Abstract
Mid1 is a putative stretch-activated Ca2+ channel component and is required for the maintenance of viability in the mating process. In response to mating pheromone, the mid1 mutant normally forms a pointed mating projection but eventually dies. This phenotype is called the mid phenotype. To identify a protein regulating Mid1 or regulated by Mid1, we isolated a multicopy suppressor that rescues the mid1-1 mutant from mating pheromone-induced death and found that it encodes a truncated Spa2 protein lacking an amino-terminal region responsible for interaction with components of the mitogen-activated protein kinase cascades. One of these SPA2 alleles was SPA2ΔN, whose product lacked the region from Ser5 to Leu230. SPA2ΔN on a multicopy plasmid (YEpSPA2ΔN) complemented the mid phenotype but not another phenotype, low Ca2+ accumulation, of the mid1-1 mutant. Neither SPA2ΔN on a low-copy plasmid nor wild-type SPA2 on a multicopy plasmid had suppressive activity. The SPA2 gene is involved in the formation of a pointed mating projection, and cells of the spa2Δ mutant lacking Spa2 are viable and develop a peanut shell-like structure when exposed to mating pheromone. Like the spa2Δ mutant, the mid1-1 spa2Δ double mutant and the mid1-1/YEpSPA2ΔN strain developed the peanut shell-like structure. The mid1-1 spa2Δ double mutant did not have the mid phenotype, indicating that SPA2 is epistatic to MID1. Overexpression of Spa2ΔN abolished the localization of Spa2-green fluorescent protein to the tip of the mating projection. These results suggest that the Spa2ΔN protein interferes with the localization of the normal Spa2 protein and thereby prevents cells from entering the mating process. Therefore, we suggest that Mid1 function is influenced by Spa2 function through polarized morphogenesis.
Polarized cell growth induced by external and internal stimuli is important for eukaryotic cells to undergo cell division, differentiation, and development (5, 16, 27). In the budding yeast Saccharomyces cerevisiae, polarized growth occurs in response to internal stimuli during budding and to external stimuli during mating (5, 52). The yeast has two haploid cell types, MATa and MATα cells. MATa cells synthesize and secrete the mating pheromone a-factor and receive the mating pheromone α-factor secreted by MATα cells. The mating pathway is activated by the binding of pheromones to specific receptors that stimulate the receptor-coupled heterotrimeric G protein, leading to the activation of a mitogen-activated protein (MAP) kinase cascade. The cascade then activates a transcription factor stimulating pheromone-responsive genes. This transcriptional activation and the functions of other factors, including the actin cytoskeleton, polarisome proteins, and cell wall integrity pathway components, lead cells to differentiate into shmoos having a pointed mating projection that emerges from an edge of the cells (28, 45).
Approximately 30 to 40 min after α-factor binding to MATa cells, when the polarized mating projection has started to elongate, Ca2+ influx is induced (19, 41, 44). When the Ca2+ influx is restricted by incubating cells in Ca2+-deficient medium, the cells die after differentiating into shmoos (19). The mid1 mutants have been isolated as those defective in Ca2+ influx and maintenance of viability in low-Ca2+ medium and are rescued from death in millimolar concentrations of extracellular Ca2+ (20). The mating pheromone-induced death phenotype is called the mid phenotype. The MID1 gene product, Mid1, is N glycosylated and present in the plasma membrane (20, 56) and the endoplasmic reticulum membrane (56) and has been shown to function as a stretch-activated Ca2+-permeable channel when expressed in mammalian cells, including Chinese hamster ovary (CHO) cells (25, 26). Therefore, Mid1 might be involved in sensing membrane stretch during projection formation and generating a Ca2+ signal.
Mid1 cooperates with Cch1, a homolog of the α1 subunit of the mammalian voltage-gated Ca2+ channel (10, 43). The putative Mid1-Cch1 channel constitutes a high-affinity Ca2+ influx system (HACS) necessary for cells incubated with mating pheromone in low-Ca2+ medium (39). The channel is also required for store-operated or capacitive Ca2+ entry (31), endoplasmic reticulum stress-induced Ca2+ uptake (4), and a hyperosmotic stress-induced increase in the cytosolic Ca2+ concentration ([Ca2+]cyt) (34). Mid1 is also required for the antiarrhythmic agent amiodarone-induced increase in [Ca2+]cyt (7, 15) and a hexose-induced transient increase in [Ca2+]cyt (54).
