Abstract
Sporisorium reilianum and Ustilago maydis are two closely related smut fungi, which both infect maize but differ fundamentally in their mode of plant invasion and site of symptom development. As a prelude to studying the molecular basis of these differences, we have characterized the mating type loci of S. reilianum. S. reilianum has two unlinked mating type loci, a and b. Genes in both loci and adjacent regions show a high degree of synteny to the corresponding genes of U. maydis. The b locus occurs in at least five alleles and encodes two subunits of a heterodimeric homeodomain transcription factor, while the a locus encodes a pheromone/receptor system. However, in contrast to that of U. maydis, the a locus of S. reilianum exists in three alleles containing two active pheromone genes each. The alleles of the a locus appear to have arisen through recent recombination events within the locus itself. This has created a situation where each pheromone is specific for recognition by only one mating partner.
Sporisorium reilianum causes head smut in maize and sorghum (41, 42). This soilborne pathogen infects the host plant at the seedling stage (25), supposedly through the roots (37). The infection is systemic, and disease symptoms become apparent only after the onset of flower development when the fungal sori replace male or female inflorescences. In its mode of colonizing the host, S. reilianum is fundamentally different from the well-studied close relative Ustilago maydis, which can infect maize plants via the leaves, stems, or flowers and rapidly causes the formation of prominent tumor-like structures on all green parts of the plant (18). For U. maydis the route of infection has been studied in detail. When haploid, yeast-like sporidia of different mating types recognize each other, they form conjugation hyphae that grow towards each other and fuse at their tips. The resulting dikaryon is filamentous and shows tip growth, leaving empty sections devoid of cytoplasm behind. On the plant surface the dikaryon differentiates into infection structures, so-called appressoria, which are characteristic swellings of the hyphal tips at the site of entry (9, 55). For S. reilianum a detailed molecular investigation of the infection process is lacking. However, it was demonstrated that haploid sporidia as well as germinated teliospores grow filamentously in the vicinity of the root (39). In an in vitro root infection system using germinated teliospores (37), roots were shown to be invaded by hyphae without the apparent formation of appressoria (37).
In U. maydis the initial stages of the infection process are regulated by two unlinked mating type loci (23). The a locus exists in two alleles and encodes a pheromone/receptor system responsible for recognition of haploid sporidia. The lipopeptide pheromones Mfa1 and Mfa2 are secreted by sporidia of the opposite a mating type and are reciprocally detected through specific seven-transmembrane pheromone receptors (Pra2 and Pra1). After detection of the pheromone signal, the cells respond by the formation of conjugation hyphae that grow towards each other and fuse at their tips. After fusion of the sporidia, a filamentous dikaryon is generated and maintained when the two nuclei also carry different alleles at the b mating type locus (8). The b locus is multiallelic and exists in at least 23 alleles (48, 54). It codes for two regulatory genes, bE and bW, whose products dimerize and become an active transcription factor only when the two subunits derive from different b alleles (26, 30). A functional b heterodimer is the key regulator for pathogenicity.
U. maydis has recently been placed in the genus Sporisorium based on sequence comparisons of the nuclear large-subunit rRNA genes (47, 57), suggesting that U. maydis and S. reilianum are closely related. We are intrigued by the differences in the infection process realized by two members of the same genus infecting the same host and would like to elucidate the underlying mechanisms. To begin this study, we have characterized the a and b mating type loci of S. reilianum at the molecular level.
MATERIALS AND METHODS
Strains and growth conditions.
S. reilianum strains were grown in 2.4% potato dextrose (PD) broth (Difco) medium on a rotary shaker at 220 rpm at 24°C or on solid PD agar at 28°C. U. maydis strains were grown in YEPSL (0.4% yeast extract, 0.4% peptone, 2% sucrose) on a rotary shaker at 220 rpm at 28°C or on solid PD agar. Spores of S. reilianum were obtained from L. Claflin (Kansas), B. Garet (Limagrain Genetics, Coussan, France), T. Lübberstedt (Tjele, Danmark), N. McLaren (Potchefstroom, South Africa), M. Piepenbring (Frankfurt, Germany), C. Roux (Castanet Tolosan, France), and X. Xianchun (Beijing, China). Haploid sporidia were isolated from individual germinated spores. All S. reilianum haploid isolates were tested for identity by discriminative PCR (62). The isolates used for functional analysis of the mating type loci were SRZ1 (a1b1), SRZ2 (a2b2), SRZ3 (a1b2) SRZ4 (a2b1), SRZCXII2 (a3b1), SRZCXI1 (a3b2), SRZCXI2 (a3b3), SRZSAW11 (a1b4), and SRZSAW21 (a3b5). U. maydis strains FB1 and FB2, as well as AB1 (a1Δb) and AB2 (a2Δb), were described previously (8, 50). Carboxin was used at 5 μg/ml for S. reilianum and 2 μg/ml for U. maydis. For cloning purposes, the Escherichia coli K-12 derivatives DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used.
Mating assays.
For mating assays, haploid isolates were grown in PD medium for 2 days at 24°C with shaking. A fraction of the culture (0.5 ml) was centrifuged and resuspended in 0.1 volume of water. To assay for the production of conjugation hyphae, 50 μl cells of each strain was added to 400 μl PD and incubated for 1 day at 24°C on a rotary shaker. Conjugation hyphae were scored by microscopic observation. To assay for the production of dikaryotic hyphae, equal amounts of cells resuspended in water were mixed and small drops of the mixtures or the haploid strains were placed on water-agar (1%) plates, which were incubated at 24°C overnight. Dikaryotic hyphae were detected under the binocular microscope as thin aerial filaments on the colony surface.
Plasmids.
Plasmids pCR2.1-TOPO, pCR4-TOPO, and pCR4Blunt-TOPO (Invitrogen) served for cloning and sequencing of fragments generated by PCR. pCR4BamHI is a derivative of pCR4-TOPO, containing a linker that introduces two BamHI sites, and was used for cloning and sequencing of genomic BamHI fragments. pNEBUC (G. Weinzierl, unpublished data) is a vector derived from pNEB (NEB) that contains a U. maydis autonomously replicating sequence (59) and a mutated version of the IP subunit of the U. maydis succinate dehydrogenase gene, which confers resistance against the antibiotic carboxin (31). Plasmids pb1 and pb2 are derivatives of pCR4BamHI and harbor 9-kb BamHI fragments containing the b locus of S. reilianum strains SRZ1 and SRZ2, respectively. pb1Cbx and pb2Cbx are derivatives of pb1 and pb2, respectively, and contain a 2-kb KpnI-SacII fragment conferring Cbx resistance cassette inserted downstream of bE. pSr-b2 is a derivative of pNEBUC and contains the 9-kb genomic BamHI fragment of the b locus of S. reilianum strain SRZ2. For b3, b4, and b5, PCR products were generated using primers oJS31 (CACATACTTCGCGGAAGCC), oJS33 (CAAGCGAGGCTTCTTGAGG), oJS57 (AGTGCTTGCGCATGTGATACG), and oJS69 (TGAGCTGTTCGTAGTTGTGCG) and used directly for sequencing.
Cloning of the a1, a2, and a3 alleles and generation of constructs for functional assays.
