Abstract
The histone deacetylase Rpd3p functions as a transcriptional repressor of a diverse set of genes, including PHO5. Here we describe a novel role for RPD3 in the regulation of phosphate transporter Pho84p retention in the cytoplasmic membrane. We show that under repressing conditions (with Pi), PHO5 expression is increased in a pho4Δ rpd3Δ strain, demonstrating PHO regulatory pathway independence. However, the effect of RPD3 disruption on PHO5 activation kinetics is dependent on the PHO regulatory pathway. Upon switching to activating conditions (without Pi), PHO5 transcripts accumulated more rapidly in rpd3Δ cells. This more rapid response correlates with a defect in phosphate uptake due to premature recycling of Pho84p, the high-affinity H+/PO43− symporter. Thus, RPD3 also participates in PHO5 regulation through a previously unidentified effect on maintenance of high-affinity phosphate uptake during phosphate starvation. We propose that Rpd3p has a negative role in the regulation of Pho84p endocytosis.
Large amounts of phosphate are required for cell growth and proliferation. In phosphate-starved yeast (Saccharomyces cerevisiae) cells, phosphate is scavenged from covalently bound sources in the environment through the action of secreted phosphatases. The PHO5 gene product constitutes the bulk of the acid phosphatase, so PHO5 regulation is key to cellular phosphate homeostasis. Many genes have been identified as being involved in phosphate scavenging (PHO genes) (22, 45, 46). Several genes in the PHO gene family participate in the regulation of PHO5 expression (Fig. 1). Transcriptional activators, Pho4p and Pho2p, are required to generate the active chromatin structure on the PHO5 promoter and stimulate transcription (14). PHO2 and PHO4 expression is not regulated by phosphate availability (4, 51). Pho80p-Pho85p is a cyclin/cyclin-dependent kinase pair that multiply phosphorylates Pho4p to negatively regulate Pho4p function. Phosphorylated Pho4p preferentially binds to Msn5p, a nuclear export protein. Pho4p phosphorylation also inhibits its interaction with the nuclear importer Pse1p/Kap21 and with the transcription factor Pho2p (20, 32). The net result is Pho4p cytoplasmic localization (nuclear exclusion). Pho81p inhibits Pho80p-Pho85p kinase activity during phosphate depletion and serves as a positive regulator of Pho4p localization and activity. Thus, Pho4p is the ultimate regulator of PHO5 expression in the PHO pathway.
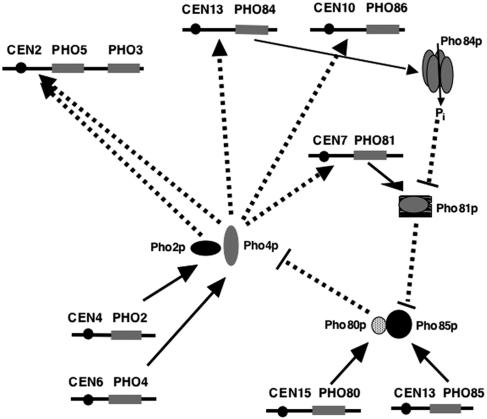
FIG. 1.
Schematic diagram of the PHO5 regulatory cascade. Pho2p and Pho4p are sequence-specific DNA binding transcription regulatory proteins. Binding sites for Pho4p are found on the PHO5, PHO81, PHO84, and PHO86 promoters. All four of these genes are activated by phosphate starvation. High intracellular phosphate represses Pho81p activity, which acts as an inhibitor of Pho80p/Pho85p Pho4p-kinase activity. Pho4p phosphorylation decreases its affinity for Pho2p and results in its cytoplasmic localization, thus inhibiting its ability to activate transcription. Phosphate depletion activates Pho81p, resulting in Pho4p nuclear localization and transcriptional activation of responsive genes (dashed arrows).
Pho84p is a high-affinity H+/PO43− symporter that is localized to the cytoplasmic membrane. Of the five transporters identified to date, Pho84p is the major contributor to phosphate uptake (50). Mutation of the other four phosphate transporter genes has no effect on PHO5 expression, and cells retained both a high Vmax and a low Km for phosphate uptake. PHO86 encodes an endoplasmic reticulum protein that is required for the exit of Pho84p out of the endoplasmic reticulum (24). Extracellular phosphate levels regulate Pho84p cytoplasmic membrane localization, and regulation of Pho84p localization is independent of the intracellular phosphate signaling pathway (24, 26, 33). During down regulation, PHO84 transcription is shut off and Pho84p is endocytosed and localized to the vacuole lumen for active degradation (26, 33). Whether sorting to the vacuole involves phosphorylation and ubiquitylation is not currently known.
In addition to serving as an excellent model for complex and combinatorial regulation of gene expression, the PHO5 gene serves as an experimental model for transcription regulation in a chromatin context (16, 44). In the presence of abundant phosphate, the promoter is repressed through formation of nucleosomes positioned over key promoter elements. When cells are starved for phosphate, the chromatin structure undergoes reconfiguration that renders the DNA in this region more accessible (1, 2, 5, 7, 13, 35, 40, 44).
Biochemical and molecular genetic studies have indicated a role for histone posttranslational modification in PHO5 regulation (3, 34, 44, 48). The reversible acetylation of lysine residues at the N termini of core histones is generally linked to transcriptional activation. The counterpart to acetylation is deacetylation. At least five genes encoding catalytic subunits of histone deacetylases have been identified in yeast. These include RPD3, HDA1, HOS1, HOS2, and HOS3 (10, 37, 47). HDA1 and RPD3 participate in determining the histone acetylation pattern on the PHO5 promoter (48). Histones over the PHO5 locus are more acetylated (at H3 lysine 9 and H4 lysine 12) in an rpd3Δ strain than in the wild-type strain (48). Thus, Rpd3p functions in regulating the histone acetylation state on the PHO5 promoter.
To properly interpret the results of experiments on PHO5 chromatin structure, the entire regulatory pathway needs to be taken into account. RPD3 regulation of PHO5 expression has not been demonstrated to be independent of the PHO pathway regulation of Pho4p activity. To test the hypothesis that RPD3 disruption affects PHO5 expression through a PHO pathway-dependent mechanism, PHO5 expression was compared in PHO4 rpd3Δ and pho4Δ rpd3Δ strains under repressing conditions (with Pi). We observed increased PHO5 expression in the pho4Δ rpd3Δ strain, demonstrating that RPD3 deletion increases basal transcription through a PHO pathway-independent mechanism.
