibit PAK kinase activity has allowed us to assess PAK function without overexpressing the kinase itself or mutant forms. Larger N-terminal regions of PAK or kinase-dead full-length PAK have been used as dominant negatives in cotransfection experiments (2, 60). Use of these larger constructs is compromised by their potential to sequester Cdc42/Rac and by the PAK N-terminal protein itself acting as an adapter potentially recruiting proteins such as Nck (3, 5, 11, 29). Expression of the PAK inhibitor in Cdc42G12V-expressing HeLa cells blocked both cell retraction and rounding and the induction of peripheral actin microspikes (Fig. 6) normally seen in epithelial, fibroblastic, and neuroblastoma cells (20, 34, 39). Similar effects of the PAK inhibitor were observed with Swiss 3T3 cells (data not shown) in terms of both reorganization of FCs and production of peripheral actin microspikes. Since coexpression of αPAK83-149 completely blocks Cdc42-mediated activation of cotransfected PAK (Fig. 5), endogenous PAK must be similarly affected. The ability of the inhibitor to block the phenotypic effects of αPAKL107F further confirms this conclusion.
Although PAK is clearly involved in Cdc42- and Rac-type morphological changes (34, 50), studies using effector mutants of Rac1 and Cdc42 that fail to interact with PAK (primarily substitutions at Y40) have been interpreted as showing that PAK is not necessary for Rac-driven membrane ruffling (15, 57) or for Cdc42 to form filopodia (21). From our results, we suggest the PAK activity is required to generate filopodia. One can reconcile these apparently contradictory observations because under some circumstances (Fig. 2), direct binding to Cdc42 is not essential for PAK recruitment to membrane-apposed Cdc42-driven focal complexes. This process does require its interaction with the focal complex protein PIX (35). Upon translocation to the plasma membrane, there is consequential activation of PAK (29). This model is supported by our data showing that PIX, the ubiquitous Rac guanine nucleotide exchange factor tightly associated with PAK, can mediate kinase activation by Cdc42(Y40C), which does not directly bind the kinase (35). The identification of this PAK-associated guanine nucleotide exchange factor might also explain the Rac-like phenotypes seen with microinjected PAK (50).
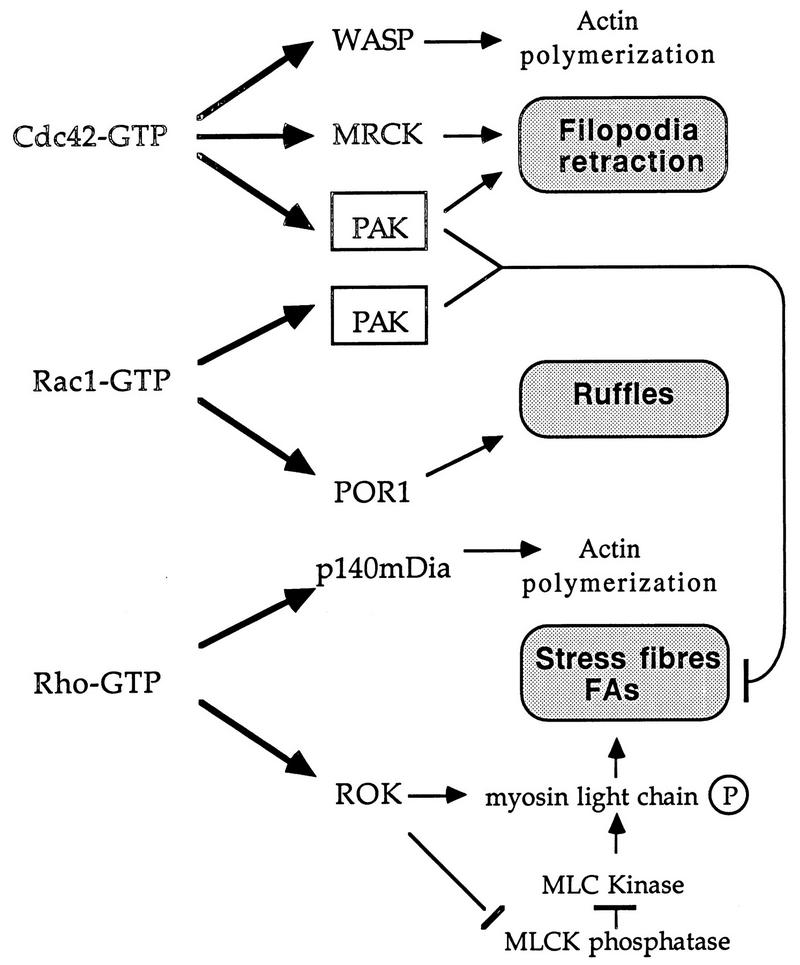
We previously showed that expression of constitutionally active PAK can antagonize Rho function. In this study, we demonstrate that PAK does indeed play such a role downstream of Cdc42 (Fig. 8). The recent identification of a ROK-like kinase lying downstream of Cdc42 termed MRCK (26) provides a Cdc42 target that promotes focal complex formation. MRCK and PAK apparently play synergistic roles in promoting filopodia and dynamic FC structures. Cdc42G12V-expressing HeLa cells form peripheral actin microspikes and become devoid of intracellular stress fibers (Fig. 6b and f), but cells coexpressing the PAK inhibitor show no such formation of microspikes and display an increased stress fiber content. In the case of Rac1G12V expression, it is clear that blocking PAK prevents the loss of stress fibers, but otherwise we do not observe any perturbation to the Rac-induced phenotype (cell spreading and ruffling). Thus, we conclude that the primary morphological role of PAK is to facilitate turnover of actin stress fibers and dissolve focal adhesions and that Cdc42 requires PAK kinase activity for production of filopodia and for cell rounding and retraction (Fig. 8).
FIG. 8.
A model of PAK function in relation to known effectors of Rho-p21s. The targets of Cdc42, Rac, and Rho which have known morphological roles are shown. MRCK is the myotonin kinase-related Cdc42-binding kinase (26); p140mDia is the mammalian homolog of Drosophila diaphanous (58); the other Rho-binding kinase, PKN (59), has no known morphological function. PAK acts downstream of both Cdc42 and Rac to break down actin cytoskeletal structures and focal adhesions (FAs). This function could be particularly important in remodeling the cytoskeleton to allow dynamic events such as filopodial extension and subsequent formation of lamellipodia, ultimately leading to cell migration or growth cone extension (20). MLC, myosin light chain; MLCK, myosin light-chain kinase.