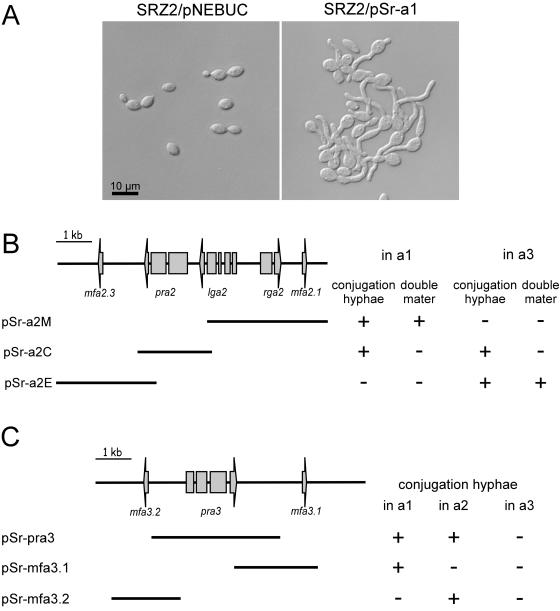
To isolate part of the a1 locus, an 8-kb genomic Sau3A library was generated from strain SRZ1 (a1b1) in pCRBamHI by published procedures (51). An a1 locus fragment was initially identified by hybridization with a probe from U. maydis a1 as described previously (7). The complete a1 locus was represented in two overlapping clones from the genomic library, designated p5211 and p527. These clones were subsequently used to identify a hybridizing 5.5-kb BamHI fragment from the a2 strain SRZ2. This fragment was cloned to yield pG10. Subsequently, an overlapping 3.5-kb genomic SacI fragment was cloned to yield pG11. The right border and the missing intervening sequences were obtained from PCR products generated using primers oJS187 (CAGCTCGACCAACTCAAATACG) and oJS188 (CGAGTCGGATGAGGATTGG), as well as oJS184 (CCTCCACCTCTTCACAAACC) and oJS200 (ACGCAGATTCAATCTCAATCCC). The a3 locus was amplified as a 9.4-kb fragment from strain SRZCXII2 (a3b1) with primers oJS161 (AGCCGTGGTTGAGCAAATCG) and oJS129 (ACATCTACGCGTCGTTCCTCG), using Phusion DNA polymerase (Finnzymes). The product was cloned in pCR4Blunt-TOPO to yield pMW3 and used for sequencing. pSr-a1 is a derivative of pNEBUC and contains the 4-kb genomic BamHI fragment of the a1 locus of S. reilianum strain SRZ1 (a1b1). The plasmids pSr-a2C, pSr-a2E, and pSr-a2M are derivatives of pNEBUC and contain a 2.0-kb Eco47III fragment, a 2.8-kb BamHI-MscI fragment, and the 3.5-kb SacI fragment from pG11, respectively, of the a2 locus of S. reilianum strain SRZ2 (a2b2) (see Fig. 6B). The plasmids pSr-pra3, pSr-mfa3.1, and pSr-mfa3.2 are derivatives of pNEBUC and contain, respectively, a 3.6-kb PCR amplified fragment carrying pra3 that was generated using oJS26 (ATGACTCTGGCTTCGATGGC) and oJS242 (CCTCAAGCTGCCATTTCTCC), a 1.9-kb EcoRI fragment of pMW3 carrying mfa3.2, and a 2.4-kb EcoRV-XbaI fragment of pMW3 carrying mfa3.1 (see Fig. 6C).
FIG. 6.
Functional analysis of the a locus genes of S. reilianum. (A) The S. reilianum strain SRZ2 (a2b2) was transformed with the self-replicating plasmid pNEBUC or with a pNEBUC derivative, pSr-a1, containing the relevant mfa1.2 gene as a 4-kb BamHI fragment of the a2 locus of S. reilianum. Cells were grown in the presence of carboxin in liquid culture and analyzed microscopically. (B) The S. reilianum strains SRZ1 (a1b1) and SRZCXII2 (a3b1) were transformed with derivatives of the self-replicating plasmid pNEBUC containing subfragments of the a2 locus of S. reilianum as indicated in the lower part of the panel. The resulting strains were analyzed for their ability to form conjugation hyphae in the absence of a mating partner and for their double mater phenotype with strains SRZ2 (a2b2) and SRZ3 (a1b2) (left columns) or SRZ2 (a2b2) and SRZCXI1 (a3b2) (right columns). +, positive reaction, −, no detectable phenotype. (C) The S. reilianum strains SRZ1 (a1b1) SRZ2 (a2b2), and SRZCXII2 (a3b1) were transformed with derivatives of the self-replicating plasmid pNEBUC, containing subfragments of the a3 locus of S. reilianum as indicated in the lower part of the panel. The resulting strains were analyzed for their ability to form conjugation hyphae in the absence of a mating partner. +, visible formation of conjugation hyphae; −, no conjugation hyphae detected.
Molecular techniques.
For cloning purposes, molecular methods followed described protocols (51). DNA isolation from S. reilianum and transformation procedures were carried out as described for U. maydis (53). Sequencing of plasmids was performed by the sequencing service of the Max-Planck Institute for Plant Breeding, Cologne, Germany. Predicted amino acid sequences were analyzed using the programs BLAST (3), PSORT (44), InterProScan (43), and MegAlign of the DNASTAR software package. RNA was isolated as described previously (35). cDNA was generated by standard reverse transcription-PCR (RT-PCR) (10). Plant infection of 5-day-old maize seedlings (variety Early Golden Bantam; Olds Seeds, Madison) was done as published earlier (13).
Stains and microscopy.
To visualize nuclei, dikaryotic filaments were transferred onto poly-l-lysine-treated coverslips, fixed for 30 min in 3% formaldehyde, and stained with DAPI (4′,6′-diamidino-2-phenylindole) as described previously (55). Microscopic analysis was performed using an Axioplan II microscope (Carl Zeiss, Jena, Germany). Samples were observed either with differential interference contrast optics or by fluorescence microscopy using the standard 4,6-diamidino-2-phenylindole filter set or a specific filter set (BP470/20, FT 493, BP 505-530) for enhanced green fluorescent protein fluorescence. Processing of images was carried out using Canvas 8.0.6 (Deneba).
Nucleotide sequence accession numbers.
The EMBL/GenBank/DDBJ accession numbers for the b1, b2, b3, b4, and b5 alleles of S. reilianum are AJ884583, AJ884584, AJ884585, AJ884586, and AJ884587, respectively. The EMBL/GenBank/DDBJ accession numbers for the a1, a2, and a3 allele of S. reilianum are AJ884588, AJ884589, and AJ884590, respectively.
RESULTS
a and b mating type alleles of S. reilianum.
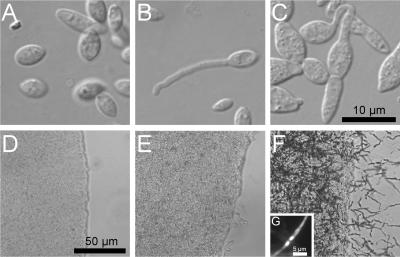
To determine the mating types of different S. reilianum strains, two mating assays were adapted and modified from methods developed for U. maydis (8, 22). The first assay examines differences in a mating type loci by scoring for the formation of conjugation hyphae in liquid (see Materials and Methods). When two haploid S. reilianum isolates of different a mating types were mixed, they responded by the production of conjugation hyphae. These were visible as long, thin hyphal extensions extruding from the lemon-shaped haploid cells (Fig. 1B). When the culture was not shaken vigorously, fusion events could also be detected (Fig. 1C).
FIG. 1.
Mating reactions of S. reilianum strains. (A to C) Single-cell morphologies of mixed sporidial cultures grown under aeration. The strains (mating types) used were as follows: (A) SRZ1 (a1b1) and SRZ3 (a1b2); (B and C) SRZ3 (a1b2) and SRZ4 (a2b1). (D to F) Colony surfaces of mixed sporidial cultures spotted on water agar plates. The strains (mating types) used were as follows: (D) SRZ2 (a2b2) and SRZ4 (a2b1); (E) SRZ2 (a2b2) and SRZ3 (a1b2); (F) SRZ1 (a1b1) and SRZ2 (a2b2). (G) Hyphae produced in (F) are dikaryotic. The hyphae were stained with DAPI and the nuclei visualized by epifluorescence. The size bar in panel C is for panels A, B, and C, and the size bar in panel D is for panels D, E, and F.