In contrast, we were surprised to observe more rapid PHO5 induction kinetics in an rpd3Δ strain shifted to activating growth medium (without Pi), suggesting that RPD3 effects PHO5 activation through a PHO signaling pathway-dependent mechanism. While investigating the mechanism, we found that RPD3 disruption results in a compromised phosphate uptake rate and that this tightly correlates with premature Pho84p recycling in the rpd3Δ strains. Therefore, RPD3 disruption affects the kinetics of PHO5 regulation through disrupting the normal retention of the high-affinity phosphate transporter, Pho84p, in the cytoplasmic membrane during phosphate starvation. We propose that Rpd3p has a negative function in the regulation of Pho84p endocytosis. Further, we propose that the resulting defect in phosphate uptake compromises the ability of cells to build up internal phosphate, causing more rapid internal phosphate depletion and activation of the PHO signaling pathway.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast strains used in this work are listed in Table 1. The original strains, YS18 and YS20, were kindly provided by B. Meyhack (Ciba-Geigy). YS20 was created by deletion of PHO4 in the YS18 background (39). These strains were used to create the rpd3Δ mutant (YS18R) and the pho4Δ rpd3Δ double mutant (YS20PR) by homologous recombination. The plasmid pMVL (37), containing a disrupted copy of the RPD3 gene locus, was digested with AatII and BglII, and the 3.5-kb fragment containing rpd3::LEU2 was gel purified. The DNA fragment was used to transform yeast cells by the conventional method. A Leu+ prototroph was selected, and the gene disruption was confirmed by Southern blotting. The strains were designated YS20PR and YS18R for rpd3 mutation in pho4 background (YS20) and PHO4 background (YS18), respectively.
TABLE 1.
Yeast strains and plasmids
| Strain | Genotype |
|---|---|
| YS18 | MATα his3-11,15 leu2-3,112 ura3Δ5 canR |
| YS18R | MATα; his3-11,15 leu2-3,112 ura3Δ5 rpd3::LEU2 canR |
| YS20 | MATα; his3-11,15 leu2-3,112 ura3Δ5 pho4::URA3 canR |
| YS20PR | MATα; his3-11,15 leu2-3,112 ura3Δ5 pho4::URA3 rpd3::LEU2 canR |
Acid phosphatase activity assay.
Synthetic complete medium with or without phosphate was prepared as described previously (41). A yeast colony was picked from yeast extract-peptone-dextrose plates of each strain and cultured in 5 ml synthetic complete medium (high phosphate) overnight at 30°C. The cultures were diluted 1:50 (to 250 ml) and grown to a density of 1 × 106 to 2 × 106. Yeast cells were centrifuged down, washed with sterile water, and resuspended in 50 ml high- or no-phosphate medium. At 3, 6, and 9 h, 1-ml samples from each strain were collected and washed once with 10 ml water before resuspension in 1 ml water.
The periplasmic acid phosphatase activity was measured in 0.5 ml containing 25 μl of 1 M sodium acetate (pH 4.0), 5 μl of 0.45-mg/ml p-nitrophenylphosphate (substrate), and various volumes of cell suspensions. The buffer and substrate were preincubated at 37°C for 10 min, the cell suspensions were added, and the reaction mixtures were incubated for an additional 10 min at 37°C. The reactions were stopped by addition of 0.12 ml of 25% trichloroacetic acid and 0.6 ml saturated Na2CO3. Cells were pelleted, and the absorbance of the supernatants at 405 nm was measured with a Beckman DU600 spectrophotometer. The units of acid phosphatase enzyme from each culture were interpolated from a standard curve, which was created by plotting known amounts of commercial phosphatase versus the absorbances at 405 nm produced by those amounts of phosphatase under the assay conditions used. One unit of acid phosphatase activity is defined as the amount of enzyme used to produce 1 μmol of p-nitrophenol in 1 min at 37°C. All experiments were conducted in triplicate and repeated as three independent experiments.
Phosphate uptake assays.
Phosphate uptake assays were performed as described previously (25). All experiments were conducted in triplicate and repeated as three independent experiments.
Preparation of RNA and cDNA synthesis.
Cells from the same cultures of each yeast strain used for the assay of acid phosphatase activity were also used for total RNA extraction. Approximately 5 × 107 cells were collected from cultures grown on phosphate-rich and no-phosphate media for 3, 6, and 9 h. RNA extractions were processed using the RNeasy minikit (QIAGEN). Samples of total RNA were quantified at optical densities of 260/280 using a UV spectrophotometer (Beckman DU600). One microgram of total RNA was subsequently treated with 1 unit of RNase-free DNase I (Roche) in digestion buffer (20 mM Tris, pH 7.5, and 10 mM MgCl2) at 37°C for 5 min. After ethanol precipitation, the DNase I-treated RNA was resuspended in 10 μl H2O. Five microliters was diluted to 400 μl in water and used in real-time PCR to check for DNA contamination. RNA in the other 5 μl was annealed with 25 pmol of oligo(dT) primers. The mixture was heated at 70°C for 15 min to remove any secondary structure. cDNAs were synthesized in a 15-μl reaction mixture containing 1 mM deoxynucleoside triphosphates, reverse transcriptase buffer, and 200 units of Moloney murine leukemia virus reverse transcriptase (Promega) at 42°C for 1 h. The final volume was adjusted with sterile H2O to 400 μl, and the amount to be used in real-time PCR was determined by titration.
Primer design for real-time PCR analysis.
Primer sets were designed for SYBR Green PCR analysis using the web-based program “Primerfinder version 0.06” (http://www.cellbiol.com/Tools.html#pick). DNA sequences from the actin gene (YFL039C/ACT1), PHO3 (YBR092C), PHO5 (YBR093C), PHO81 (YGR233C), PHO84 (YML123C), and PHO86 (YJL117W) were retrieved from the Saccharomyces Genome Database. The sequences of primers used are listed in Table 2. Since the coding regions of PHO3 and PHO5 are almost (87%) identical, forward primers for PHO3 and PHO5 were designed to be complementary to the 5′ untranslated region. Primers for the rest of the genes examined were designed from coding sequences. The melting temperatures are in the range of 60 to 65°C for all primer sets. The amplicon length was kept between 99 and 112 bp.
TABLE 2.