ACKNOWLEDGMENT
This work was supported by the Glaxo Singapore Research Fund.
REFERENCES
- 1.Aspenstrom P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr Biol. 1995;6:70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- 2.Bagrodia S, Derijard B, Davis R J, Cerione R A. Cdc42 and PAK-mediated signalling leads to JNK and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 3.Bagrodia S, Taylor S J, Creasy C L, Chernoff J, Cerione R A. Identification of a mouse p21cdc42/rac activated kinase. J Biol Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 4.Benner G E, Dennis P B, Masaracchia R A. Activation of an S6/H4 kinase (PAK 65) from human placenta by intramolecular and intermolecular autophosphorylation. J Biol Chem. 1995;270:21121–21128. doi: 10.1074/jbc.270.36.21121. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 6.Brown J L, Stowers L, Baer M, Trejo J A, Coughlin S, Chant J. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 7.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 8.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signalling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 9.Coussens L, Parker P J, Rhee L, Yang-Feng T L, Chen E, Waterfield M D, Francke U, Ullrich A. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signalling pathways. Science. 1986;233:859–866. doi: 10.1126/science.3755548. [DOI] [PubMed] [Google Scholar]
- 10.Edelman A M, Takio K, Blumenthal D K, Hansen R S, Walsh K A, Titani K, Krebs E G. Characterization of the calmodulin-binding and catalytic domains in skeletal muscle myosin light chain kinase. J Biol Chem. 1985;260:11275–11285. [PubMed] [Google Scholar]
- 11.Galisteo M L, Chernoff J, Su Y-C, Skolnik E Y, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 12.House C, Kemp B E. Protein kinase C contains a pseudosubstrate prototype in its regulatory domain. Science. 1987;238:1726–1728. doi: 10.1126/science.3686012. [DOI] [PubMed] [Google Scholar]
- 13.Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. The small GTP-binding protein Rho binds to and activates a 160 kda Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 14.Ito M, Guerriero V, Jr, Chen X M, Hartshorne D J. Definition of the inhibitory domain of smooth muscle myosin light chain kinase by site-directed mutagenesis. Biochemistry. 1991;30:3498–3503. doi: 10.1021/bi00228a021. [DOI] [PubMed] [Google Scholar]
- 15.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 16.Kemp B E, Parker M W, Hu S, Tiganis T, House C. Substrate and pseudosubstrate interactions with protein kinases: determinants of specificity. Trends Biochem Sci. 1994;19:440–444. doi: 10.1016/0968-0004(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 17.Kennelly P J, Edelman A M, Blumenthal D K, Krebs E G. Rabbit skeletal muscle myosin light chain kinase. The calmodulin binding domain as a potential active site-directed inhibitory domain. J Biol Chem. 1987;262:11958–11963. [PubMed] [Google Scholar]
- 18.Kolluri R, Tolias K F, Carpenter C L, Rosen F S, Kirchhausen T. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc Natl Acad Sci USA. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 22.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Steo20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 25.Leung T, Chen X-Q, Manser E, Lim L. The p160 RhoA-binding kinase ROKα is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol. 1996;16:5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung T, Chen X-Q, Tan I, Manser E, Lim L. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol. 1998;18:130–140. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation pathways by p21 GTPases. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Styles C, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 29.Lu W, Katz S, Gupta R, Mayer B J. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 30.Manser E, Leung T, Monfries C, Teo M, Hall C, Lim L. Diversity and versatility of GTPase activating proteins for the p21rho subfamily of ras G proteins detected by a novel overlay assay. J Biol Chem. 1992;267:16025–16028. [PubMed] [Google Scholar]