The second assay scores the formation of dikaryotic filaments, which can be detected as aerial hyphae emanating from a colony after cospotting. If the two mating partners differ in both a and b mating type loci, dikaryotic filaments are produced, as is well established for U. maydis (8). When different mixtures of S. reilianum isolates were spotted on water-agar plates, three different colony morphologies could be observed (Fig. 1D, E, and F). If strains were identical in a, the colony surface as well as the edge were smooth (Fig. 1D). When the colony had a smooth surface but a ragged edge (Fig. 1E), the mixed strains were identical in b but different in a. Only when the strains were different in a and b, the colony surface was covered by aerial filaments (Fig. 1F), which were shown to be dikaryotic (Fig. 1G).
Using these assays, we tested haploid isolates generated from a spore sample collected in Germany. Four mating types could be differentiated, suggesting the presence of two mating type loci, a and b, existing in two alleles each. To test linkage of these loci, single spores of S. reilianum were germinated and used to isolate haploid progeny. Mating type analysis revealed that from one spore four mating types could be isolated in roughly equal numbers (not shown). This shows that the a and b loci are unlinked in S. reilianum.
The b locus of the close relative U. maydis is estimated to exist in at least 23 alleles (48, 54). To find out whether the b locus of S. reilianum exists in more than two alleles in nature, spore samples derived from different locations in Germany, France, China, the United States, and South Africa were analyzed. Using the mating assays described, five different b alleles could be identified. Surprisingly, mating assays also indicated the presence of three different a alleles (Table 1). This is at variance to all other members of the Ustilaginales investigated, where the a locus exists in no more than two alleles (14, 21, 40, 48, 52). If the a loci of S. reilianum also code for pheromones and receptors (as we show below), this indicates that the a loci code for either multiple receptor or pheromone genes. Alternatively, the a loci might code for receptors that can recognize more than one pheromone. Precedence for this exists in Schizophyllum commune and Coprinopsis cinerea (16, 24).
TABLE 1.
Distribution of mating type loci of S. reilianuma
| Strain genotype | No. of isolates from:
|
||||
|---|---|---|---|---|---|
| Germany | France | China | United States | South Africa | |
| a1b1 | 47 | 25 | 0 | 15 | 0 |
| a1b2 | 12 | 20 | 3 | 3 | 0 |
| a1b4 | 0 | 0 | 0 | 0 | 19 |
| a1b5 | 0 | 0 | 0 | 0 | 14 |
| a2b1 | 2 | 26 | 23 | 7 | 0 |
| a2b2 | 59 | 36 | 26 | 10 | 0 |
| a2b3 | 0 | 0 | 22 | 4 | 0 |
| a3b1 | 0 | 0 | 25 | 0 | 0 |
| a3b2 | 0 | 0 | 31 | 8 | 0 |
| a3b3 | 0 | 0 | 7 | 3 | 0 |
| a3b4 | 0 | 0 | 0 | 0 | 6 |
| a3b5 | 0 | 0 | 0 | 0 | 28 |
| Total haploid progeny analyzed (samplesb) | 120 (>2) | 107 (7) | 137 (4) | 50 (>4) | 67 (3) |
Haploid sporidia were isolated from individual germinated spores and tested for their mating type using the assays described in the text.
Number of spore samples investigated. > indicates that spores were a mixture collected from several infected plants.
Cloning of the b loci of S. reilianum.
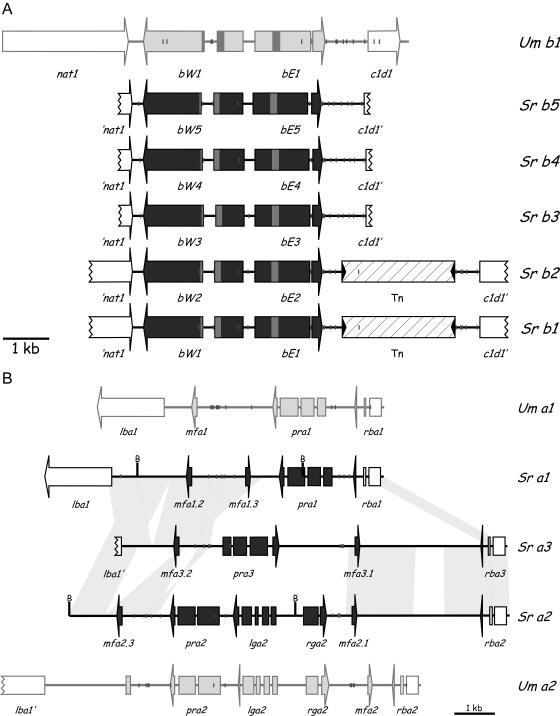
To clone the b locus of S. reilianum, a 450-bp fragment was generated by PCR using primers designed to amplify a conserved region of the b locus of Ustilago scitaminea (2). This PCR product was used as a probe and specifically hybridized to 9-kb BamHI fragments of b1 and b2 strains, which were cloned and sequenced. Using this sequence information, the corresponding regions of b3, b4, and b5 were amplified by PCR and sequenced. Sequence analysis revealed that all alleles contained two divergent open reading frames (ORFs) (Fig. 2A) whose deduced amino acid sequence showed high degrees of identity to the bE and bW proteins of U. maydis and Ustilago hordei. For example, bE1 and bW1 of S. reilianum showed 48.2% and 36.1% amino acid identity, respectively, to bE1 and bW1 of U. maydis and 47.5% and 32.8% amino acid identity, respectively, to bE1 and bW1 of U. hordei. The positions of introns were inferred by sequencing RT-PCR products obtained from RNA isolated from a mixture of compatible strains. Both the bW and the bE genes contained one intron at a position identical to the introns in the b genes of U. hordei and U. maydis (Fig. 2A).
FIG. 2.
Comparison of the mating type genes of S. reilianum to those of U. maydis. (A) Schematic representation of the gene order in the S. reilianum b1 to b5 loci in comparison to U. maydis b1. Open reading frames are indicated by arrows; interruptions mark introns. The bW and bE genes contain a homeodomain motif (indicated by shading in the respective ORFs). The flanking ORFs encode a potential N-terminal acetyltransferase (nat1) and a proposed nuclear regulator, c1d1, related to human C1D, which is implicated in transcription and DNA maintenance (17). The b1 and b2 loci of S. reilianum contain a transposon insertion of 2.5 kb (Tn) (hatched box), which is absent in b3, b4, and b5. The inverted repeats flanking the transposon are indicated by black triangles. (B) Schematic representation of the organization and gene order in the a1, a2, and a3 loci of S. reilianum in comparison to the a1 and a2 loci of U. maydis. Genes are indicated by horizontal arrows interrupted by verified introns. ORFs within the nonhomologous sequences are indicated by closed arrows; flanking ORFs are indicated by open arrows. Relevant BamHI sites (B) are indicated. The pheromone pseudogene in the U. maydis a2 locus is indicated by a single gray box. Nucleotide sequences showing more than 90% sequence identity are indicated by gray areas connecting a loci of S. reilianum. Sr, S. reilianum; Um, U. maydis. Short vertical lines indicate the positions of potential Prf1 binding sites (ACAAAGGGA, allowing at most one mismatch). Figures are to scale.
