Abstract
Establishment of skeletal muscle lineages is controlled by the MyoD family of basic helix-loop-helix (bHLH) transcription factors. The ability of these factors to initiate myogenesis is dependent on two conserved amino acid residues, alanine and threonine, in the basic domains of these factors. It has been postulated that these two residues may be responsible for the initiation of myogenesis via interaction with an essential myogenic cofactor. The myogenic bHLH proteins cooperatively activate transcription and myogenesis through protein-protein interactions with members of the myocyte enhancer factor 2 (MEF2) family of MADS domain transcription factors. MyoD-E12 heterodimers interact with MEF2 proteins to synergistically activate myogenesis, while homodimers of E12, which lack the conserved alanine and threonine residues in the basic domain, do not interact with MEF2. We have examined whether the myogenic alanine and threonine in the MyoD basic region are required for interaction with MEF2. Here, we show that substitution of the MyoD basic domain with that of E12 does not prevent interaction with MEF2. Instead, the inability of alanine-threonine mutants of MyoD to initiate myogenesis is due to a failure to transmit transcriptional activation signals provided either from the MyoD or the MEF2 activation domain. This defect in transcriptional transmission can be overcome by substitution of the MyoD or the MEF2 activation domain with the VP16 activation domain. These results demonstrate that myogenic bHLH-MEF2 interaction can be uncoupled from transcriptional activation and support the idea that the myogenic residues in myogenic bHLH proteins are essential for transmission of a transcriptional activation signal.
During skeletal muscle development and differentiation, numerous muscle-specific genes are expressed. Members of the myocyte enhancer factor 2 (MEF2) family of transcription factors play an important role in the activation of muscle-specific genes. There are four members of the MEF2 family in vertebrates which are the products of separate genes, mef2a, mef2b, mef2c, and mef2d (6, 9, 18, 26–28, 31, 37, 47). MEF2 factors belong to the MADS (MCM1-Agamous-Deficiens-serum response factor) box family of transcription factors (41). The MADS domain and the adjacent MEF2 domain are highly conserved and comprise the first 86 amino acids of each MEF2 protein. These two domains mediate DNA binding and dimerization and together define the MEF2 subclass of MADS domain proteins (36). MEF2 proteins bind as homo- and heterodimers to an A/T-rich DNA consensus sequence present in the control regions of nearly all muscle-specific genes (10, 14).
Targeted disruption of the mef2c gene in the mouse results in embryonic lethality due to severe cardiac defects (21). Because of this early lethality and because of the genetic redundancy resulting from four mef2 genes in the mouse, the requirement for mef2 gene products in skeletal myogenesis in vivo has been difficult to assess. However, Drosophila melanogaster contains only a single mef2 gene product (19, 34). P-element-mediated disruption of this mef2 gene results in the loss of differentiated muscle cells from all muscle lineages in the fly, demonstrating the requirement for MEF2 protein in the differentiation of skeletal muscle (5, 20, 38).
The MyoD family of myogenic basic helix-loop-helix (bHLH) transcription factors is also essential for the control of the myogenic program. There are four myogenic bHLH proteins, which are all capable of conversion of nonmuscle cells into terminally differentiated myotubes when transfected into nonmuscle cells in culture (35). The myogenic bHLH factors function as heterodimers with a second class of ubiquitously expressed bHLH proteins known as E proteins (such as E12). These factors heterodimerize through their helix-loop-helix (HLH) domains, and their basic domains mediate binding to a consensus DNA sequence, CANNTG, known as an E box (8, 17, 33).
While E12-MyoD heterodimers are capable of converting nonmuscle cells to differentiated myotubes, E12 homodimers are incapable of this effect. Numerous studies have been conducted to determine the protein sequences responsible for the myogenic activity of the myogenic bHLH factors. Substitution of the basic domain of MyoD with that of E12, in a mutant known as MyoD-E12basic, renders MyoD nonmyogenic (11, 12, 44). Similar results have also been obtained with myogenin and myf-5 (7, 46). This myogenic activity has been mapped to two amino acid residues in the core of the MyoD basic domain and one amino acid residue in the junction region of the first helix of MyoD (12). Substitution of the myogenic alanine and threonine in the basic domains of the myogenic bHLH factors with the corresponding two asparagines of E12 renders the myogenic bHLH factors inactive (7, 12, 46). The myogenic activity of these two amino acid residues is also clearly demonstrated by the mutation of the asparagine residues in the MyoD-E12basic mutant back to the alanine and threonine residues normally found in MyoD. This revertant mutant, known as MyoD-E12basic(AT), has full myogenic activity restored (12). Since the MyoD-E12basic mutant binds DNA, it has been postulated that its inability to activate myogenesis is due to a failure of this mutant to interact with an essential myogenic cofactor (11, 12, 40, 44). According to this model, the alanine and threonine would be required for the myogenic bHLH proteins to adopt a conformation compatible with the recruitment of an essential myogenic cofactor.
Recent evidence has suggested that MEF2 proteins may be the cofactors required for myogenic activation by MyoD. This hypothesis stems from the observations that MEF2 proteins serve as transcriptional cofactors for members of the myogenic and neurogenic bHLH families (1, 15, 25, 29, 31). MEF2 and MyoD family members associate through direct physical interaction to synergistically activate transcription and myogenesis (15, 29, 31). This interaction occurs via association of these two heterologous classes of transcription factors through their DNA-binding and dimerization motifs (15, 29). Synergistic activation of transcription by these two factors requires only one factor to be bound to DNA. The bound factor is then capable of recruiting the other factor through protein-protein interactions (29). While MEF2 proteins can potently synergize with myogenic bHLH-E12 heterodimers, these proteins cannot activate transcription in collaboration with E12 homodimers (1, 29).
In this study, we investigated the role of the MyoD basic region in mediating interaction with MEF2 and in transcriptional activation. We show that the myogenic residues, alanine and threonine, in the basic domain are required for MyoD to synergistically activate transcription with MEF2, but they are not required for interaction with MEF2. These findings suggest a two-step model for transcriptional synergy. In step 1 of this model, MyoD and MEF2 must form a complex which then acquires transcriptional competence through a mechanism dependent on the MyoD basic region. The requirement of the MyoD basic region for step 2, transmission of the transcriptional activation signal, can be bypassed through substitution of the MyoD or the MEF2 activation domain with the constitutive activation domain of herpesvirus viral protein 16 (VP16). These results demonstrate that the myogenic amino acid residues in the basic region of the myogenic bHLH factors are essential for transmission of a transcriptional activation signal and support the notion that cofactor binding alone cannot mediate myogenesis in the absence of transmission of the transcriptional activation signal. Furthermore, these results support a model of transcriptional activation in which a cofactor activates transcription through protein-protein interactions by relaying its activation signal through its DNA-bound partner.
MATERIALS AND METHODS
Plasmids.