Primer sequences for SYBR Green real-time PCR analysis
| Amplified gene | Directiona and sequence (5′→3′) | Product size (bp) |
|---|---|---|
| Actin gene | F, CTGTCGAGAGATTTCTCTTTTACC | 108 |
| R, GCCCCTATTTATTCCAATAATATCG | ||
| PHO2 | F, CGTCAGAATTTGGTTTCAGAACAG | 112 |
| R, GATCGTAATCGTTGGCAATATCAC | ||
| PHO3 | F, GTAAAGAAAGGGCCATTCCAAATTACC | 99 |
| R, CTCTCCGAGGGGAATTGTACCTG | ||
| PHO4 | F, AGAGCAGCATTCTTGATAAAGTCGG | 91 |
| R, CTCGTCGTTTTGCTCGTTGAAG | ||
| PHO5 | F, GCAAGCAAATTCGAGATTACCAATG | 99 |
| R, GCTAGTTTGCCTAAGGGAATGGTACC | ||
| PHO81 | F, CCATTCCCACGCTAAAGGCTAG | 103 |
| R, GGCAGCTTTATTTTCTTGCAGTCTC | ||
| PHO84 | F, TGATGTCCTACGTTTACTGGCACG | 103 |
| R, ACCAAAACCAAATTGACCAATAACAG | ||
| PHO86 | F, ACCACTTGATATTGATGCTCCTCC | 103 |
| R, GTTCTTGTCTGATGAAATCCGC | ||
| INO1 | F, ATATTGCTCCAATCACCTCCG | 256 |
| R, GTGCTTATTCGCCAATACCG |
F, forward; R, reverse.
SYBR Green real-time PCR.
Real-time PCR experiments were performed using the iCycler iQ multicolor real-time PCR detection system (Bio-Rad). Several sets of cDNA templates were prepared from two sets of RNA to ensure the reproducibility of real-time PCR. SYBR Green analysis-based PCRs were performed in duplicate in each experiment. The transcript levels of the genes of interest (PHO2, PHO3, PHO4, PHO5, PHO81, PHO84, and PHO86) were compared to that of the actin gene (ACT1). The total volume of each reaction mixture was 25 μl. The PCR mixture contained 200 nM each of forward and reverse primer, 200 μM deoxynucleoside triphosphates, 2.5 mM MgCl2, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, SYBR Green diluted 50,000-fold from the manufacturer's stock solution (Molecular Probes), 1 nM fluorescein (Bio-Rad), 1 unit of JumpStart Taq DNA polymerase (Sigma), and 5 to10 μl of cDNA template. All primer sets were tested with genomic DNA from strain YS18R to check for the linearity of amplification and optimize PCR parameters. For all primer sets, thermocycler conditions were used as follows: stage 1, initial denaturation at 95°C for 3 min for 1 cycle; stage 2, denaturation at 95°C for 30 s and annealing and elongation at 60° for 45 s for 40 cycles. After amplification, the PCR products were also checked for nonspecific amplification by 2% agarose gel electrophoresis and melting curve analysis. All experiments were conducted in triplicate and repeated as three independent experiments.
Fluorescence microscopy.
Wild-type and rpd3Δ cells containing the plasmid pPHO84-GFP were cultured in SC medium containing phosphate to mid-log phase before being switched to no-phosphate medium. The plasmid pPHO84-GFP (EB0666), consisting of the intact PHO84 locus fused with green fluorescent protein (GFP)-coding sequence at the C terminus, was kindly provided by E. K. O'Shea (24). The expression of chimeric Pho84p-GFP was analyzed by fluorescence microscopy at 2, 4, 6, and 8 of h growth on no-phosphate medium. Total cells in each field were counted (with no fluorescent filter) from 20 fields containing approximately the same number of cells (six to eight). In each of the same fields the number of cells with a green cytoplasmic membrane ring (with the fluorescent filter) was scored, providing the number of total cells and cells with a green ring from each field. Cells with GFP in the intracellular compartment but no green ring were scores as non-membrane-localized Pho84p-GFP cells. The fraction of cells with cytoplasmic membrane Pho84p-GFP out of the total cells at 2, 4, 6, and 8 h of growth on no-phosphate medium for wild-type and rpd3Δ cultures was calculated. The experiment was repeated five times, and the results were averaged and plotted, with the standard deviation indicated.
RESULTS
Considering the complexity of the PHO5 regulatory pathway, investigating the PHO pathway dependence of effects of RPD3 disruption on PHO5 expression is important. To test the hypothesis that RPD3 functions in PHO5 repression through PHO pathway regulation of Pho4p activity, four yeast strains (wild type, pho4Δ, rpd3Δ, and pho4Δ rpd3Δ) (Table 1) were compared with regard to levels of acid phosphatase activity and PHO5 transcript. Since Pho4p is the ultimate regulator of PHO5 activity, deletion of PHO4 results in the loss of the PHO5 response to phosphate starvation through the PHO regulatory pathway (see the introduction). The first question addressed here is whether deletion of RPD3 affects PHO5 repression in cells grown on high-phosphate medium when regulation through the phosphate level is disconnected through PHO4 disruption. The second question addressed is whether RPD3 disruption has any effect on PHO5 induction, by determining the kinetics of PHO5 activation in the rpd3Δ strain versus the wild-type strain grown in the absence of phosphate.
PHO5 derepression in rpd3Δ strains is independent of Pho4p.
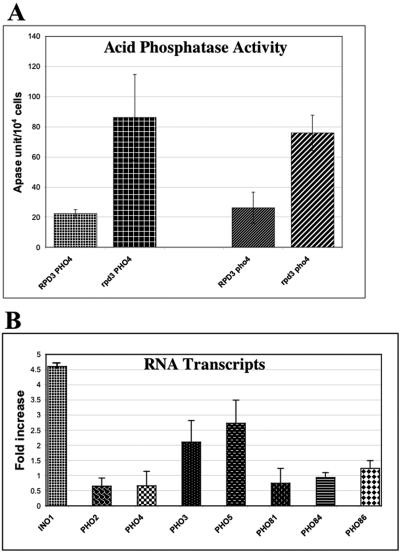
The acid phosphatase activities from both RPD3 deletion strains (PHO4 and pho4Δ) grown on high-phosphate medium were two- to threefold higher (derepressed) than those from the RPD3 wild-type strains (PHO4 and pho4Δ) (Fig. 2A). These results suggest that RPD3 disruption affects PHO5 repression independently of the PHO regulatory pathway and Pho4p activity.
FIG. 2.