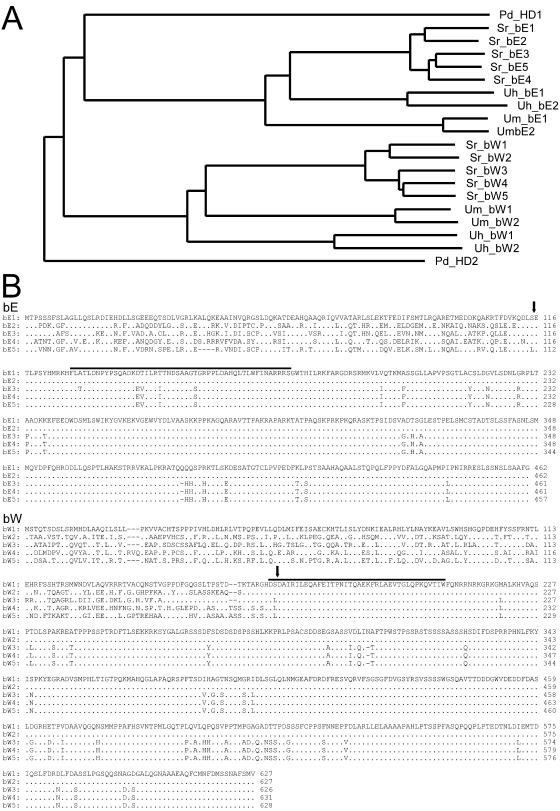
Phylogenetic analysis (Fig. 3A) of the b proteins indicates that the five bE proteins of S. reilianum are more closely related to each other than to the bE proteins of either U. hordei or U. maydis. The same holds true for the S. reilianum bW proteins (Fig. 3A). The b proteins carry a homeodomain motif (Fig. 2A and 3B) and can be divided into two domains, a conserved C-terminal domain and a variable N-terminal domain. The U. maydis b proteins are characterized by a similar domain structure, and in this system it has been shown that the variable domains provide for dimerization and are responsible for self/nonself recognition (30). The C-terminal domains of the bW proteins of S. reilianum are nearly 100% identical, and a similar degree of conservation is apparent between the C termini of bE1 and bE2, while N-terminal regions show at most 59.1% amino acid identity between any two bE or bW alleles (Fig. 3B). The conserved domains of bW1 and bE1 of S. reilianum show 41.5% and 56.6% identity to the respective conserved domains of bW1 and bE1 of U. maydis and 37.8% and 52.9% identity to the respective conserved domains of bW1 and bE1 of U. hordei, while the variable regions are 23.4% and 24.3% identical to the respective variable regions of U. maydis bW1 and bE1 and 23.1% and 32.7% identical to the respective variable regions of U. hordei bW1 and bE1. Thus, bE and bW proteins in S. reilianum display the same overall structure with variable and constant domains as found in other basidiomycetes (16).
FIG. 3.
bE and bW proteins of S. reilianum and orthologues from other fungi. (A) Phylogenetic analysis of bE and bW proteins. Compared are bE1, bE2, bE3, bE4, and bE5 of S. reilianum (Sr); bW1, bW2, bW3, bW4, and bW5 of S. reilianum; bE and bW of the U. maydis b alleles 1 and 2 (Um) (GenBank accession numbers EAK81226 and B32696, both with the additional exon described [53], EAK81227, AAA34221); bE and bW of the U. hordei b alleles 1 and 2 (Uh) (GenBank accession numbers CAA79218, CAA79216, CAA79219, and CAA79217); and the homeodomain proteins HD1 and HD2 of Pleurotus djamor (Pd) (accession numbers AAS46746, AAS46747). The alignment of the protein sequences was done with ClustalX version 1.18 (58) using default values; the tree was generated with NJplot (46). (B) Amino acid alignment of the bE1, bE2, bE3, bE4, and bE5 proteins of S. reilianum (upper panel) and of the respective bW proteins (lower panel). Amino acids identical to bE1 or bW1 are indicated by dots. The end of the variable domain was arbitrarily assigned (vertical arrow) for amino acid comparisons. The region encompassing the homeodomain motif is indicated by a line above the sequence.
The bE and bW genes of S. reilianum are flanked by two genes whose sequences were only partially determined, ′nat1 and c1d1′ (Fig. 2A). These two genes, respectively, encode a potential N-terminal acetyltransferase and a proposed nuclear regulator related to human C1D, which has been implicated in transcription and DNA maintenance (17). Interestingly, these two genes occupy the same genomic position in S. reilianum and U. maydis and show 60% and 72% amino acid identity, respectively, to their counterparts in U. maydis. However, in S. reilianum b1 and b2, a 2.4-kb transposon of the FotI family (20) is inserted between bE and the c1d1 gene (Fig. 2A).
Functionality of the cloned b genes from S. reilianum.
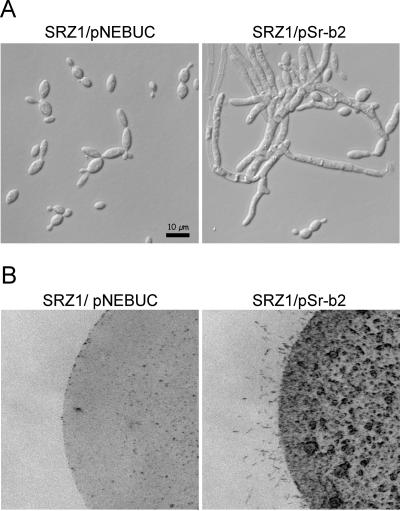
To test for functionality of the bE and bW genes of S. reilianum, we adapted molecular techniques developed for U. maydis for use in S. reilianum. The 9-kb BamHI fragment containing the entire b2 locus of S. reilianum was cloned into the U. maydis self-replicating vector pNEBUC. The resulting plasmid, pSr-b2, was introduced into an a1b1 strain (SRZ1) of S. reilianum by chemical transformation of protoplasts. The resulting strain grew filamentously in liquid culture (Fig. 4A). The filaments formed were thicker than hyphae generated upon pheromone stimulation (compare Fig. 1B), developed branches, and contained septa. When spotted on a water agar plate, the a1b1 strain containing pSr-b2 produced aerial filaments (Fig. 4B). This shows that upon introducing pSr-b2, a functional b heterodimer that triggers filamentation is expressed.
FIG. 4.
Functional assay of the b genes of S. reilianum. The S. reilianum strain SRZ1 (a1b1) was transformed with the self-replicating plasmid pNEBUC or with a pNEBUC derivative, pSr-b2, containing the b2 locus of S. reilianum as a 9-kb BamHI fragment. Cells were grown in the presence of carboxin in liquid culture and analyzed microscopically (A) or spotted on water agar plates (B).
To test whether the b heterodimer of S. reilianum can replace the function of the b proteins in U. maydis, we constructed two plasmids for integration into U. maydis. Plasmids pb1Cbx and pb2Cbx carry the cloned 9-kb b1 and b2 loci of S. reilianum, respectively, as well as a mutated version of the IP subunit of the U. maydis succinate dehydrogenase gene (cbx) conferring resistance against the antibiotic carboxin (31). Both plasmids were integrated in single copy into the cbx locus (36) of the U. maydis strains AB1 (a1Δb) and AB2 (a2Δb) (50), respectively. When the resulting strains, AB1cbx::pb1Cbx and AB2cbx::pb2Cbx, were cospotted on charcoal-containing PD plates, they developed aerial filaments, indicative of dikaryon formation (not shown). When the same strains were coinoculated into maize seedlings, 23 of 74 infected maize plants developed tumors on leaves and stems within 7 days after infection. Control infections with mixtures of AB1 and AB2 failed to induce tumors, while infections with a mixture of compatible wild-type U. maydis strains FB1 and FB2 resulted in 92% tumor formation. This shows that the b proteins from S. reilianum can functionally replace the b proteins from U. maydis, albeit with somewhat reduced efficiency.