- 31.Manser E, Leung T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 32.Manser E, Leung T, Salihuddin H, Zhao Z-S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 33.Manser E, Chong C, Zhao Z-S, Leung T, Michael G, Hall C, Lim L. Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J Biol Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- 34.Manser E, Huang H-Y, Loo T-H, Chen X-Q, Dong J-M, Leung T, Lim L. Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manser E, Loo T-H, Koh C-G, Zhao Z-S, Tan I, Leung T, Lim L. A family of guanine nucleotide exchange factors (PIXs) is directly coupled to the p21(Cdc42 and Rac)-activated kinase αPAK. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 36.Martin G A, Bollag G, McCormick F A, Abo A. A novel serine kinase activated by rac/Cdc42Hs-dependent autophosphorylation is related to PAK65 and Ste20. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsui T, Amano M, Yamamoto T, Chilhara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamamatsu A, Kaibuchi K. Rho-associated kinase, a novel serine threonine kinase, as a putative target for the small GTP-binding protein Rho. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 38.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signalling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 39.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibres, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 40.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 42.Parker P J, Coussens L, Totty N, Rhee L, Young S, Chen E, Stabel S, Waterfield M D, Ullrich A. The complete primary structure of protein kinase C—the major phorbol ester receptor. Science. 1986;233:853–859. doi: 10.1126/science.3755547. [DOI] [PubMed] [Google Scholar]
- 43.Peppelenbosch M P, Qiu R-G, de Vries-Smits A M M, Tertoolen L G J, de Laat S W, McCormick F, Hall A, Symons M H, Bos J L. Rac mediates growth factor-induced arachidonic acid release. Cell. 1995;81:849–856. doi: 10.1016/0092-8674(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 44.Peter M, Neiman A M, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 45.Polverino A, Frost J, Yang P, Hutchison M, Neiman A M, Cobb M H, Marcus S. Activation of mitogen-activated protein kinase cascades by p21-activated protein kinases in cell-free extracts of Xenopus oocytes. J Biol Chem. 1995;270:26067–26070. doi: 10.1074/jbc.270.44.26067. [DOI] [PubMed] [Google Scholar]
- 46.Ridley A J, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibres in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 47.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 48.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 49.Sells M A, Chernoff J. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 50.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 51.Simon M N, Devirgilio C, Souza B, Pringle J R, Abo A, Reed S I. Role for the Rho-family GTPase Cdc42 in yeast mating pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 52.Soderling T R. Protein kinases. Regulation by autoinhibitory domains. J Biol Chem. 1990;265:1823–1826. [PubMed] [Google Scholar]
- 53.Symons M, Derry J M J, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase Cdc42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 54.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 55.Teo M, Manser E, Lim L. Identification and molecular cloning of a p21cdc42/rac1-activated serine/threonine kinase that is rapidly activated by thrombin in platelets. J Biol Chem. 1995;270:26690–26697. doi: 10.1074/jbc.270.44.26690. [DOI] [PubMed] [Google Scholar]
- 56.Van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of the small GTPase Rho. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B M, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westwick J K, Lambert Q T, Clarke G J, Symons M, Van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Z-S, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziman M, O’Brien J M, Ouellette L A, Church W R, Johnson D I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]