The expression plasmids for E12 (29), myogenin (3), MyoD (3), MEF2C (3), MyoD-E12basic (29), MyoD-E12basic(AT) [also called MyoD-E12(AT)] (29), mutant MEF2C KR23,24ID (30), and MEF2C-VP16 (also called 1-117/VP16) (1) are described elsewhere. Plasmids pCITE.E12 and pCITE.myogenin were used for in vitro translation of the E12 and myogenin cDNAs and contain the full-length open reading frames of each of the cDNAs cloned as fusion proteins into the translational enhancement vector pCITE-2A (Novagen). The reporter plasmid 4RtkCAT contains four tandem copies of the right E box from the muscle creatine kinase (MCK) enhancer (43). The GAL4-MEF2C bait plasmid, GAL(DBD)-MEF2C, used in trihybrid analyses encodes amino acids 1 to 143 of MEF2C fused at the amino terminus to amino acids 1 through 147 of yeast GAL4 (30). The GAL4-E12 bait plasmid, GAL(DBD)-E12, and the GAL4-dependent reporter plasmid, pG5E1bCAT, which contains five tandem copies of the GAL4 binding site, have been described elsewhere (29). Plasmid E12-VP16 was a gift from Richard Baer and contains the E12 cDNA sequence from the E2-5 gene fused to the VP16 activation domain. The MyoD-VP16, MyoD-E12basic-VP16, and MyoD-E12basic(AT)-VP16 fusion plasmids encode the initiating methionine and bHLH domains of MyoD or the MyoD mutants fused to the activation domain of VP16. The MyoD bHLH fragments were generated by PCR from the full-length expression constructs encoding each of the cDNAs, using the PCR primers 5′-GCGCGAATTCAAGCTTATGGAGAAGCGCAAGACCACCAAC-3′ and 5′-GCGCGCGAATTCGGGCGCGGCGTCCTGGTC-3′. PCRs were conducted with the TaKaRa (Takara Shuzo Co., Ltd.) high-fidelity polymerase to reduce the potential for PCR-generated errors. The PCR primers used contained HindIII and EcoRI restriction enzyme clamps to facilitate subsequent cloning steps into the expression plasmid pCDNAI/amp (Invitrogen). Each of the constructs was sequenced on both strands, using an ABI 373 automated DNA sequencer, to confirm that the intended fusion constructs were correctly generated and that no unintentional mutations were introduced by the PCR.
DNA binding and immunoprecipitation assays.
The ability of myogenin-E12 and MEF2C to associate with each other while both factors were bound to DNA was examined by a coprecipitation assay. For this assay, the MCK right E box (5′-CCCAACACCTGCTGCCTGAG-3′) or the MCK MEF2 site (5′-CTCTAAAAATAACCCT-3′) was labeled with biotin. Oligonucleotides were labeled with biotin by incubating the following together at 37°C for 1 h in a 30-μl reaction volume: 500 pmol of sense-strand oligonucleotide, 8 μl of biotin-16-dUTP, 2 μl of terminal deoxythymidine transferase (25 U/μl), and 6 μl of 5× tailing buffer. Likewise, the MCK right E-box and MCK MEF2 sites were labeled with 32P by incubating the following together at 37°C for 30 min in a 30-μl reaction volume: 60 pmol of sense-strand oligonucleotide, 3 μl of 10× polynucleotide kinase buffer, 1 μl of polynucleotide kinase (10 U/μl), and 12 μl of [γ-32P]rATP. After labeling, oligonucleotides were purified by using a Qiagen tip-5 column and were annealed to unlabeled antisense oligonucleotides to make double-stranded DNA probes. Unlabeled MEF2C and myogenin-E12 cDNAs were transcribed and translated in vitro by using a TNT kit (Promega) for 2 h at 30°C according to the manufacturer’s recommendations. Briefly, plasmids pCITE.E12 and pCITE.myogenin (400 ng of each) were cotranscribed and translated in a 12.5-μl reaction volume, while 300 ng of MEF2C or mutant MEF2C cDNA in plasmid pCDNAI/amp (Invitrogen) was transcribed and translated in a 12.5-μl reaction volume. Transcription reactions were conducted with the bacteriophage T7 RNA polymerase. Binding reactions were conducted by mixing 1 μl of biotin-labeled E box (16 pmol) and 2 μl of 32P-labeled MEF2 site (4 pmol; 105 cpm) (or vice versa) with 5 μl of TNT lysate containing the appropriate proteins or unprogrammed lysate and electrophoretic mobility shift assay buffer in a 30-μl reaction mixture and incubating the mixture at 25°C for 30 min (the buffer and binding conditions have been described elsewhere [32]). Following binding, the reaction mixtures were immunoprecipitated in a 200-μl reaction mixture by using streptavidin-conjugated agarose for 1 h at 4°C. Immunoprecipitates were washed three times with immunoprecipitation buffer (29), and pellets were analyzed for total radioactivity in a scintillation counter.
Cell culture and transfections.
10T1/2 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). Transfections were performed by calcium phosphate precipitation as described elsewhere (3). Briefly, for transient transfections, 60-mm-diameter dishes were seeded at 25% confluence in DMEM plus 10% FCS and without antibiotics 16 h prior to transfection. The cells were then transfected for 16 h, washed once with phosphate-buffered saline, and incubated in DMEM plus 10% FCS for 24 h before harvesting. In each transfection, 6 μg of plasmid DNA was transfected by mixing it with 0.167 ml of 0.25 M CaCl2 and 0.167 ml of 2× BBS (50 mM BES, 250 mM NaCl, 1.5 mM Na2HPO4 [pH 6.95]) and adding this mixture to the cells.
CAT assays.
Transfected cells were harvested, and cellular extracts were prepared by sonication and heat inactivation as described previously (2). Cell lysates were then quantitated for total protein (23), and an equivalent amount of cell lysate (normalized for total protein) from each transfection was assayed for chloramphenicol acetyltransferase (CAT) activity as described elsewhere (2). Reactions were conducted for 5 h at 37°C. Conversion to acetylated forms was analyzed by thin-layer chromatography and quantitated by PhosphorImager (Molecular Dynamics) analysis.
RESULTS
Myogenin-E12 associates with MEF2 while both are bound to DNA.
Previous results have shown that myogenin interacts with MEF2C to synergistically activate transcription and to augment myogenic conversion (29). Likewise, the neuron-specific bHLH transcription factor MASH1 interacts with MEF2 to synergistically activate transcription (1). These previous studies demonstrated that bHLH heterodimers and MEF2 can interact when neither factor is bound to DNA or when only one factor is bound to DNA. When only one factor is bound to DNA, it can recruit the other factor via protein-protein interactions to synergistically activate transcription (1, 29, 31).