Cloning of the a loci of S. reilianum.
A 4-kb BamHI fragment of the S. reilianum a1 strain SRZ1 was identified, which hybridized to a probe derived from the a1 locus of U. maydis (7). The fragment was cloned by screening a size-fractionated library. When used as a probe, this fragment hybridized specifically with a 5.5-kb BamHI fragment of the a2 strain SRZ2. This 5.5-kb fragment was also cloned, and both fragments were sequenced completely. Sequence analysis showed that the cloned fragments contained parts of the a loci of S. reilianum (Fig. 2B). To obtain the complete loci, fragments overlapping the borders were cloned either by screening genomic libraries or by PCR (see Materials and Methods). Based on the sequence information for the S. reilianum a1 and a2 loci, the complete a3 locus was amplified from a3 strain SRZCXII2 by PCR and sequenced.
The a1, a2, and a3 loci of S. reilianum are idiomorphs, which contain 6.2 kb, 11.0 kb, and 9.1 kb of unique sequences, respectively. These heterologous parts are flanked on either side by conserved open reading frames named, respectively, lba and rba (for left border a and right border a) (Fig. 2B). Interestingly, and in contrast to the situation in U. maydis, two pheromone precursor genes, designated mfa1.2 and mfa1.3, could be identified within the unique region of the a1 locus, in addition to a single pheromone receptor gene, pra1 (Fig. 2B). The pra1 ORF is interrupted by three introns, and these were confirmed by sequencing of RT-PCR products derived from RNA isolated from an a1/a2 mating mixture. In the region unique to the a2 locus of S. reilianum, five ORFs were identified (Fig. 2B): the pheromone receptor gene, pra2, which contains two introns; the lga2 ORF, containing four introns; the rga2 ORF, containing one intron; and two pheromone precursor genes, designated mfa2.1 and mfa2.3 (Fig. 2B). The presence of all introns was verified by RT-PCR (not shown). In the region unique to the a3 locus of S. reilianum, one pheromone receptor gene, pra3, and two pheromone precursor genes, designated mfa3.1 and mfa3.2, could be identified (Fig. 2B). The pra3 ORF is interrupted by three introns, which were confirmed by sequencing of RT-PCR products derived from RNA isolated from mating mixtures. The six identified pheromone precursor genes contained one intron each, located 9, 22, and 37 bp, respectively, downstream of the ORF in the 3′ untranslated region (not shown). The presence of introns downstream of open reading frames is unusual but appears to be typical for pheromone precursor genes. Identified examples include the pheromone precursor genes of U. maydis (11), U. hordei (4), and Rhodosporidium toruloides (1). The pheromone receptors Pra1 and Pra2 are most closely related to the Pra1 and Pra2 receptors of U. maydis (69.7% and 63.4% amino acid identity, respectively), while the pheromone receptor Pra3 shows only 24.6% and 24.8% identity, respectively, to Pra1 and Pra2 of U. maydis. Among each other, Pra1, Pra2, and Pra3 of S. reilianum show at most 25.5% amino acid identity. Lga2 and Rga2 are exclusively encoded by the a2 locus of S. reilianum and share 26.9% and 30.0% amino acid identity to the respective proteins of the U. maydis a2 locus. Homologs of Lga2 and Rga2 seem to be absent from the genomes of a1 or a3 strains of S. reilianum (data not shown). This is in accordance with their apparent dispensability for mating or pathogenicity in U. maydis (60). The U. maydis Lga2 and Rga2 proteins localize to mitochondria and are implicated in mitochondrial fusion processes (12). All six identified pheromone genes from S. reilianum are expected to encode pheromone precursors, which are characterized by a CAAX motif (C, cysteine; A, aliphatic amino acid; X, any amino acid) (19) at the C terminus (Fig. 5). This motif serves as signal for isoprenylation and carboxymethylation at the cysteine residue, which becomes the C-terminal residue in the secreted pheromone (15). In addition to the modifications at the C terminus, there is N-terminal processing, which yields mature pheromones, consisting of 9 and 13 amino acids for a1 and a2 pheromones of U. maydis, respectively (11). The three a loci of S. reilianum contain two different pheromone precursor genes each. Mfa1.2 and Mfa3.2 are completely identical in sequence and are most closely related to Mfa1 of U. maydis and Mfa1 of U. hordei (58.5% and 52.4% amino acid identity, respectively) (Fig. 5). The Mfa2.1 and Mfa3.1 precursors are almost identical in sequence, except for two conservative amino acid substitutions at position 6 (serine instead of threonine) and position 20 (aspartate instead of glutamate) (Fig. 5). Mfa2.1 and Mfa3.1 are most closely related to Mfa2 of U. hordei (48.7% and 53.8% amino acid identity, respectively), and to Mfa2 of U. maydis (41.0% amino acid identity each). Mfa1.3 and Mfa2.3 are identical in amino acid sequence but are only weakly related to pheromone precursors of U. hordei, U. maydis, Schizosaccharomyces pombe, and the other pheromone precursors of S. reilianum (between 14.6% and 20.5% amino acid identity).
FIG. 5.
Comparison of the S. reilianum mating pheromone precursors. The pheromone precursors of S. reilianum (Sr) were aligned to the respective pheromone precursors of U. maydis (Um) and U. hordei (Uh). The potential mature pheromone peptide sequences are in boldface (33, 56). The processing sites of the S. reilianum pheromone precursors have not been verified.
The left border of the S. reilianum a locus is situated in the promoter of a putative ORF, lba1 (left border a locus) (Fig. 2B). Lba1 shows 86.1% amino acid identity to the corresponding Lba1 protein of U. maydis. lba1 encodes a conserved protein with unknown function. However, it has a polysaccharide deacetylase domain and a probable signal peptide of 20 amino acids. Near its C terminus a transmembrane domain is predicted, making it likely that Lba1 is associated with the cell membrane. The right border of the nonhomologous sequences is located within the rba1/2/3 genes, which show more than 75% amino acid identity to the Rba1/2 proteins of U. maydis. rba1 encodes a small peptide of 121 amino acids that is predicted to localize to the inner mitochondrial membrane. Rba1 appears to be specific for U. maydis and S. reilianum, since homologs cannot be found in other organisms. Rba1, Rba2, and Rba3 show about 86% amino acid identity to each other, with the N-terminal 104 amino acids being identical and the differences manifesting themselves only in the last 17, 39, and 28 C-terminal residues, respectively. The N-terminal 104 amino acids of Rba1, Rba2, and Rba3, which comprise the predicted membrane-spanning domain, are also conserved in Rba1 and Rba2 of U. maydis, suggesting a common function of this domain.
Functional analysis of pheromone and pheromone receptor genes of S. reilianum.
To determine the functionality of the a genes, the 4-kb BamHI fragment containing the mfa1.2 and mfa1.3 pheromone precursor genes (Fig. 2B) was introduced into the U. maydis self-replicating vector pNEBUC. When the resulting plasmid, pSr-a1, was introduced into S. reilianum SRZ2 (a2b2), the strain responded by the formation of conjugation hyphae in the absence of a mating partner (Fig. 6A). Assay of SRZ2 containing pSr-a1 in the plate mating assay described above revealed that it can form aerial filaments with both an a1b1 strain (SRZ1) and an a2b1 strain (SRZ3) (not shown). This double mater phenotype is characteristic of strains expressing two pheromone genes that can stimulate both a1 and a2 mating partners (11). Since the a2 strain used as recipient of pSr-a1 carries the mfa2.3 pheromone precursor gene that is identical to the introduced mfa1.3 gene, the observed morphological response must be due to the introduction of the mfa1.2 gene. This illustrates that the mfa1.2 gene leads to the expression of a functional pheromone responsible for the induction of conjugation hyphae and the ability to mate with a2 strains.