Expression of acid phosphatase activity and PHO5 transcript level are derepressed in rpd3Δ cells. (A) Periplasmic acid phosphatase (Apase) activity was measured for wild-type, rpd3Δ, pho4Δ, and rpd3Δ pho4Δ cells at 9 h of growth in medium containing phosphate as described in Materials and Methods. (B) Reverse transcription and real-time PCR were used to quantitate the transcript levels of PHO2, PHO3, PHO4, PHO5, PHO81, PHO84, PHO86, and INO1. The fold derepression is determined by the transcript level in rpd3Δ PHO4 cells over that in RPD3 PHO4 cells under repressing condition (with Pi). INO1 served as a positive control for derepression in rpd3Δ cells (38, 42). All experiments were conducted in triplicate and repeated as three independent experiments. Error bars indicate standard deviations.
Several steps lay between PHO5 transcription and acid phosphatase expression. To verify that the function for RPD3 is through PHO5 transcriptional repression, we quantified PHO5 transcript levels under repressing (high-phosphate) conditions in the same four yeast strains. INO1 transcription repression is known to be highly dependent on RPD3 and therefore served as a positive control (38, 42). In addition, since PHO3 is located directly downstream of PHO5 on chromosome II, we compared the relative levels of PHO3 transcripts in the four strains as a measure of the size of the chromosomal locus affected by RPD3 disruption. As shown in Fig. 2B, derepression was observed from INO1 (4.6-fold), PHO5 (2.7-fold), and PHO3 (2.1-fold). RPD3 disruption had no effect on PHO4 expression, nor did it affect the expression of the Pho4p positive regulatory genes PHO2, PHO81, PHO84, and PHO86, indicating that RPD3 disruption affects repression of the PHO5 transcription through a Pho4p- and, therefore, PHO pathway-independent mechanism (Fig. 2B).
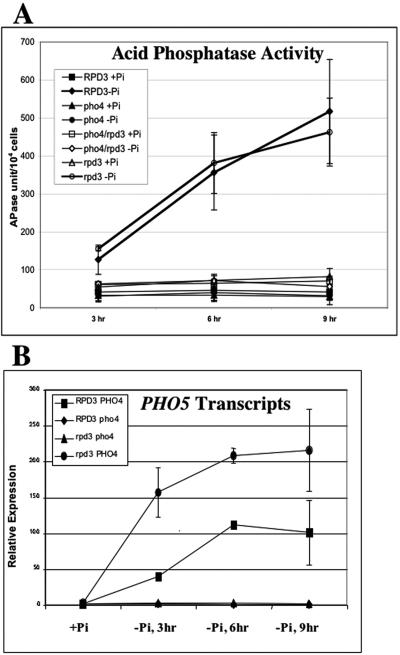
PHO5 activation is potentiated in rpd3Δ strains.
The induction kinetics of acid phosphatase activity are not significantly different between the wild-type and rpd3Δ strains (Fig. 3A). Thus, we were surprised to find that the PHO5 transcript level increases more quickly and to a higher level during phosphate depletion in the rpd3Δ strain (Fig. 3B). This finding is consistent with the role of RPD3 in determining the histone acetylation state on the PHO5 locus. However, we could not rule out the possibility that RPD3 disruption affects PHO5 expression through Pho4p activity, since activation cannot be studied in a pho4Δ strain. To test the hypothesis that RPD3 disruption increased expression of one or more of the other PHO genes in the PHO5 regulatory cascade, we analyzed the effect of RPD3 disruption on PHO81 activation kinetics.
FIG. 3.
PHO5 transcription is potentiated in rpd3Δ cells. (A) Acid phosphatase (APase) activity on cells from each of the four yeast strains (RPD3 PHO4, RPD3 pho4, rpd3 PHO4, and rpd3 pho4) grown in the presence of phosphate (+Pi) or in the absence of phosphate (−Pi). (B) Reverse transcription and real-time PCR were used to quantitate the PHO5 mRNA levels in RPD3 PHO4, rpd3 PHO4, RPD3 pho4, and rpd3 pho4 cells grown in the absence of phosphate for 3, 6, and 9 h as described in Materials and Methods. All experiments were conducted in triplicate and repeated as three independent experiments. Error bars indicate standard deviations.
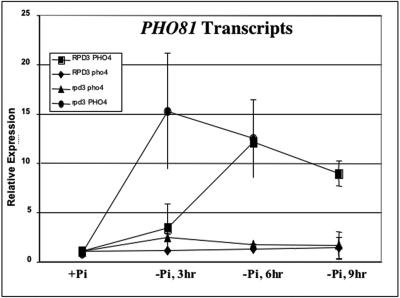
Activation of PHO81 is potentiated by RPD3 disruption.
Pho81p and Pho2p positively regulate Pho4p activity. PHO81 expression is regulated by phosphate availability, and its promoter contains Pho4p binding sites (12, 31; this study). This is not true of PHO2 (4, 51). Therefore, we only tested whether RPD3 disruption affects the activation of transcript accumulation for PHO81. Interestingly, we observed a more rapid induction of PHO81 in the rpd3Δ strain than in the RPD3 wild-type strain (Fig. 4). Since PHO81 is not derepressed in the rpd3Δ strains grown in high-phosphate medium, these findings suggest that RPD3 affects transcriptional activation of PHO81 through a PHO pathway-dependent mechanism only. Our results thus far suggest that RPD3 disruption affects PHO5 induction kinetics through PHO pathway control of Pho4p activity, which responds to the intracellular phosphate level. Therefore, we tested the idea that RPD3 disruption affects phosphate uptake.
FIG. 4.
Activation of PHO81 is potentiated in rpd3Δ cells. Reverse transcription and real-time PCR were used to quantitate the PHO81 mRNA levels in RPD3 PHO4, rpd3 PHO4, RPD3 pho4, and rpd3 pho4 cells grown in the absence of phosphate for 3, 6, and 9 h as described in Materials and Methods. All experiments were conducted in triplicate and repeated as three independent experiments. Error bars indicate standard deviations.
RPD3 disruption results in compromised phosphate uptake.