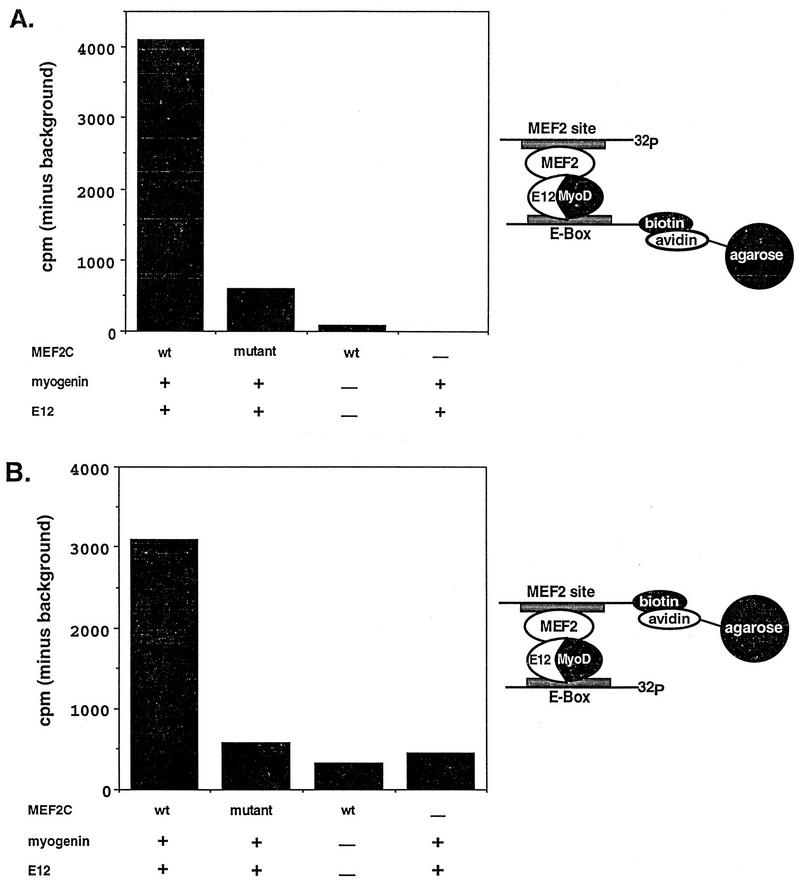
Since the DNA-binding domains of myogenin and MEF2 mediate their interaction with each other, we were interested in whether the two proteins could physically interact while both factors were bound to DNA. To test this, we designed a coprecipitation strategy where a positive signal could be obtained only if both factors were bound to each other and to DNA at the same time. To do this, we labeled an E box with biotin and a MEF2 site with 32P. The biotinylated E box could then be efficiently precipitated by using avidin-conjugated agarose. However, the MEF2 site labeled with 32P could not be precipitated with avidin-conjugated agarose since it lacked a biotin moiety. We reasoned that if myogenin-E12 heterodimers and MEF2 molecules were added to the reaction mixture containing both labeled DNA probes, each would bind to its respective site. If 32P-labeled probe was coprecipitated with the avidin-conjugated agarose, this would indicate that MEF2 and myogenin-E12 were physically associated with each other while bound to their DNA-binding sites. A schematic representation of the experiment is shown in Fig. 1A. The results showed that the 32P-labeled MEF2 site was precipitated only when both myogenin-E12 and MEF2C were present in the reaction mixture. Essentially no counts above background were precipitated if only myogenin-E12 or only MEF2C was included in the assay. Likewise, if a MEF2C mutant (KR23,24ID) that is deficient in its ability to bind DNA (30) but still capable of interaction with myogenin-E12 (4) was included in the reaction mixture, few counts above background were coprecipitated. Furthermore, if a mutant MEF2 site which fails to bind MEF2 was used in this assay, no counts above background were precipitated (data not shown).
FIG. 1.
Double-DNA binding assay for MEF2C and myogenin. Biotin-labeled E-box probe and 32P-labeled MEF2 site probe (A) or biotin-labeled MEF2 site probe and 32P-labeled E-box probe (B) were mixed with either wild-type (wt) or mutant MEF2C and myogenin-E12 proteins transcribed and translated in vitro. A dash indicates the use of unprogrammed reticulocyte in place of MEF2, E12, or myogenin-containing lysate. Complexes were immunoprecipitated with avidin-conjugated agarose and were washed three times. Radioactive counts from the 32P-labeled probes were measured in a scintillation counter. 32P-labeled probe can be precipitated by the avidin-conjugated agarose only if protein-protein interaction occurs. The data are expressed as the counts per minute precipitated minus the background counts per minute precipitated in the presence of unprogrammed lysate alone. The background in panel A was 1,174 cpm, and the background in panel B was 385 cpm. The results shown are from representative experiments. For the experiments shown in both panels, similar results were obtained in three separate immunoprecipitations using three separate preparations of in vitro-translated proteins and labeled probes. The MEF2C mutant used (KR23,24ID) interacts with myogenin-E12 heterodimers (4) but is incapable of binding DNA (30). The schematic representations at the right show how the 32P-labeled probes are coimmunoprecipitated by the avidin-conjugated agarose in these experiments.
In this assay, the 32P- and biotin-labeled DNA-binding sites were used in molar excess to the in vitro-translated proteins. In this regard, the efficiency of the coprecipitation was high since approximately 5% of the total counts from the 32P-labeled sites included in the assay were specifically precipitated. We estimate this amount of precipitated counts to indicate that between 50 and 100% of the MEF2 and MyoD-E12 proteins included in the assay were associated with each other and with the DNA-binding-site probes.
We also reversed the probes and examined the ability of avidin-conjugated agarose to coprecipitate a 32P-labeled E box along with a biotinylated MEF2 site in the presence of myogenin-E12 heterodimers and MEF2C protein (Fig. 1B). In this experiment, a significant number of counts from 32P-labeled sites were precipitated only in the presence of wild-type MEF2C and myogenin-E12. Together, these results demonstrate that MEF2 and myogenin-E12 can physically associate with each other while both factors are bound to DNA.
Analysis of MyoD-E12 and MEF2 interactions in vivo.