To functionally analyze the genes of the a2 locus of S. reilianum, different fragments of the locus (Fig. 6B) were cloned into pNEBUC and introduced in the a1b1 strain SRZ1. The resulting strains were analyzed for their ability to form conjugation hyphae in the absence of a mating partner and for a double mater phenotype. Introducing pSr-a2M (Fig. 6B), containing the putative mfa2.1 pheromone gene, into the a1b1 strain SRZ1 caused a double mater phenotype. This shows that the mfa2.1 pheromone gene is functional. The introduction of pSr-a2C, coding for the putative Pra2 receptor, into SRZ1 caused the formation of conjugation hyphae without a mating partner, but the strain did not show a double mater phenotype (Fig. 6B). This phenotype is expected if pSr-a2C contains a functional pheromone receptor gene and if pheromone stimulation of each mating partner is required for successful mating. On these grounds we conclude that the pra2 gene of S. reilianum encodes a functional pheromone receptor. Introducing pSr-a2E (Fig. 6B), carrying the putative mfa2.3 gene, produced no morphological response in SRZ1, indicating that Mfa2.3 is not recognized by the pheromone receptor Pra1. To test whether Mfa2.3 was specifically detected by the Pra3 pheromone receptor, the constructs pSr-a2M, pSr-a2C, and pSr-a2E were also introduced into the a3b1 strain SRZCXII2. In this strain, the introduction of pSr-a2M did not lead to a morphological response, suggesting that Pra3 cannot recognize Mfa2.1. Introducing pSr-a2C, encoding the pheromone receptor Pra2, lead to the formation of conjugation hyphae without a mating partner, confirming that the a3 strain secretes a pheromone that can be recognized by Pra2. When the a3b1 strain SRZCXII2 was transformed with pSr-a2E, encoding Mfa2.3, the resulting strain formed conjugation hyphae without a mating partner and displayed a double mater phenotype. This shows that Mfa2.3 can specifically be recognized by Pra3 but not by Pra1 or Pra2.
A similar set of experiments was done for genes carried on the a3 locus of S. reilianum. To this end, plasmids pSr-pra3, pSr-mfa3.1, and pSr-mfa3.2, which contain, respectively, the pra3, mfa3.1, and mfa3.2 genes, were introduced into an a1 strain (SRZ1), an a2 strain (SRZ2), and an a3 strain (SRZCXII2) (Fig. 6C). As a control, pSr-pra3 was introduced into SRZCXII2, and as expected, this did not lead to the formation of conjugation hyphae. However, when pSr-pra3 was introduced into either SRZ1 or SRZ2, the resulting strains formed conjugation hyphae without a mating partner. This shows that Pra3 can recognize a pheromone encoded in the a1 locus and a pheromone encoded in the a2 locus. Since Mfa1.2 is identical to Mfa3.2, the a1 pheromone recognized by Pra3 must be Mfa1.3. Likewise, the a2 pheromone recognized by Pra3 must be Mfa2.3, since the mature pheromones Mfa2.1 and Mfa3.1 are identical. Plasmid pSr-mfa3.1 induced the formation of conjugation hyphae after introduction into SRZ1 but failed to do so when transformed into SRZ2 or SRZCXII2. This confirms that Pra1 specifically recognizes Mfa3.1. Likewise, introduction of pSr-mfa3.2 into SRZ2 led to strains forming conjugation hyphae without a mating partner, while the same plasmid in SRZ1 or SRZCXII2 failed to do so. This confirms that Pra2 specifically recognizes Mfa3.2. Accordingly, we named the pheromone precursor gene mfa3.2 (i.e., mating factor a3 specific for recognition by Pra2).
DISCUSSION
In this study we have molecularly characterized the a and b mating type loci of S. reilianum. We demonstrate that the b locus of this organism is multiallelic and displays the same features as the b locus of U. maydis. In contrast to the case for the biallelic a locus present in other members of the Ustilaginales, we have uncovered that the a locus of S. reilianum exists in three alleles and displays the unique feature of having two pheromone genes which are alternatively used to stimulate strains carrying the other two a mating types.
We have identified, cloned, and sequenced five different b alleles of S. reilianum. All five alleles show perfect synteny to the b locus of U. maydis with respect to gene order, orientation, and intron position. This holds true also for the adjacent regions, where the same genes are present in U. maydis and S. reilianum and are even more conserved than the b genes themselves. This highlights the highly polymorphic nature of the mating type genes and represents a general feature of self/nonself discrimination systems thought to be maintained by balancing selection (49). Interestingly, the b1 and b2 alleles, which were found in S. reilianum isolates from France and Germany, carried a transposon insertion downstream of the bE gene. The transposon is absent in the b3, b4, and b5 strains from China, the United States, and South Africa. The presence of the transposon exclusively in the European strains indicates that it was acquired only recently. It is possible that the disease, which became prominent in Europe only since the beginning of the 1980s (38), has spread to Europe from the United States or China by just one infected plant. The b4 and b5 alleles were found only in the spore samples from South Africa. This could indicate that b4 and b5 specificities are also of recent origin and have not yet spread.
We have shown by exchanging the b genes in U. maydis with those of S. reilianum that that the b proteins from S. reilianum can functionally replace the b proteins from U. maydis. A similar set of experiments has been carried out with the b proteins of U. hordei in U. maydis (5). However, in that study it was analyzed whether bE or bW of U. hordei can function in combination with the bW or bE protein of U. maydis. The chimeric b proteins triggered filamentous growth, while pathogenicity symptoms were weak or absent (5). These experiments are not directly comparable to our experiments, where we analyzed the ability of the bE/bW heterodimer of S. reilianum to trigger the regulatory cascade in U. maydis. The finding that normal pathogenicity symptoms occur in about 30% of the infected plants demonstrates that the bE/bW complex of S. reilianum functions efficiently in U. maydis despite the fact that the proteins show only 48% and 36% amino acid identity, respectively, to the U. maydis bE and bW proteins. Thus, the regulatory programs initiated by these protein complexes must be very similar or identical in both organisms.
In U. maydis, genes in the a and b loci are all subject to control by the pheromone response factor Prf1. Prf1 is an HMG domain protein that binds to the conserved sequence element ACAAAGGGA (28). Clusters of putative Prf1 binding sites are found in intergenic regions of the a and b loci of S. reilianum as well, although the exact locations of sites differ in U. maydis (Fig. 2A and B). This suggests that the genes of the a and b loci of S. reilianum are under the same kind of control as the a and b genes in U. maydis (28, 29).
Quite unexpectedly, S. reilianum could be shown to contain three a mating type alleles, with each allele encoding two functional pheromones and one pheromone receptor. The six encoded pheromone precursors are all expected to be both N- and C-terminally processed. The presence of a CAAX motif (19) at the C terminus suggests that they are isoprenylated as well as carboxymethylated at their C-terminal cysteine residues. Based on the alignment of the pheromone precursor genes with those from U. maydis and U. hordei, we propose N-terminal processing of the S. reilianum pheromone precursors to yield mature pheromones of 14 (Mfa1.2 and Mfa3.2), 13 (Mfa1.3 and Mfa2.3), and 9 (Mfa2.1and Mfa3.1) amino acids (Fig. 5). When making this assumption, the two pheromones with the same specificity are predicted to have exactly the same sequence. This is supported by biological tests, which show that the mature pheromones encoded by mfa1.2 and mfa3.2 are specific for recognition by Pra2. Similarly, the mature pheromones specified by mfa1.3 and mfa2.3 are specifically recognized by Pra3, while the genes mfa2.1 and mfa3.1 both encode pheromones activating Pra1 (Fig. 6 and data not shown).