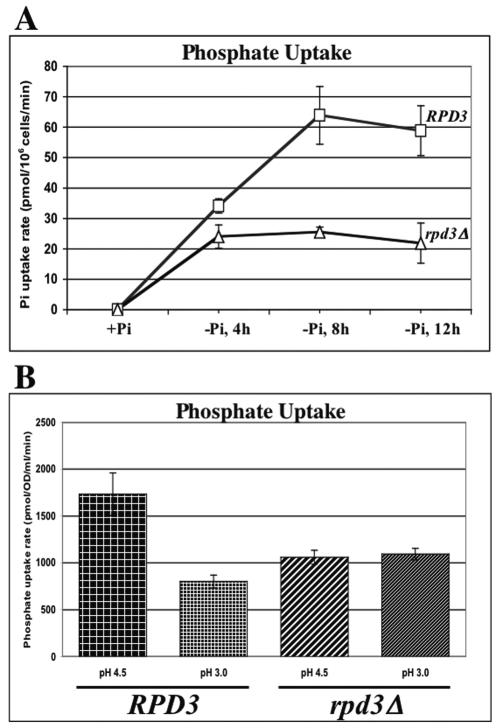
As shown in Fig. 5A, the phosphate uptake rate in rpd3Δ cells is significantly compromised. It is worth noting that the initial inductions of uptake rates (0 to 4 h) in the RPD3 wild-type and deletion strains are quite similar. However, the fully activated rate (low-phosphate adapted, 4 to 12 h) is about threefold lower in the rpd3Δ strain than in the RPD3 wild-type strain. We hypothesized that the mechanism of this decreased uptake rate is decreased transporter activity. As described in the introduction, Pho84p is the primary high-affinity phosphate transporter in yeast cells. A decrease in transporter activity could occur through a loss of proton motive force (PMF) across the plasma membrane or through a decreased Pho84p membrane protein level.
FIG. 5.
Phosphate uptake is defective in rpd3Δ cells but is not rescued by restoration of the proton motive force across the plasma membrane. (A) The rate of phosphate uptake into RPD3 PHO4 (□), and rpd3 PHO4 (▵) cells grown in no-phosphate medium for 4, 8, and 12 h was measured as described in Materials and Methods. The uptake rates are quite comparable in the first 4 hours. However, at 8 h and 12 h the uptake rate observed in the rpd3Δ strain was over twofold lower than that in the wild type. (B) The phosphate uptake rate defect in the rpd3Δ cells cannot be rescued by lowering extracellular pH. At pH 4.5 (normal pH of culture in SC medium), rpd3Δ cells exhibited a phosphate uptake rate almost twofold lower than that observed in wild-type cells. Lower extracellular pH (pH 3) restores a proton gradient across the plasma membrane but does not increase the phosphate uptake rate. The lower extracellular pH does decrease the phosphate uptake rate of the RPD3 wild-type cells to that of the rpd3Δ cells. All experiments were conducted in triplicate and repeated as three independent experiments. Error bars indicate standard deviations.
The proton pump function of Pma1p is essential for phosphate transport. Thus, the reduced phosphate uptake rate observed in the rpd3Δ strain could result from effects on Pma1p activity or expression. A defect in proton pumping is rescued by providing a more acidic extracellular environment to restore the proton motive force across the plasma membrane (25). Thus, we could test the hypothesis that phosphate uptake is compromised by insufficient transmembrane PMF by performing phosphate uptake assays at pH 3.0 and pH 4.5 with the RPD3 wild-type and rpd3Δ strains. As shown in Fig. 5B, increasing the proton gradient across the plasma membrane by lowering the extracellular pH did not rescue the uptake defect. Phosphate uptake in the RPD3 wild-type strain was decreased twofold at pH 3.0 compared to at pH 4.5. A lowered pH does not further inhibit phosphate uptake in the rpd3Δ strain. These results are consistent with the differences in pH optima of the high-affinity (Pho84p) and low-affinity phosphate transporters (43). We will return to the effects of lower pH on phosphate transporter activity in Discussion.
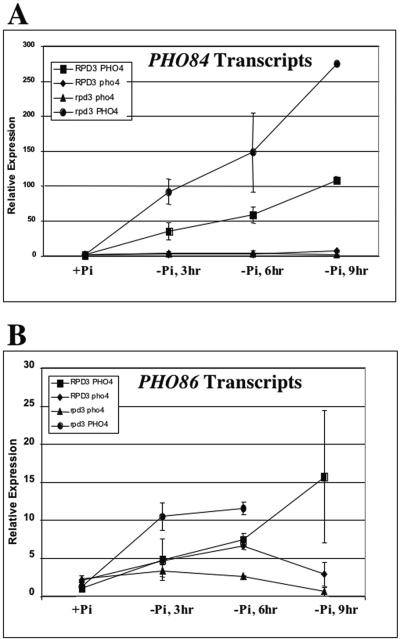
Next, we tested whether RPD3 disruption affects the repression and activation of transcript accumulation for PHO84 and PHO86. Both genes are regulated by phosphate availability and have promoters containing Pho4p binding sites (9, 52; this study). In cells grown in high-phosphate medium PHO84 is unaffected and PHO86 is only slightly derepressed (1.5-fold or less [Fig. 2B]). Paradoxically, we did observe a more rapid induction of PHO84 and PHO86 in the RPD3 deletion strain than in the wild-type strain, and PHO84 was activated to a higher level (Fig. 6A and B). To understand how the RPD3 disruption could result in higher PHO84 transcription and in lower Pho84p activity, we investigated the impact of RPD3 disruption on Pho84p protein level and cellular localization by using fluorescence microscopy of Pho84p-GFP.
FIG. 6.
Activation of PHO84 and PHO86 is potentiated in rpd3Δ cells. Reverse transcription and real-time PCR were used to quantitate the PHO84 (A) and PHO86 (B) mRNA levels in RPD3 PHO4, rpd3 PHO4, RPD3 pho4, and rpd3 pho4 cells grown in the absence of phosphate for 3, 6, and 9 h as described in Materials and Methods. All experiments were conducted in triplicate and repeated as three independent experiments. Error bars indicate standard deviations.
Pho84p is prematurely recycled in rpd3Δ cells.