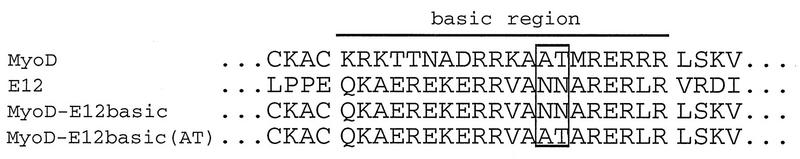
It has also been demonstrated previously that myogenin-E12 heterodimers can synergistically activate transcription in collaboration with MEF2C. We were interested in whether the MyoD-E12 interaction with MEF2 and transcriptional synergy in collaboration with MEF2 could be uncoupled. To investigate this question, we examined the ability of heterodimers of E12 with myogenin, MyoD, and two mutants of MyoD to associate with MEF2 and to activate transcription in collaboration with MEF2. The sequences of the basic domains of these mutants are shown in Fig. 2. The first mutant which we examined, MyoD-E12basic, contains the basic domain of E12 substituted for the basic domain of MyoD (11, 12, 44). The failure of this mutant to activate myogenic transcription has been mapped to two amino acid residues (alanine and threonine) in the core of the basic domain (7, 12). It has been postulated that the failure of this mutant to activate myogenic transcription may be due to its inability to associate with an essential myogenic cofactor (11, 12, 40, 44). Since we have shown previously that MEF2 serves as a cofactor for myogenic bHLH factors (29), we examined the ability of the MyoD-E12basic mutant to associate and collaborate with MEF2. The second mutant which we examined, MyoD-E12basic(AT), contains the basic domain of E12 substituted for the MyoD basic domain as in MyoD-E12basic except that the two asparagine residues in the E12 basic domain have been replaced with the alanine and threonine residues normally found in the MyoD basic domain (12). This mutant serves as a myogenic revertant since it regains the ability to activate myogenic transcription (12).
FIG. 2.
Amino acid sequences of the basic regions examined in this study. The basic domains of MyoD, MyoD-E12basic, MyoD-E12basic(AT), and E12 are indicated at the top. The myogenic residues alanine-114 and threonine-115 of the MyoD basic region and the corresponding asparagine residues of the E12 basic domain are boxed. The junction sequence of the first helix follows the basic domain.
We used an in vivo trihybrid analysis to test the abilities of myogenin, MyoD, and the MyoD mutants to interact with MEF2C and to synergistically activate transcription with MEF2C (Fig. 3A). The results of this analysis are shown in Fig. 3B. Myogenin, MyoD, and MyoD-E12basic(AT) interacted strongly with MEF2C and MEF2C-VP16 to activate transcription, as predicted. Surprisingly, the positive control mutant MyoD-E12basic also interacted with MEF2C-VP16 (lane 15) to activate transcription nearly as well as wild-type myogenin, MyoD, and MyoD-E12basic(AT) (lanes 13, 14, and 16). However, the MyoD-E12basic mutant was incapable of activating transcription in collaboration with wild-type MEF2C (lane 10), whereas myogenin, MyoD, and MyoD-E12basic(AT) strongly activated transcription through interaction with MEF2C (lanes 8, 9, and 11). We interpret these results to indicate that MEF2C interacts with the MyoD-E12basic mutant but fails to transmit its activation signal through this mutant basic domain.
FIG. 3.
Interaction between myogenic bHLH proteins and MEF2C detected by an in vivo trihybrid assay. (A) Schematic representation of the trihybrid assay used in these experiments. In panel B, 10T1/2 cells were transfected with the GAL4-dependent CAT reporter plasmid pG5E1bCAT and the indicated expression plasmids. Plasmids included are indicated by name or with a plus sign and are described in Materials and Methods. A minus sign indicates that the parent expression vector without a cDNA insert was used. The results in panel B show the fold activation in CAT activity compared to that for reporter plus GAL(DBD)-E12 bHLH alone. Extracts were serially diluted such that each sample yielded activity in the linear range of the assay. Total extract was held constant in the serial dilutions by using lysate from untransfected 10T1/2 cells. Results of a representative experiment are shown; similar results were achieved in three independent transfections and analyses. MyoD-wt, wild-type MyoD.
These results support a model whereby the MEF2 activation signal must be transmitted through the bound bHLH heterodimer and not through direct activation of the transcription initiation complex. MyoD-E12basic is incapable of transmitting the transcriptional activation signal provided by MEF2 when MEF2 is bound only through protein-protein interactions. This defect in activation can be overcome by the addition of the acidic activation domain of VP16, which can directly activate the basal transcriptional machinery when associated with the GAL(DBD)-E12 bait through the trihybrid interaction (16, 22, 39). The synergy between MyoD-E12basic, GAL(DBD)-E12, and MEF2C-VP16 observed in lane 15 was due to protein-protein interactions mediated by the MADS and MEF2 domains of MEF2C. The observed synergy was not a result of interaction between E12 and the VP16 sequences in MEF2C-VP16 since E12-E12 homodimers failed to interact with MEF2C-VP16 (lane 12) and since VP16 when expressed alone or when fused to other unrelated proteins failed to activate transcription in association with E12–MyoD-E12basic heterodimers (data not shown). The hypothesis that the MyoD-E12basic mutant is defective in transmission of an activation signal is consistent with the results shown in lanes 3 through 6, where myogenin, MyoD, and the MyoD-E12 basic(AT) revertant were capable of activating transcription when associated with the GAL(DBD)-E12 bait in the absence of MEF2C or MEF2C-VP16 whereas the MyoD-E12basic mutant was dramatically reduced in its ability to activate transcription in this analysis. The inability of the MyoD-E12basic mutant to activate transcription in the presence of GAL(DBD)-E12 is not due to a failure to dimerize since fusion of the VP16 activation domain directly to MyoD-E12basic results in transcriptional activation to a level similar to that observed with MyoD and MyoD-E12basic(AT) fused to VP16 (data not shown).