The gene order in the a1 locus of S. reilianum shows nearly perfect synteny to that in U. maydis a1, with the exception that the a1 locus of S. reilianum carries the additional pheromone precursor gene (Fig. 2B). The same is true for the a2 locus of S. reilianum, where the gene order is highly conserved in comparison to that in the a2 locus of U. maydis. The spacing between genes seems also largely conserved, with the exception of the region between the pheromone gene mfa2.1 and the right border gene rba2, which spans 0.5 kb in U. maydis a2 and 3 kb in the a2 locus of S. reilianum. The additional region seems to be devoid of obvious open reading frames and shows no homology to the U. maydis genome. Interestingly, the second S. reilianum pheromone precursor gene, mfa2.3, occupies a position in the a2 locus at which in U. maydis a pheromone precursor pseudogene is located (60). This might indicate that in U. maydis, like in S. reilianum, formerly three different a alleles existed. In this case one of these a alleles must have disappeared during evolution of the fungus, and the second unused pheromone precursor genes in the remaining a1 and a2 loci either were lost or accumulated mutations rendering them nonfunctional. As a consequence, the a1 and a2 loci of U. maydis do not share significant regions of sequence identity. Alternatively, the presence of the pseudogene in the U. maydis a2 locus might indicate an early step towards a3 locus evolution in this species. If the invading pheromone precursor gene led to self-stimulated strains, then its conversion into a pseudogene might have been selected for.
When the three a alleles of S. reilianum are compared, it becomes clear that large segments between them must have been exchanged by recombination. The region adjacent to the right border of the a3 locus contains two blocks of sequences, which show 89% and 95% nucleotide sequence identity, respectively, to the a2 locus of S. reilianum (Fig. 2B). The 2.2 kb of sequence adjacent to the left border and including the mfa3.2 gene is also present (with >95% nucleotide sequence identity) near the left border (interrupted by 0.8 kb of sequence) of the a1 locus of S. reilianum (Fig. 2B). Interestingly, the interspersed 0.8-kb segment present in the a1 locus in addition to an adjoining 1.2 kb of sequence including the mfa1.3 ORF are also present (with 99% nucleotide sequence identity) near the left border of the a2 locus, with the 1.2-kb sequence present in inverted orientation (Fig. 2B). It appears that the regions containing the pheromone genes have been exchanged by recombination during evolution of the three a alleles. This also explains why nearly identical pheromone genes can be present in more than one a locus. While it is possible that the a3 locus has acquired its pheromone genes from a1 and a2, the pra3 gene shows only weak homology to either pra1 or pra2 and is expected to have a different origin. Based on the fact that interspecies hybridization within the genus Ustilago has been described (6, 32), it appears feasible that the receptor has been introduced from a close relative not yet identified. In such a scenario, the mfa2.3 and mfa1.3 pheromone genes must have been introduced also from this relative and subsequently distributed to a1 and a2 via homologous recombination.
In the homobasidiomycete mushroom species C. cinerea and S. commune, even more complex mating type loci are known. In these fungi the B locus consists of up to three functionally redundant gene sets, with each encoding one pheromone receptor and carrying up to eight pheromone genes (24, 27, 45, 61). However, in these cases, the pheromones have distinct specificities (with some of them being able to activate more than one receptor) and are unrelated in primary amino acid sequence (24). The multiallelic mating type genes of mushrooms are thought to be generated by gene duplications before becoming divergent (16, 34). Locus integrity is maintained by dissimilarity in primary sequence between allelic versions of the genes. This excludes the possibility that recombination occurs within a group of genes which would bring together compatible gene combinations. In these examples compatibility is generated if mating partners bring together different alleles of just one group of genes. This scenario is very different from the situation uncovered in S. reilianum, where a third allele of the a locus can in part be traced back to a recent recombination event occurring within the locus itself. Thus, while mushrooms increase their number of mating type alleles through recombination in conserved flanking regions, the hemibasidiomycetes achieve the same goal via recombination within loci.
Acknowledgments
We thank L. Claflin, B. Garet, T. Lübberstedt, N. McLarren, M. Piepenbring, C. Roux, and X. Xianchun for gifts of S. reilianum spores.
This work was supported by the German Science Foundation DFG via grant SFB395.
REFERENCES
- 1.Akada, R., K. Minomi, J. Kai, I. Yamashita, T. Miyakawa, and S. Fukui. 1989. Multiple genes coding for precursors of rhodotorucine A, a farnesyl peptide mating pheromone of the basidiomycetous yeast Rhodosporidium toruloides. Mol. Cell. Biol. 9:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, H. H., and S. Schenck. 1996. PCR amplification from a homolog of the bE mating-type gene as a sensitive assay for the presence of Ustilago scitaminea DNA. Plant Dis. 80:1189-1192. [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, C. M., D. A. Willits, P. J. Kosted, E. J. Ford, A. D. Martinez-Espinoza, and J. E. Sherwood. 1999. Molecular analysis of the pheromone and pheromone receptor genes of Ustilago hordei. Gene 240:89-97. [DOI] [PubMed] [Google Scholar]
- 5.Bakkeren, G., and J. W. Kronstad. 1993. Conservation of the b mating-type gene complex among bipolar and tetrapolar smut fungi. Plant Cell 5:123-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakkeren, G., and J. W. Kronstad. 1996. The pheromone cell signaling components of the Ustilago a mating-type loci determine intercompatibility between species. Genetics 143:1601-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakkeren, G., B. Gibbard, A. Yee, E. Froeliger, S. Leong, and J. Kronstad. 1992. The a-loci and b-loci of Ustilago maydis hybridize with DNA sequences from other smut fungi. Mol. Plant-Microbe Interact. 5:347-355. [DOI] [PubMed] [Google Scholar]
- 8.Banuett, F., and I. Herskowitz. 1989. Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banuett, F., and I. Herskowitz. 1994. Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and b loci. Exp. Mycol. 18:247-266. [Google Scholar]
- 10.Beverley, S. 2001. Enzymatic amplification of RNA by PCR (RT-PCR), p. 15.5.1-15.5.6. In M. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 11.Bölker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 12.Bortfeld, M., K. Auffarth, R. Kahmann, and C. W. Basse. 2004. The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1. Plant Cell 16:2233-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brachmann, A., J. Schirawski, P. Müller, and R. Kahmann. 2003. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22:2199-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler, G., H. Boughey, and H. Cauwood. 1978. Mating system of Ustilago longissima in vitro. Trans. Br. Mycol. Soc. 71:203-208. [Google Scholar]
- 15.Caldwell, G. A., F. Naider, and J. M. Becker. 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59:406-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casselton, L. A. 2002. Mate recognition in fungi. Heredity 88:142-147. [DOI] [PubMed] [Google Scholar]
- 17.Chen, E. S., T. Sutani, and M. Yanagida. 2004. Cti1/C1D interacts with condensin SMC hinge and supports the DNA repair function of condensin. Proc. Natl. Acad. Sci. USA 101:8078-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen, J. J. 1963. Corn smut caused by Ustilago maydis. Monograph 2. American Phytopathological Society, St. Paul, Minn.