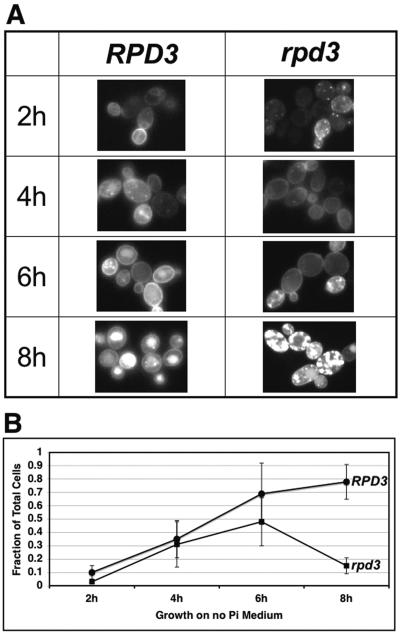
RPD3 wild-type and deletion strains containing the expression plasmid pPHO84-GFP were cultured in phosphate-containing medium to mid-log phase then switched to no-phosphate medium. The Pho84p-GFP chimeric protein expressed from this plasmid complements the pho84 phenotype (24). In both strains the fractions of cells expressing Pho84p-GFP, as measured by the presence of fluorescence, were comparable (80 to 90%). As shown in Fig. 7, RPD3 wild-type and deletion strains harboring the pPHO84-GFP plasmid exhibited a fluorescent signal that was seen as early as 2 h after switching to no-phosphate medium. At 2 h most of the fluorescent signal was associated with newly synthesized protein located in an intracellular compartment (likely the rough endoplasmic reticulum [33]), and the percentage of cells exhibiting Pho84p-GFP localization to the plasma membrane was higher in wild-type (10%) than in rpd3Δ (3%) cells. At 4 h the fraction of cells with plasma membrane-localized Pho84p-GFP increased in both strains (35% and 31%, respectively). Between 6 and 8 h, wild-type cells reached a steady state of Pho84p-GFP plasma membrane localization (69% and 78%, respectively). In contrast, at 6 h and 8 h in rpd3Δ cells the proportion with plasma membrane-localized Pho84p-GFP peaked (48% at 6 h) and then significantly decreased (15% at 8 h). The GFP-Pho84p fluorescence microscopy results correlate well with the effect of RPD3 disruption on phosphate uptake (Fig. 5A and 7). At 4 h the phosphate uptake rate in the rpd3Δ cells was only slightly lower than that in wild-type cells, and the patterns of localization of Pho84p-GFP on the plasma membrane from both RPD3 wild-type and deletion strains were essentially the same, while at 8 h both phosphate uptake and the proportion of cells with membrane-associated Pho84p-GFP were much lower in rpd3Δ cells than in wild-type cells. These findings indicate that RPD3 disruption results in a phosphate uptake defect through an effect on the regulation of Pho84p endocytosis that results in premature internalization in cells starved for phosphate.
FIG. 7.
Pho84p-GFP is prematurely endocytosed in rpd3Δ cells grown on no-phosphate medium. Fluorescence microscopy of chimeric Pho84p-GFP was used to monitor Pho84p expression and cellular localization in RPD3 PHO4 and rpd3 PHO4 cells grown in medium without Pi for 2, 4, 6, and 8 h. (A) Fluorescence micrographs of example fields of cells from the two yeast strains at the four time points. (B) The fraction of cells in which Pho84p-GFP is localized on the plasma membrane is plotted as the average for five independent experiments, with error bars indicating the standard deviation between experiments. The rpd3Δ cells exhibited a significant decrease in Pho84p-GFP localized to the cytoplasmic membrane, compared to no decrease in the wild-type cells, after 8 h of growth in no-phosphate medium.
DISCUSSION
We set out to test the hypothesis that RPD3 disruption affects PHO5 transcriptional regulation through both PHO regulatory pathway-dependent and -independent mechanisms. We found that the increased basal expression of PHO5 and PHO3 observed in this study and others is independent of PHO regulatory pathway control through Pho4p activity. A sin4 mutation also causes derepression of a PHO5 promoter construct lacking the two upstream activation sequences (27). SIN4 encodes a protein component of the Rpd3p complex that is required for transcriptional repression. Our findings are consistent with these earlier results and demonstrate that derepression was not due to the deletion of an upstream repressing sequence upstream of the TATA box.
Accelerated PHO5 activation kinetics in rpd3Δ cells is PHO pathway dependent.
PHO5 reaches half-maximal activity after 1.5 h of phosphate starvation in the rpd3Δ strain, compared to 3 h in the wild-type strain. The growth rates for the RPD3 wild-type and rpd3Δ strains on no-phosphate medium are the same (results not shown); thus, acceleration of activation kinetics does not result from faster depletion of phosphate simply through more rapid growth. Another possibility is feed-forward activation acting through the potentiation of PHO81 activation. However, PHO81 overexpression or constitutive hyperactivity alone has no significant effect on PHO5 expression (11, 19, 21, 30). A third possibility is an effect on phosphate accumulation, which involves phosphate storage as polyphosphate in vacuoles and phosphate uptake (29).
RPD3 disruption does not affect the expression of the polyphosphate synthesis genes (6). In addition, disruption of the polyphosphate synthesis genes results in a plateauing of phosphate uptake in 5 to 10 min rather than after 4 hours as we observed in the rpd3Δ strain, and a partial defect in polyphosphate synthesis has little to no effect on phosphate uptake rates (29). Thus, in contrast to repression, these results suggest that RPD3 disruption affects Pho4p activity through a PHO-signaling pathway-dependent mechanism.
Phosphate uptake is compromised in rpd3Δ cells.
We observe a phosphate uptake defect in the rpd3Δ strain (Fig. 5). This defect is not a result of decreased PHO84 or PHO86 transcription. To the contrary, we see accelerated activation kinetics for mRNA accumulation from these genes, as well.
PHO84 encodes the high-affinity H+/PO43− symporter (8). Therefore, its function is dependent on the proton motive force across the cytoplasmic membrane. For example, if the −ΔG of the proton motive force across the cytoplasmic membrane was reduced to half normal, one would expect a concomitant decrease in the maximum, but not the initial, rate of phosphate uptake when the +ΔG of pumping phosphate across the cytoplasmic membrane reaches equilibrium with the −ΔG of the proton electrochemical gradient. This idea is supported by the finding that extracellular pH affects phosphate uptake kinetics in yeast cells (28, 43). Pma1p, an ATP-dependent H+ pump, maintains the proton motive force. In addition, it has been shown that PMA1 is required for high-affinity phosphate uptake through Pho84p (25). We tested the hypothesis that RPD3 disruption affects phosphate uptake through a reduction in proton motive force across the cytoplasmic membrane by performing phosphate uptake assays at lower (pH 3.0) and higher (pH 4.5) extracellular pHs with RPD3 wild-type and deletion strain cells. Reducing the extracellular pH, to artificially provide a greater proton gradient across the membrane, does not rescue the phosphate uptake defect in the rpd3Δ strain. This indicates that the impact of RPD3 disruption on phosphate uptake is not through depletion of the proton motive force.