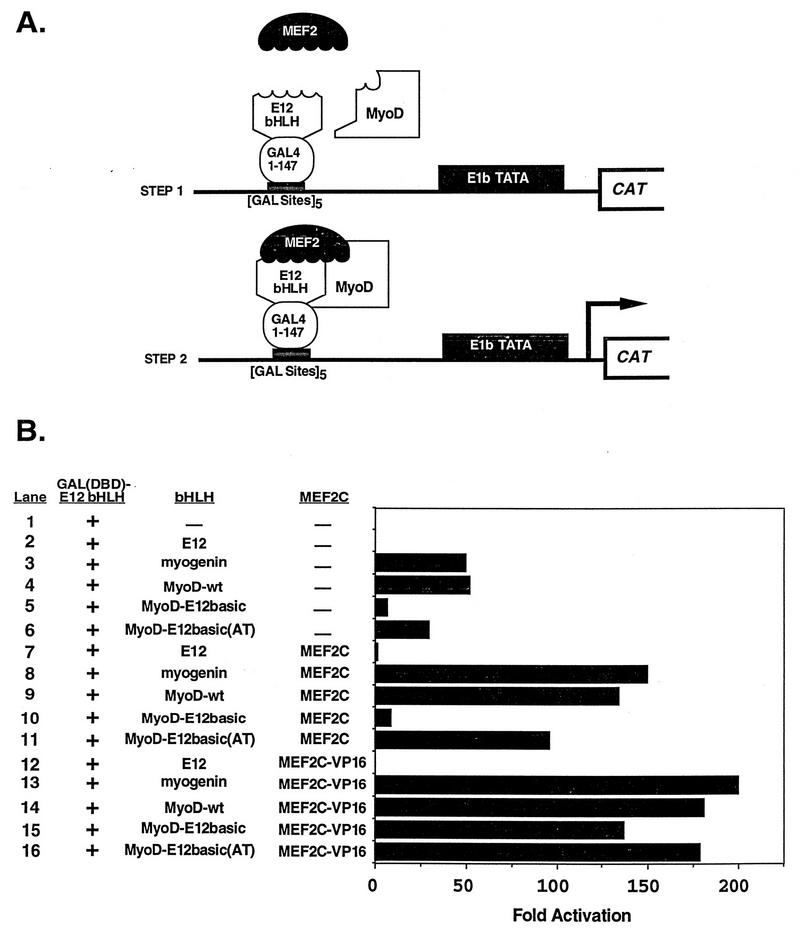
To further examine the ability of the MyoD-E12basic mutant to interact with MEF2C and to determine whether MyoD and the mutants of MyoD could transmit an activation signal through a MEF2 factor bound to DNA via the GAL4 DNA-binding domain, we performed an additional trihybrid analysis using GAL(DBD)-MEF2C as the bait (Fig. 4A). The GAL4 fusion encoded amino acids 1 to 143 of MEF2C, which lacks a transactivation domain (30). Thus, in this trihybrid analysis, the only transactivation domains present were provided by the MyoD proteins interacting with MEF2C. The results presented in Fig. 4B clearly show that the MyoD-E12basic mutant was incapable of activation in collaboration with MEF2C (lane 4), while this mutant fused to the activation domain of VP16 (lane 8) synergized with MEF2C to activate transcription. As predicted, wild-type MyoD and the revertant mutant, MyoD-E12basic(AT), were capable of transcriptional synergy in collaboration with MEF2C both as full-length proteins and as VP16 fusions. Again, we interpret these results to indicate that the MyoD-E12basic mutant interacts with MEF2C but cannot activate transcription due to a defect in transmission of the activation event; this defect can be overcome by fusion to the VP16 activation domain. The interaction and activation observed for the wild-type and mutant MyoD molecules in lanes 7 through 9 were due to interaction of the GAL(DBD)-MEF2C bait with the bHLH portion of MyoD and not due to direct interaction with VP16 since E12-VP16 did not activate transcription (lane 6). The MyoD-E12basic mutant failed to synergize with this full-length MEF2C fused to GAL4, but it did not prevent full-length MEF2C from activating transcription on its own (data not shown). Taken together with the results of Fig. 3, this result indicates that the MyoD-E12basic mutant can block transcriptional transmission provided by the activation domain of MEF2C only when MyoD-E12basic is bound to DNA and not when MEF2 is the DNA-bound factor.
FIG. 4.
In vivo trihybrid analysis of transcriptional synergy mediated by the MEF2C amino terminus. (A) Schematic representation of the trihybrid assay used in these experiments. In panel B, 10T1/2 cells were cotransfected with the GAL4-dependent CAT reporter plasmid pG5E1bCAT, GAL(DBD)-MEF2C, which encodes amino acids 1 to 143 of MEF2C, and E12 expression plasmid. Also included were the indicated bHLH expression plasmids. The presence of GAL(DBD)-MEF2C and E12 is indicated with a plus sign. The absence of a cDNA-encoding plasmid and the presence of the parent expression vector are denoted by a minus sign. The mean fold activation in CAT activity compared to that for reporter alone from four independent transfections and analyses is shown.
Transcriptional activation occurs through MyoD-E12 bound to the E box.
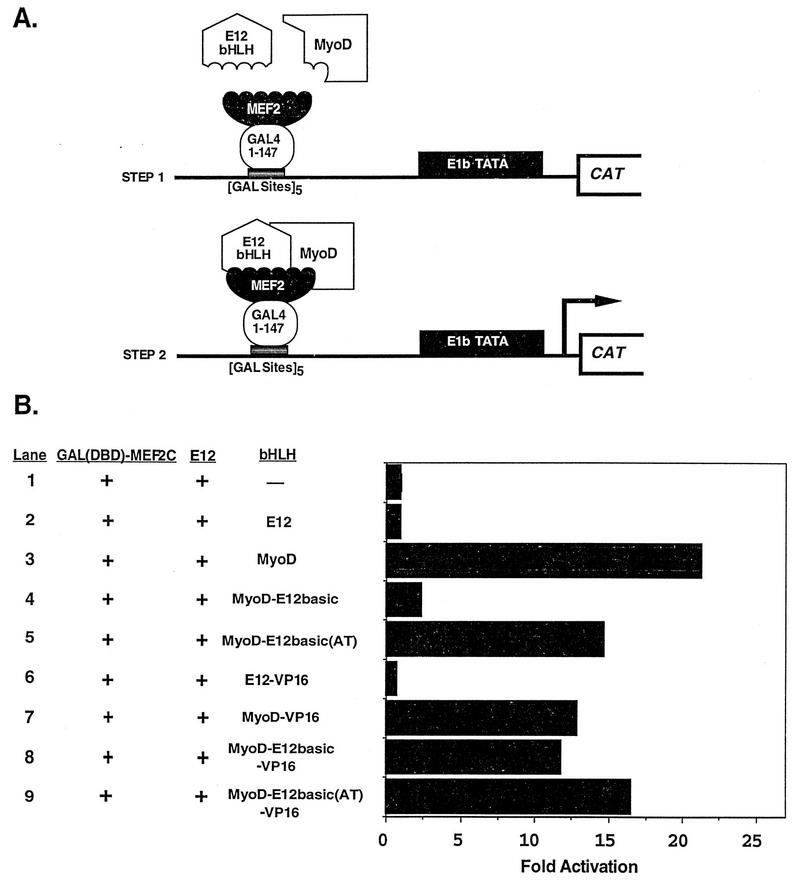
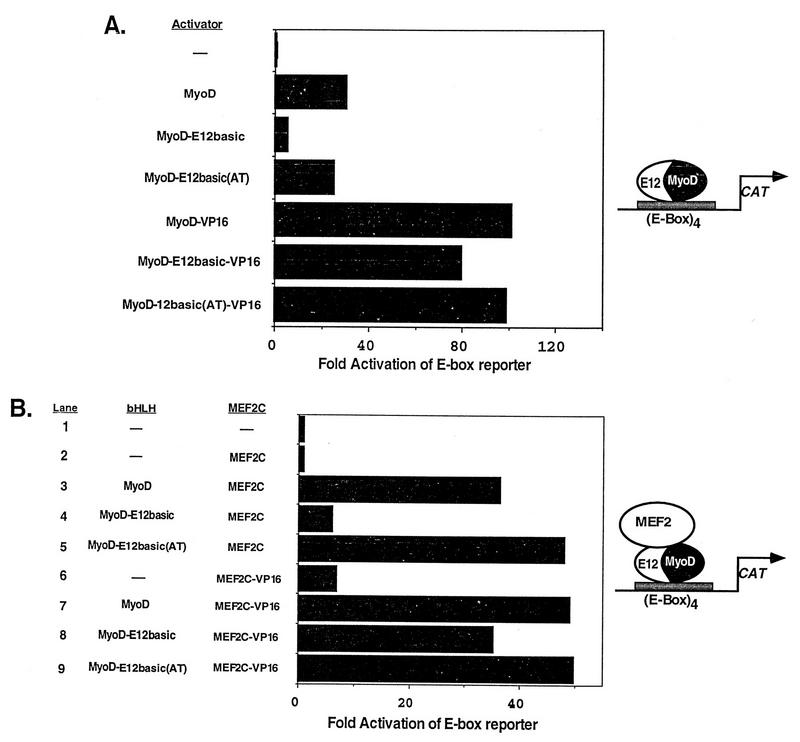
Next, we examined the ability of MyoD and the MyoD mutants to activate the transcription of an E-box-dependent reporter plasmid (Fig. 5A). Each of these proteins has been shown previously to bind to the MCK E box present in this plasmid (12). MyoD and MyoD-E12basic(AT) both activated the reporter greater than 25-fold over the activity of the reporter alone, while the MyoD-E12basic mutant activated the reporter only 6-fold, indicating that this mutant was largely defective in transcriptional activation. However, when fused to the VP16 activation domain, the MyoD-E12basic mutant strongly activated transcription of the reporter to a level similar to those seen for MyoD-VP16 and MyoD-E12basic(AT)-VP16 (Fig. 5A). These observations are consistent with previous reports indicating that the MyoD-E12basic mutant was inefficient in activation of this reporter plasmid whereas MyoD-E12basic-VP16 was capable of transcriptional activation (12, 44).