Spa2 is a polarisome protein (or a polarity-determining protein) involved in polarized morphogenesis induced by mating pheromone, localizes at the polarized growth site (called the shmoo tip), and is required for efficient mating (13). Polarisome proteins form a 12S complex, in which Spa2, Pea2, Bud6, and Bni1 assemble (23, 49). The polarisome proteins function as an apical scaffold for Cdc24-Cdc42 during apical shmoo formation and regulate the organization of the actin cytoskeleton (45). Cdc42 is a Rho GTPase and has an essential function in polarizing the actin cytoskeleton (8). Cdc24 is the guanine nucleotide exchange factor of Cdc42. Bni1 in the polarisome proteins interacts with profilin, a regulator of actin polymerization (11), and with Bud6, an actin binding protein. Bni1 also interacts with Rho1, a regulator of polarized growth in an actin-dependent manner (11, 45). Thus, the polarisome proteins function in shmoo formation mediated by actin reorganization; the actin patch localizes at the shmoo tip, the actin cable is reorganized toward the shmoo tip orientationally, and Bni1 is at the center of the polarisome proteins. In the shmoo, after the reorganization of actin, proteins necessary for mating and new cell wall materials are synthesized and carried to the shmoo tip using the secretory pathway and/or actin cable. The amino-terminal region of Spa2 spanning from Met1 to Arg120, called the Spa2 homology domain I (SHD-I) region, also interacts with the MAP kinase kinase kinase Ste11 and the MAP kinase kinases Ste7 and Mkk1/2 (49). Thus, Spa2 regulates the MAP kinase cascades. By sensing membrane stretch during shmoo formation, the protein kinase C cell integrity pathway containing the MAP kinase cascade is activated (46, 57).
These cumulative observations have led us to speculate that the polarized growth regulated by the polarisome proteins may stretch the plasma membrane to activate Mid1, resulting in the generation of a Ca2+ signal during the mating process, although no evidence has been presented so far. In the present study, we isolated a multicopy suppressor of the mid1 mutations and found that it encodes an amino-terminally truncated Spa2 protein. Our study on the relationship between Mid1 and Spa2 in terms of Ca2+ uptake and polarization suggests that Spa2 regulates a Ca2+ channel composed of Mid1 and Cch1 through polarized morphogenesis.
MATERIALS AND METHODS
Strains, plasmids, and media.
The yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea | Source or reference(s) |
|---|---|---|
| Isogenic derivatives of H207 | ||
| H207 | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 | 20 |
| H301 | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 mid1-1 | 20 |
| H311 | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 mid1-Δ5::HIS3 | 53 |
| H207-SD | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 spa2Δ::HIS3 | This study |
| H301-SD | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 mid1-1 spa2Δ::HIS3 | This study |
| H313 | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 cch1Δ::TRP1 | 21 |
| H321b | MATahis3-Δ1 leu2-3,112 trp1-289 ura3-52 sst1-2 cnb1Δ::HIS3 | This study |
| Isogenic derivatives of YKT38 | ||
| YKT38 | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 | 2, 36 |
| YKT455 | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 BNI1-EGFP::KanMX6 | 2, 36 |
| YKT512 | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 MYO2-ARG-GFP::HIS3 | 2, 36 |
| YKT570 | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 URA3::SPA2-EGFP | 2, 36 |
| YKM14 | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 [PACTI-GFP-BUD6 CEN4 URA3] | 1, 36 |
| Another strain, TYSH1 | MATahis3 leu2 trp1 ade2 ura3 spa2Δ::HIS3 | 11 |
SST1 encodes a protease that degrades the mating pheromone α-factor, and the sst1-2 mutation thus renders cells hypersensitive to α-factor without affecting the mating reaction (6, 32, 51).