Pho84p transport activity is suboptimal at pH 3.5, while the other low-affinity, phosphate transporter activities are near their optimal pH of 4.0 (43). We observe a decrease in phosphate uptake rate in wild-type cells at pH 3.5 to the level seen in the rpd3Δ cells, but the phosphate uptake rate in the rpd3Δ cells is unaffected by lowered pH. These observations are consistent with the idea that the difference in phosphate uptake rates between the RPD3 wild-type and rpd3Δ cells is due to a difference in Pho84p level or activity and that the low-affinity transporters continue to function.
Pho84p cytoplasmic membrane localization is deregulated in rpd3Δ cells.
Expression and recycling of Pho84p are regulated (24, 26, 33). RPD3 disruption potentiates PHO84 transcription, which is not consistent with a decrease in phosphate uptake rate. We tested whether the Pho84p level and plasma membrane localization are affected in the rpd3Δ strain by using the fusion protein Pho84p-GFP and fluorescence microscopy. Disruption of RPD3 affects the timing of Pho84p-GFP endocytosis. In the rpd3Δ strain, localization of Pho84p-GFP to the plasma membrane increases at 4 h after culture in the absence of phosphate. However, the number of cells with Pho84p-GFP localized on the plasma membrane peaks at 6 h and then significantly decreases at 8 h in the rpd3Δ strain cells. In contrast, membrane-localized Pho84p-GFP increases at 6 h and remains high at 8 h in the wild-type cells. These findings correlate well with the defect in phosphate uptake that we observed in rpd3Δ cells. The rate of phosphate transport is more than twofold lower in rpd3Δ cells than in wild-type cells after 8 h of culture in no-phosphate medium. Therefore, we have identified cytoplasmic membrane localization of the high-affinity phosphate transporter Pho84p as the step in phosphate transport affected by RPD3 disruption.
Proposed mechanism for the effect of RPD3 disruption on PHO5 activation kinetics.
Regulation of the timing of Pho84p internalization is aberrant in rpd3Δ cells. Under high-phosphate conditions both transcription and membrane protein levels are down regulated. The intracellular phosphate level regulates PHO84 transcription through a PHO pathway-dependent mechanism. Pho84p recycling in RPD3 wild-type cells has been shown to occur in late-log-phase growth and in response to high extracellular phosphate levels (24, 26, 33). In rpd3Δ cells only membrane localization is abnormal. We propose that aberrant regulation of Pho84p cytoplasmic membrane localization results in deficient phosphate uptake and reduced intracellular accumulation. As a result, rpd3Δ cells deplete their phosphate stores more rapidly and exhibit accelerated PHO5 activation kinetics.
Rpd3p may have a negative function in the regulation of Pho84p endocytosis.
Membrane-associated Pho84p recycling occurs through endocytosis and targeting to the vacuole, where it is degraded (24, 33). Recycling of other membrane-associated proteins in Saccharomyces cerevisiae, such as pheromone receptors and nutrient transporters, involves ubiquitylation- and/or phosphorylation-regulated endocytosis and vacuolar degradation (15, 17, 18, 36, 49). The mechanism of cytoplasmic membrane receptor and transporter protein endocytosis is thought to begin with serine/threonine phosphorylation by casein kinase I (Yck1p/Yck2p), which activates Rsp5p (E3 enzyme) lysine ubiquitylation. PEST-like and “SINNDAKSS” recognition sequences required for the phosphorylation and ubiquitylation have been identified in yeast membrane proteins. Endocytosis and vacuolar targeting require actin (End3p) and actin cytoskeleton-organizing proteins (End4p/Sla2p, End5p, and End6p). Proper Rsp5p cytoplasmic membrane localization is dependent on END4 as well. Finally, ubiquitylated protein binding receptors are involved. Pho84p-GFP internalization and degradation are blocked in an end4ts strain grown at the restrictive temperature (24). However, “SINNDAKSS” sequence mutations have no effect on the regulation of Pho84p localization (23).
While the regulatory mechanism of Pho84p endocytosis is not known, the finding that RPD3 disruption affects the timing of Pho84p endocytosis suggests that Rpd3p has a role in its regulation. We propose that Rpd3p has a negative role in the regulation of Pho84p endocytosis and that RPD3 disruption results in early Pho84p turnover. For example, Rpd3p may inhibit RSP5, END4, END5, or END6 expression or function, and their overexpression or hyperactivity in rpd3Δ cells could result in premature Pho84p endocytosis.
Prior to this study, RPD3 was known to function in regulation of the histone acetylation state on the PHO5 locus. While it has been generally assumed, here we demonstrate that RPD3 disruption results in derepression of PHO5 basal transcription through a mechanism independent of the PHO signaling pathway and Pho4p activity. More importantly, our findings demonstrate that RPD3 disruption affects the kinetics of PHO5 activation through a previously unknown effect on the regulation of Pho84p recycling. Therefore, Rpd3p not only functions to repress PHO5 transcription in the presence of abundant phosphate but also functions in the maintenance of cytoplasmic membrane localization of the high-affinity phosphate transporter Pho84p when free extracellular phosphate in limiting. This finding directs future experimental avenues for establishing the Pho84p endocytic pathway and the role of RPD3 in its regulation.
Acknowledgments
We thank B. Meyhack (Ciba Geigy) for yeast strains YS18 and YS20. We also express our gratitude to E. O'Shea (UCSF) for the plasmid pPHO84-GFP (EB0666). Finally, we thank Laurie Stargell and David Goldstrohm for critical reading of the manuscript and for providing useful and insightful suggestions.