FIG. 5.
Transcriptional activation of an E-box-dependent reporter by MyoD and MEF2. 10T1/2 cells were transiently transfected with the E-box-dependent reporter 4RtkCAT along with the indicated expression vectors. Plasmids are described in Materials and Methods. (A) Ability of MyoD or the MyoD mutants to activate the reporter as either full-length or VP16 fusion proteins. The data show that the activation defect in MyoD-E12basic is overcome by the fusion of the VP16 activation domain. Values are expressed as the fold induction of CAT activity over the activity of the reporter alone. The results shown represent the mean fold activation obtained in four independent transfections and analyses. (B) Ability of either MEF2C or MEF2-VP16 to activate transcription in collaboration with MyoD or the MyoD mutants bound to the E boxes in the reporter. Wild-type MEF2C was unable to activate transcription through the MyoD-E12basic mutant bound to DNA (lane 4), whereas MEF2-VP16 activated transcription in collaboration with MyoD-E12basic to the same extent as with wild-type MyoD (lane 8). The results shown are from a representative experiment. Similar results were obtained in two independent transfections and analyses. The diagrams at the right illustrate how we envision that transcriptional activation occurs.
Using this same E-box-dependent reporter plasmid, we analyzed the ability of MEF2C or MEF2C-VP16 to activate transcription in collaboration with MyoD, using the bound MyoD-E12 heterodimer as a platform for activation (Fig. 5B). As predicted, strong transcriptional activation was observed for MyoD and MyoD-E12basic(AT) in the presence of MEF2C (lanes 3 and 5). However, only weak activation was observed in the presence of MyoD-E12basic and MEF2C (lane 4). Once again, the lack of transcriptional activation by MEF2C in the presence of MyoD-E12basic was overcome by fusion of the VP16 activation domain to MEF2C (lane 8). In the presence of MEF2C-VP16, all three of the MyoD molecules activated transcription to similar levels (lanes 7 through 9). While fusion of the VP16 activation domain was able to overcome the transactivation defect in the MyoD-E12basic mutant, MyoD-E12basic-VP16 is incapable of activating the myogenic program (12, 44). The failure of MyoD-E12basic-VP16 to activate myogenesis indicates that while this fusion protein is capable of interaction and activation in collaboration with MEF2, it is incapable of initiating myogenesis. These results again support the notion that the MyoD-E12basic mutant is incapable of transmitting a transcriptional activation signal provided by MEF2 when MEF2 is bound only through protein-protein interactions and suggest that the MEF2 activation signal must be transmitted through the bound bHLH heterodimer.
DISCUSSION
The role of the basic regions of the myogenic bHLH factors has been the focus of intense interest because these factors can initiate myogenesis in a wide variety of cell types, while other bHLH factors cannot initiate myogenesis even though they bind to the same DNA sequence. Since this myogenic activity has been mapped to the basic regions of these factors, detailed analysis of this domain in the myogenic bHLH factors has provided insight into the mechanisms of cell-type-specific transcription mediated by protein-protein interactions and protein conformation. The results of this study demonstrate that the myogenic defect in the MyoD-E12basic mutant is not due to a failure to interact with MEF2 factors. This mutant interacts with MEF2C to activate transcription as well as wild-type MyoD if either MEF2C or MyoD-E12basic has the constitutive VP16 activation domain substituted for its own activation domain (Fig. 3 to 5). However, despite the fact that MyoD-E12 basic interacts with MEF2C, it cannot activate transcription on its own or in collaboration with MEF2C due to a defect in transmission of transcriptional activation signals (Fig. 3 to 5). The observation that the alanine and threonine present in the MyoD basic domain are not essential for MEF2 interaction is consistent with previously published results that MASH1, which lacks the alanine and threonine, effectively interacts with MEF2 factors to synergistically activate transcription (1, 25).
While MyoD-E12basic/E12 heterodimers efficiently interact with MEF2C, E12 homodimers fail to interact with MEF2C even if the VP16 activation domain is present. This result suggests that the basic domains alone are not sufficient to mediate the interaction with MEF2 since the basic domains are the same in these two dimers. The interaction of MyoD with MEF2 probably requires sequences present in both the basic and HLH domains of MyoD. We know that an HLH domain alone is not sufficient to mediate the interaction with MEF2 since neither Id, an HLH protein which lacks a basic domain, nor a MyoD mutant with the basic domain deleted will interact with MEF2C even though these factors efficiently dimerize with E12 (4).
Previously published work has shown that myogenic bHLH factors can collaborate with MEF2 to synergistically activate transcription when only one factor is bound to DNA (29, 31). However, many muscle-specific promoters and enhancers contain MEF2 sites and E boxes in proximity to one another, suggesting that both classes of transcription factors may be bound to DNA at the same time while interacting with each other. The results of Fig. 1 show that MEF2C and myogenin can interact with each other while both are bound to DNA. This result demonstrates the multifunctional nature of the DNA-binding and dimerization motifs of these transcription factors since these domains mediate DNA binding at the same time that they are facilitating protein-protein interactions with heterologous classes of transcription factors. The notion that MEF2 and myogenic bHLH factors associate with each other while they are bound to DNA is also interesting since this result suggests that the proximity of MEF2 sites and E boxes in muscle-specific regulatory regions may serve to bring these two classes of DNA-binding factors into high local concentration at or near muscle-specific promoters. Alternatively, both MEF2 and myogenic bHLH factors bound to DNA may stabilize the protein-protein interactions between them to more efficiently activate transcription.