Strain H321 was constructed by using the gene disruption plasmid pBS3 (gift from T. Miyakawa), and successful disruption of the CNB1 gene was confirmed by tetrad analysis, PCR, and phenotypic analysis.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| YEp13 | 2μm-ori LEU2 | 48 |
| pMID106 | Portion of SPA2 YLL020C KNS1 COX19 on YEp13 | This study |
| pMID107 | Same as pMID106 | This study |
| YEp351 | 2μm-ori LEU2 | 17 |
| YEpSPA2a | SPA2 on YEp351 | 11 |
| YEpSPA2ΔN | SPA2ΔN on YEp351 | This study |
| YCplac22 | ARS1 CEN4 TRP1 | 14 |
| YCpSPA2ΔN | SPA2ΔN on YCplac22 | This study |
| YEplac181 | 2μm-ori LEU2 | 14 |
| YEpMID1-GFP | MID1-GFP on YEplac181 | C. Ozeki-Miyawaki et al.b |
| YEpGFP | GFP on YEplac181 | C. Ozeki-Miyawaki et al.b |
The original name is YEp351-SPA2 (11).
C. Ozeki-Miyawaki, Y. Moriya, H. Tatsumi, H. Iida, and M. Sokabe, submitted for publication. YEpMID1-GFP complements the lethality and low Ca2+ uptake activity of the mid1-1 mutant.
Rich media and the synthetic medium SD were prepared as described by Sherman et al. (48). Yeast nitrogen base without calcium sources (CaCl2 and calcium pantothenate) was prepared according to the Difco manual (9) and supplemented with 1.7 μM sodium pantothenate. SD.Ca100 medium was prepared by adding 100 μM CaCl2 to Ca2+-deficient SD-Ca medium (20). The Escherichia coli strain XL1-Blue was purchased from Stratagene (La Jolla, CA). Competent cells were prepared with CaCl2 or by the method of Inoue et al. (22). Luria-Bertani medium and Terrific broth were prepared as described by Sambrook et al. (47) and Ausubel et al. (3).
Screening for multicopy suppressors of the mid1-1 mutant.
Strain H301 (MATa mid1-1 leu2-3,112) was transformed with 10 μg of a genomic library carried on YEp13, a multicopy vector marked with LEU2 (gift from Y. Ohya). After incubation for 3 days at 30°C on SD plates, about 12,000 Leu+ transformants (about 500 colonies per plate) were obtained, and 0.1 ml of 1 mM α-factor and 6 ml of SD medium containing 0.5% agar, 0.01% methylene blue, and no leucine were overlaid on the colonies on each plate. The plates were further incubated for several days. Eight methylene blue-negative, viable colonies, namely, Mid+ Leu+, were isolated and retested. To ensure that the suppression of the mid1-1 mutation was caused by the plasmids designated pMID101 to pMID108, the candidate suppressor plasmids were recovered from each transformant, propagated in E. coli, and reintroduced into the mid1-1 mutant, and the transformants were tested for cell viability in the liquid medium SD.Ca100 containing 6 μM α-factor. The results showed that the plasmids, except for pMID102, suppressed the mid phenotype of the mid1-1 mutant.
PCR using a set of primers that hybridize to the MID1 open reading frame (ORF), restriction analysis of the products, and DNA sequencing of the inserts showed that five out of the seven candidate suppressor plasmids carried DNA fragments containing the MID1 gene itself. The remainder, pMID106 and pMID107, carried an identical DNA fragment containing no MID1 gene, and thus pMID106 was investigated further. DNA sequences were compared with those in the S. cerevisiae genome using the Saccharomyces Genome Database World Wide Web site (http://genome-www.Stanford.edu/Saccharomyces/). Because pMID106 contained multiple ORFs, individual or clustered ORFs from the plasmid were subcloned into multicopy vectors to identify the specific gene responsible for the suppression of the mid1 mutation.
Construction of SPA2ΔN.