This research was supported in part by a grant from the NSF. S.W. was supported through a scholarship from the Royal Thai Government.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Almer, A., H. Rudolph, A. Hinnen, and W. Horz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berben, G., M. Legrain, and F. Hilger. 1988. Studies on the structure, expression and function of the yeast regulatory gene PHO2. Gene 66:307-312. [DOI] [PubMed] [Google Scholar]
- 5.Bergman, L. W., and R. A. Kramer. 1983. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J. Biol. Chem. 258:7223-7227. [PubMed] [Google Scholar]
- 6.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 8.Borst-Pauwels, G. W. F. H. 1993. Kinetical parameters of monovalent cation uptake in yeast calculated on accounting for the mutual interaction of cation uptake and membrane potential. Biochim. Biophys. Acta 1152:201-206. [DOI] [PubMed] [Google Scholar]
- 9.Bun-Ya, M., M. Nishimura, S. Harashima, and Y. Oshima. 1991. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11:3229-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmen, A. A., S. E. Rundlett, and M. Grunstein. 1996. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271:15837-15844. [DOI] [PubMed] [Google Scholar]
- 11.Creasy, C. L., S. L. Madden, and L. W. Bergman. 1993. Molecular analysis of the PHO81 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 21:1975-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creasy, C. L., D. Shao, and L. W. Begman. 1996. Negative transcriptional regulation of PH081 expression in Saccharomyces cerevisiae. Gene 168:23-29. [DOI] [PubMed] [Google Scholar]
- 13.Ebbert, R., A. Birkmann, and H. J. Schuller. 1999. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 32:741-751. [DOI] [PubMed] [Google Scholar]
- 14.Fascher, K. D., J. Schmitz, and W. Horz. 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 9:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graschopf, A., J. A. Stadler, M. K. Hoellerer, S. Eder, M. Sieghardt, S. D. Kohlwein, and R. J. Schweyen. 2001. The yeast plasma membrane protein Alr1 controls Mg2+ homeostasis and is subject to Mg2+-dependent control of its synthesis and degradation. J. Biol. Chem. 276:16216-16222. [DOI] [PubMed] [Google Scholar]
- 16.Han, M., U. J. Kim, P. Kayne, and M. Grunstein. 1988. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 7:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hicke, L. 1999. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9:107-112. [DOI] [PubMed] [Google Scholar]
- 18.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 19.Huang, S., D. A. Jeffery, M. D. Anthony, and E. K. O'Shea. 2001. Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol. Cell. Biol 21:6695-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaffman, A., N. M. Rank, and E. K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman, P. D., and M. R. Botchan. 1994. Assembly of nucleosomes: do multiple assembly factors mean multiple mechanisms? Curr. Opin. Genet. Dev. 4:229-235. [DOI] [PubMed] [Google Scholar]
- 22.Kramer, R. A., and N. Andersen. 1980. Isolation of yeast genes with mRNA levels controlled by phosphate concentration. Proc. Natl. Acad. Sci. USA 77:6541-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagerstedt, J. O., R. Zvyagilskaya, J. R. Pratt, J. Pattison-Granberg, A. L. Kruckeberg, J. A. Berden, and B. L. Persson. 2002. Mutagenic and functional analysis of the C-terminus of Saccharomyces cerevisiae Pho84 phosphate transporter. FEBS Lett. 526:31-37. [DOI] [PubMed] [Google Scholar]
- 24.Lau, W. T., R. W. Howson, P. Malkus, R. Schekman, and E. K. O'Shea. 2000. Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proc. Natl. Acad. Sci. USA 97:1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau, W. W., K. R. Schneider, and E. K. O'Shea. 1998. A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics 150:1349-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, P., R. Zvyagilskaya, P. Allard, and B. L. Persson. 1998. Physiological regulation of the derepressible phosphate transporter in Saccharomyces cerevisiae. J. Bacteriol. 180:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuno, T., and S. Harashima. 2000. Activation of basal transcription by a mutation in SIN4, a yeast global repressor, occurs through a mechanism different from activator-mediated transcriptional enhancement. Mol. Gen. Genet. 263:48-59. [DOI] [PubMed] [Google Scholar]
- 28.Nieuwenhuis, B. J., and G. W. Borst-Pauwels. 1984. Derepression of the high-affinity phosphate uptake in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 770:40-46. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa, N., K. Noguchi, H. Sawai, Y. Yamashita, C. Yompakdee, and Y. Oshima. 1995. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa, N., K. Noguchi, Y. Yamashita, T. Yasuhara, N. Hayashi, K. Yoshida, and Y. Oshima. 1993. Promoter analysis of the PHO81 gene encoding a 134 kDa protein bearing ankyrin repeats in the phosphatase regulon of Saccharomyces cerevisiae. Mol. Gen. Genet. 238:444-454. [DOI] [PubMed] [Google Scholar]
- 32.O'Neill, E. M., A. Kaffman, E. R. Jolly, and E. K. O'Shea. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209-212. [DOI] [PubMed] [Google Scholar]
- 33.Petersson, J., J. Pattison, A. L. Kruckeberg, J. A. Berden, and B. L. Persson. 1999. Intracellular localization of an active green fluorescent protein-tagged Pho84 phosphate permease in Saccharomyces cerevisiae. FEBS Lett. 462:37-42. [DOI] [PubMed] [Google Scholar]
- 34.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 35.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 36.Riballo, E., M. Herweijer, D. H. Wolf, and R. Lagunas. 1995. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J. Bacteriol. 177:5622-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 39.Sengstag, C., and A. Hinnen. 1987. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 15:233-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 41.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 42.Slekar, K. H., and S. A. Henry. 1995. SIN3 works through two different promoter elements to regulate INO1 gene expression in yeast. Nucleic Acids Res. 23:1964-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamai, Y., A. Toh-e, and Y. Oshima. 1985. Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J. Bacteriol. 164:964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terrell, A. R., S. Wongwisansri, J. L. Pilon, and P. J. Laybourn. 2002. Reconstitution of nucleosome positioning, remodeling, histone acetylation, and transcriptional activation on the PHO5 promoter. J. Biol. Chem. 277:31038-31047. [DOI] [PubMed] [Google Scholar]
- 45.Toh-e, A., Y. Kaneko, J. Akimaru, and Y. Oshima. 1983. An insertion mutation associated with constitutive expression of repressible acid phosphatase in Saccharomyces cerevisiae. Mol. Gen. Genet. 191:339-346. [DOI] [PubMed] [Google Scholar]
- 46.Ueda, Y., A. Toh-e, and Y. Oshima. 1975. Isolation and characterization of recessive, constitutive mutations for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J. Bacteriol. 122:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal, M., and R. F. Gaber. 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:6317-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 49.Wesp, A., L. Hicke, J. Palecek, R. Lombardi, T. Aust, A. L. Munn, and H. Riezman. 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell. 8:2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wykoff, D. D., and E. K. O'Shea. 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159:1491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, K., Z. Kuromitsu, N. Ogawa, and Y. Oshima. 1989. Mode of expression of the positive regulatory genes PHO2 and PHO4 of the phosphatase regulon in Saccharomyces cerevisiae. Mol. Gen. Genet. 217:31-39. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida, K., N. Ogawa, and Y. Oshima. 1989. Function of the PHO regulatory genes for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 217:40-46. [DOI] [PubMed] [Google Scholar]