Previously, the explanations that have been postulated to account for the activation defect in the MyoD-E12basic mutant have focused on the idea that a myogenic cofactor is essential for myogenic bHLH-mediated activation of transcription and that basic domain mutants are unable to interact with such a cofactor (7, 11, 12, 40, 44). Fusion of the VP16 activation domain to MyoD-E12basic overcomes the transactivation defect in that mutant, suggesting that the constitutive activation domain of VP16 may circumvent the need for an essential myogenic cofactor, thereby allowing transcriptional activation (12, 44). The results of this study support the notion that the defect in MyoD-E12basic is due to a conformational change negatively influencing the conformation of MyoD rather than its ability to interact with an essential cofactor. This idea is supported further by the crystal structure of MyoD-E12 heterodimers bound to DNA, which shows that the alanine and threonine of the basic domain are not exposed on the surface of the molecule, making the direct contact of these two residues with a cofactor unlikely (24). The crystal structure suggests instead that the alanine and threonine are required for the proper conformation of the MyoD protein such that when they are mutated, the overall conformation of the molecule is affected, rendering it transcriptionally inactive (24). The results of this study support the idea that the MyoD-E12basic mutant contains a conformational defect preventing transcriptional activation regardless of whether it is bound to DNA since GAL4 fusions of MyoD-E12basic (Fig. 3) and MyoD-E12basic associated with MEF2 bound to DNA (Fig. 4) are also incapable of transmitting a transcriptional activation signal. This transactivation defect in the MyoD-E12basic mutant likely results from a defective conformation due to sequences in both the basic and HLH domains since the E12 basic domain is not transcriptionally deficient in the context of native E12. However, the E12 basic region when juxtaposed with the MyoD HLH domain results in defective transcriptional transmission.
The failure of MyoD-E12basic to activate transcription in collaboration with MEF2C originally suggested to us that MyoD-E12basic probably failed to interact with MEF2C (29). The results of this study confirm that MyoD-E12basic cannot collaborate with MEF2C to activate transcription but show that the failure to activate transcription is not due to an inability to interact with MEF2 proteins but rather is due to a failure to transmit a signal required for the activation of transcription. The ability of VP16 fused to MEF2C to overcome this activation defect likely is due to the constitutive nature of VP16 activation. VP16 can directly stimulate activated transcription through direct contact with the transcription initiation complex (16, 22, 39). Even though fusion of the VP16 activation domain to basic domain mutants of the myogenic bHLH factors overcomes the transcriptional transmission defect, these VP16 fusions are still unable to initiate myogenesis in transfected cells (12, 40, 44, 46). There are several possible explanations to account for this observation. The first is that MyoD-E12basic cannot interact with another myogenic cofactor other than MEF2 which is required to initiate myogenesis. Another possibility is that while MyoD-E12basic-VP16 can activate transcription through the multimerized E boxes present in the reporter plasmid 4RtkCAT, it may be unable to efficiently bind to or activate transcription through all E boxes present in essential muscle-specific genes. Finally, specific repression of MyoD-E12basic may occur on muscle-specific enhancers preventing activation of the myogenic program due to specific cis-acting repressor sequences which target the E12 basic domain and inhibit activation of those genes (45).
In addition to MEF2 proteins, there may be other factors which interact with MyoD and with MyoD-E12basic. Some of these factors may be able to overcome the activation defect present in MyoD-E12basic through protein-protein interactions and direct activation of the basal transcriptional machinery. This notion stems from the observation that while MyoD-E12basic is defective in transcriptional activation in 10T1/2 and most other cell types, there are some cell types in which MyoD-E12basic has the ability to activate transcription of reporter genes without a constitutive activation domain fused to MyoD-E12basic (44). If this is the case, the absence of such a factor still cannot account for the entire defect in the MyoD-E12basic mutant since this mutant remains nonmyogenic even in cell types where it is transcriptionally active (44).
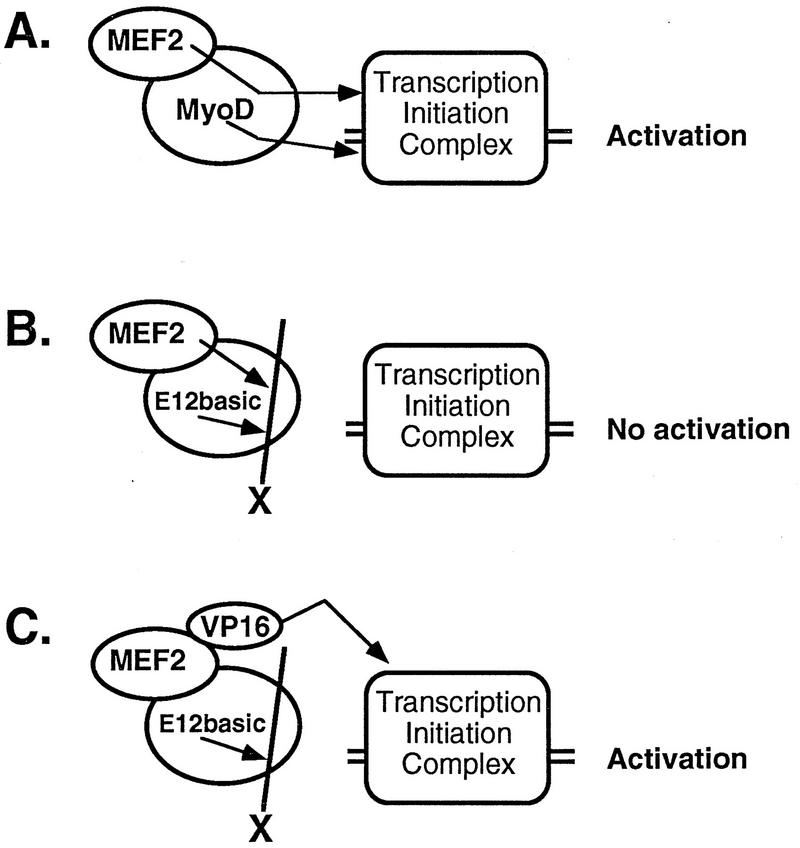
The results of the present study suggest a model for activation in which MEF2 functions as a transcriptional cofactor for MyoD while MyoD is bound to DNA (Fig. 6). Both the MyoD and MEF2 transactivation signals are transmitted to the transcription initiation complex via a mechanism dependent on the myogenic residues in the MyoD basic domain (Fig. 6A). Mutation or substitution of the MyoD basic domain with the basic domain of E12 allows interaction to occur but blocks the transmission of the activation signal (Fig. 6B). This block in activation can be overcome by the addition of the VP16 activation domain (Fig. 6C) which can directly associate with components of the transcription initiation complex to activate transcription (16, 22, 39, 42). The block in transcriptional transmission likely occurs as a result of a conformational change in MyoD which prevents activation. This may result from a failure of the MyoD-E12basic mutant to properly remodel chromatin in such a way that transcriptional activation occurs. This hypothesis is supported by recent observations that wild-type MyoD initiates extensive chromatin remodeling when bound to muscle-specific control regions (13). Alternatively, MyoD-E12basic may support interaction with a repressor protein which can block the activation signals provided by both MEF2 and MyoD but not the activation signal transmitted by VP16 when MyoD-E12basic is the DNA-bound factor, or the MyoD basic domain may mediate a covalent modification of the basal transcriptional machinery required for activation to occur. These results suggest a novel mechanism for the activation of transcription in which a transcriptional activator synergistically activates transcription through protein-protein interaction and relies on its DNA-bound cofactor to relay its activation signal to the basal transcription initiation complex.