- 19.Clarke, S. 1992. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu. Rev. Biochem. 61:355-386. [DOI] [PubMed] [Google Scholar]
- 20.Daboussi, M.-J., T. Langin, and Y. Brygoo. 1992. FotI, a new family of fungal transposable elements. Mol. Gen. Genet. 232:12-16. [DOI] [PubMed] [Google Scholar]
- 21.Day, A. W. 1979. Mating type and morphogenesis in Ustilago violacea. Bot. Gaz. 140:94-101. [Google Scholar]
- 22.Day, P. R., and S. L. Anagnostakis. 1971. Corn smut dikaryon in culture. Nat. New Biol. 231:19-20. [DOI] [PubMed] [Google Scholar]
- 23.Feldbrügge, M., J. Kämper, G. Steinberg, and R. Kahmann. 2004. Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7:666-672. [DOI] [PubMed] [Google Scholar]
- 24.Fowler, T. J., M. F. Mitton, L. J. Vaillancourt, and C. A. Raper. 2001. Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics 158:1491-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fredericksen, R. A. 1977. Head smuts of corn and sorghum. Proc. Corn Sorghum Res. Conf. 32:89-104. [Google Scholar]
- 26.Gillissen, B., J. Bergemann, C. Sandmann, B. Schroeer, M. Bölker, and R. Kahmann. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647-657. [DOI] [PubMed] [Google Scholar]
- 27.Halsall, J. R., M. J. Milner, and L. A. Casselton. 2000. Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics 154:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann, H. A., R. Kahmann, and M. Bölker. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 29.Kaffarnik, F., P. Müller, M. Leibundgut, R. Kahmann, and M. Feldbrügge. 2003. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22:5817-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kämper, J., M. Reichmann, T. Romeis, M. Bölker, and R. Kahmann. 1995. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73-83. [DOI] [PubMed] [Google Scholar]
- 31.Keon, J. P., G. A. White, and J. A. Hargreaves. 1991. Isolation, characterization and sequence of a gene conferring resistance to the systemic fungicide carboxin from the maize smut pathogen, Ustilago maydis. Curr. Genet. 19:475-481. [DOI] [PubMed] [Google Scholar]
- 32.Kniep, H. 1926. Über Artkreuzungen bei Brandpilzen. Z. Pilzkunde 5:217-247. [Google Scholar]
- 33.Kosted, P. J., S. A. Gerhardt, C. M. Anderson, A. Stierle, and J. E. Sherwood. 2000. Structural requirements for activity of the pheromones of Ustilago hordei. Fungal Genet. Biol. 29:107-117. [DOI] [PubMed] [Google Scholar]
- 34.Kothe, E., S. Gola, and J. Wendland. 2003. Evolution of multispecific mating-type alleles for pheromone perception in the homobasidiomycete fungi. Curr. Genet. 42:268-275. [DOI] [PubMed] [Google Scholar]
- 35.Krüger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 36.Loubradou, G., A. Brachmann, M. Feldbrügge, and R. Kahmann. 2001. A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40:719-730. [DOI] [PubMed] [Google Scholar]
- 37.Martinez, C., A. Jauneau, C. Roux, C. Savy, and R. Dargent. 2000. Early infection of maize roots by Sporisorium reilianum f. sp. zeae. Protoplasma 213:83-92. [Google Scholar]
- 38.Martinez, C., C. Roux, and R. Dargent. 1999. Biotrophic development of Sporisorium reilianum f. sp. zeae in vegetative shoot apex of maize. Phytopathology 89:247-253. [DOI] [PubMed] [Google Scholar]
- 39.Martinez, C., M. Buée, A. Jauneau, G. Becard, R. Dargent, and C. Roux. 2001. Effects of a fraction from maize root exudates on haploid strains of Sporisorium reilianum f. sp zeae. Plant Soil 236:145-153. [Google Scholar]
- 40.Martinez-Espinoza, A. D., K. J. Dugan, M. E. Bjarko, and J. E. Sherwood. 1992. Improved media for testing the mating reaction and genetic complementation of Ustilago hordei. Can. J. Bot. 70:788-793. [Google Scholar]
- 41.Matyac, C. A., and T. Kommedahl. 1985. Factors affecting the development of head smut caused by Sphacelotheca reiliana on corn. Phytopathology 75:577-581. [Google Scholar]
- 42.Matyac, C. A., and T. Kommedahl. 1985. Occurrence of chlorotic spots on corn seedlings infected with Sphacelotheca reiliana and their use in evaluation of head smut resistance. Plant Dis. 69:251-254. [Google Scholar]
- 43.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bradley, P. Bork, P. Bucher, L. Cerutti, R. Copley, E. Courcelle, U. Das, R. Durbin, W. Fleischmann, J. Gough, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, J. McDowall, A. Mitchell, A. N. Nikolskaya, S. Orchard, M. Pagni, C. P. Ponting, E. Quevillon, J. Selengut, C. J. A. Sigrist, V. Silventoinen, D. J. Studholme, R. Vaughan, and C. H. Wu. 2005. InterPro, progress and status in 2005. Nucleic Acids Res. 33:(Database Issue)D201-D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 45.O'Shea, S. F., P. T. Chaure, J. R. Halsall, N. S. Olesnicky, A. Leibbrandt, I. F. Connerton, and L. A. Casselton. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 47.Piepenbring, M., M. Stoll, and F. Oberwinkler. 2002. The generic position of Ustilago maydis, Ustilago scitaminea, and Ustilago esculenta (Ustilaginales). Mycol. Prog. 1:71-80. [Google Scholar]
- 48.Puhalla, J. E. 1970. Compatibility reactions on solid medium and interstrain inhibition in Ustilago maydis. Genetics 60:461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richman, A. 2000. Evolution of balanced genetic polymorphism. Mol. Ecol. 12:1953-1963. [DOI] [PubMed] [Google Scholar]
- 50.Romeis, T., A. Brachmann, J. Kämper, and R. Kahmann. 2000. Identification of a target gene for the bE-bW homeodomain protein complex in Ustilago maydis. Mol. Microbiol. 37:54-66. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Saxena, K. M. S., and K. Singh. 1966. The mating pattern of Ustilago scitaminea. Indian Phytopathol. 19:286-289. [Google Scholar]
- 53.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schäfer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 54.Silva, J. 1972. Alleles at the b incompatibility locus in Polish and North American populations of Ustilago maydis (DC) Corda. Physiol. Plant Pathol. 2:333-337. [Google Scholar]
- 55.Snetselaar, K. M., and C. W. Mims. 1992. Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84:193-203. [Google Scholar]
- 56.Spellig, T., M. Bölker, F. Lottspeich, R. W. Frank, and R. Kahmann. 1994. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoll, M., M. Piepenbring, D. Begerow, and F. Oberwinkler. 2003. Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Can. J. Bot. 81:976-984. [Google Scholar]
- 58.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukuda, T., S. Carleton, S. Fotheringham, and W. K. Holloman. 1988. Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8:3703-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urban, M., R. Kahmann, and M. Bölker. 1996. The biallelic a mating type locus of Ustilago maydis: remnants of an additional pheromone gene indicate evolution from a multiallelic ancestor. Mol. Gen. Genet. 250:414-420. [DOI] [PubMed] [Google Scholar]
- 61.Vaillancourt, L. J., M. Raudaskoski, C. A. Specht, and C. A. Raper. 1997. Multiple genes encoding pheromones and a pheromone receptor define the B beta 1 mating-type specificity in Schizophyllum commune. Genetics 146:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, M. L., A. E. Melchinger, and T. Lübberstedt. 1999. Species-specific detection of the maize pathogens Sporisorium reiliana and Ustilago maydis by dot blot hybridization and PCR-based assays. Plant Dis. 83:390-395. [DOI] [PubMed] [Google Scholar]