FIG. 6.
Hypothetical model of transcriptional activation mediated by MEF2 bound to MyoD. (A) MEF2 relays its activation signal to the myogenic bHLH factor, which then transmits both its own activation signal and that of MEF2 to the transcription initiation complex. This transcriptional transmission is dependent on the myogenic residues, alanine and threonine, in the myogenic factor’s basic domain. (B) MEF2 sends its activation signal to the basic domain of E12 substituted into the myogenic bHLH factor (E12basic), but the E12 basic domain substitution is unable to transmit that activation signal or its own activation signal to the initiation complex due to a conformational defect. (C) VP16 directly activates the transcription initiation complex, thus bypassing the transcriptional block caused by E12 basic domain. The E12basic mutant is unable to transmit its own activation signal because it lacks the myogenic residues.
ACKNOWLEDGMENTS
We thank Andrew Lassar (Harvard University) and Richard Baer (UT Southwestern) for plasmid constructs and Mike Perry for critical review of the manuscript. We appreciate the technical assistance provided by Kathy Kunkle.
This work was supported by grants from the National Institutes of Health, the Muscular Dystrophy Association, the Robert A. Welch Foundation, and the Human Frontiers Science Foundation (to E.N.O.). B.L.B. and J.D.M. were supported by postdoctoral fellowships from the American Cancer Society and the National Institutes of Health, respectively.
REFERENCES
- 1.Black B L, Ligon K L, Zhang Y, Olson E N. Cooperative transcriptional activation by the neurogenic bHLH protein MASH1 and members of the MEF2 family. J Biol Chem. 1996;271:26659–26663. doi: 10.1074/jbc.271.43.26659. [DOI] [PubMed] [Google Scholar]
- 2.Black B L, Lyles D S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black B L, Martin J F, Olson E N. The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J Biol Chem. 1995;270:2889–2992. doi: 10.1074/jbc.270.7.2889. [DOI] [PubMed] [Google Scholar]
- 4.Black, B. L., and E. N. Olson. Unpublished observations.
- 5.Bour B A, O’Brien M A, Lockwood W L, Goldstein E S, Bodmer R, Taghert P H, Abmayr S M, Nguyen H T. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 6.Breibart R, Liang C, Smoot L B, Laheru D, Mahdavi V, Nadal-Ginard B. A fourth human MEF-2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- 7.Brennan T J, Chakraborty T, Olson E N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan T J, Olson E N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990;4:582–595. doi: 10.1101/gad.4.4.582. [DOI] [PubMed] [Google Scholar]
- 9.Chambers A E, Kotecha S, Towers N, Mohun T J. Muscle-specific expression of SRF-related genes in the early embryo of Xenopus laevis. EMBO J. 1992;11:4981–4991. doi: 10.1002/j.1460-2075.1992.tb05605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cserjesi P, Olson E N. Myogenin induces muscle-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol Cell Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R L, Cheng P-F, Lassar A B, Weintraub H. The MyoD binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 12.Davis R L, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 13.Gerber A N, Klesert T R, Bergstrom D A, Tapscott S J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 14.Gossett L A, Kelvin D J, Sternberg E A, Olson E N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaushal S, Schneider J W, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic bHLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 18.Leifer D, Krainc D, Yu Y T, McDermott J C, Breibart R, Heng J, Neve R L, Kosofsky B, Nadal-Ginard B, Lipton S A. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lilly B, Galewski S, Firulli A B, Schulz R A, Olson E N. mef2: a MADS gene expressed in the differentiating mesoderm and the somatic muscle lineage during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 21.Lin Q, Schwarz J J, Bucana C, Olson E N. Control of mouse cardiac morphogenesis and myogenesis by the myogenic transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Y-S, Green M R. Mechanisms of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 23.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Ma P C M, Rould M A, Weintraub H, Pabo C O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 25.Mao Z, Nadal-Ginard B. Functional and physical interactions between mammalian achaete-scute homolog 1 and myocyte enhancer factor 2A. J Biol Chem. 1996;271:14371–14375. doi: 10.1074/jbc.271.24.14371. [DOI] [PubMed] [Google Scholar]
- 26.Martin J F, Miano J M, Hustad C M, Copeland N G, Jenkins N A, Olson E N. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin J F, Schwarz J J, Olson E N. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDermott J C, Cardoso M C, Yu Y T, Andres V, Leifer D, Krainc D, Lipton S A, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin J D, Black B L, Martin J F, Olson E N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molkentin J D, Firulli A B, Black B L, Martin J F, Hustad C M, Copeland N, Jenkins N, Lyons G, Olson E N. MEF2B is a potent transactivator expressed in early myogenic lineages. Mol Cell Biol. 1996;16:3814–3824. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molkentin J D, Kalvakolanu D, Markham B E. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the α-myosin heavy chain gene. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan J N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen H T, Bodmer R, Abmayr S M, McDermott J C, Spoerel N A. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci USA. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson E N. The MyoD family, a paradigm for development? Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 36.Olson E N, Perry M, Schulz R A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 37.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 38.Ranganayakulu G, Zhao B, Dokodis A, Molkentin J D, Olson E N, Schulz R A. A series of mutations in the D-MEF2transcription factor reveals multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 39.Roberts S G E, Ha I, Maldanado E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz J J, Chakraborty T, Martin J, Zhou J, Olson E N. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol Cell Biol. 1992;12:266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shore P, Sharrocks A D. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 42.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 43.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weintraub H, Dwarki V J, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 45.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 46.Winter B, Braun T, Arnold H-H. Co-operativity of functional domains in the muscle-specific transcription factor Myf-5. EMBO J. 1992;11:1843–1855. doi: 10.1002/j.1460-2075.1992.tb05236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y T, Breibart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]