To construct the plasmid YEpSPA2ΔN, a gene product which lacks the region spanning from Ser5 to Leu230 of the Spa2 protein, two XhoI sites were generated stepwise in the SPA2 coding region on YEpSPA2 with the QuikChange site-directed mutagenesis kit (Stratagene) using the two sets of primer pairs shown in Table 3: SPA2/XhoI-1F and SPA2/XhoI-1R, and SPA2/XhoI-2F and SPA2/XhoI-2R. The mutagenized plasmid was digested with XhoI, and the resulting DNA fragment lacking 15% of the SPA2 coding region was subjected to self-ligation using Ligation kit version 2 (TaKaRa, Kyoto, Japan). Successful construction of YEpSPA2ΔN was confirmed by DNA sequencing.
TABLE 3.
Primers used in this study
| Primer name | DNA sequencea |
|---|---|
| For construction of SPA2ΔN | |
| SPA2/XhoI-1F | 5′-GGG TAC GTC ACT CGA GGT TTC TCT CGC ACA TCA TAG-3′ |
| SPA2/XhoI-1R | 5′-CTA TGA TGT GCG AGA GAA ACC TCG AGT GAC GTA CCC-3′ |
| SPA2/XhoI-2F | 5′-GAC TCC CAA GTG CTC GAG CGT GAT ATC ACA TCA ATG GC-3′ |
| SPA2/XhoI-2R | 5′-GCC ATT GAT GTG ATA TCA CGC TCG AGC ACT TGG GAG TC-3′ |
| For disruption of the SPA2 locus | |
| SPA2-HIS3-F2 | 5′-GTC AAG CGA GGT TTC TCT CGC ACA TCA TAG AG-3′ |
| SPA2-HIS3-R2 | 5′-CCA GAG CCA AGC TGT TGA AGC TAT CTT CTT CG-3′ |
The boldface indicates nucleotides that have been changed to generate an XhoI site (underlined).
Disruption of the SPA2 locus in strains H207 and H301.
Yeast genomic DNA was prepared from a SPA2-deleted strain, TYSH1 (MATa ura3 leu2 trp1 his3 ade2 spa2::HIS3) (gift from Y. Takai), in which nucleotides 1089 to 3099 of the 4,401-bp SPA2 ORF were replaced with a HIS3 cassette (11). Amplification of the spa2::HIS3 region from TYSH1 was carried out by PCR with the primer pair SPA2-HIS3-F2 and SPA2-HIS3-R2 (Table 3), and the PCR-amplified spa2::HIS3 DNA fragment was introduced into strains H207 (MID1) and H301 (mid1-1). Successful disruption of the SPA2 gene was confirmed by PCR as well as tetrad analysis showing that the Spa− and His+ phenotypes always cosegregated.
Measurement of the complementing ability of plasmids.
Plasmids were introduced into the mid1-1 strain (H301) according to the method of Mount et al. (38). Each transformant was grown to 2 × 106 cells/ml in SD.Ca100 selection medium and received 6 μM α-factor. The viability of each transformant was determined by the methylene blue liquid method as described by Iida et al. (19) to estimate the ability of plasmids to complement the mid1-1 mutation.
DNA sequencing.
DNA sequencing was performed by an ABI Prism automated sequencing kit and an ABI 310 automated sequencer (Applied Biosystems, Foster City, CA).
Determination of Ca2+ accumulation.
The method described by Iida et al. (19) was generally followed. Briefly, yeast cells were grown to 2 × 106 cells/ml in SD.Ca100 medium and incubated with 6 μM α-factor and 5 μCi/ml 45CaCl2 (equivalent to 185 kBq/ml; 1.8 kBq/nmol). Samples (200 μl) were filtered with a filter (type HA, 0.45 μm; Millipore, Billerica, MA) presoaked in 5 mM CaCl2 and washed five times with 5 ml of the same solution. The radioactivity retained on the filter was counted with a liquid scintillation counter.
Immunoblotting.
Immunoblotting was performed according to the method described previously (20). Rabbit anti-Spa2-trpE polyclonal antibodies (50) were used to detect the wild-type Spa2 and Spa2ΔN proteins.
Fluorescence microscopy.
Cells expressing green fluorescent protein (GFP)-tagged proteins at the exponential phase of growth were exposed to 6 μM α-factor for 2 h, harvested, and placed on slides. The slides were then sealed under a coverslip. GFP images were observed using a fluorescence microscope (Olympus IX71) with UPlanApo 100/1.35 (Olympus) as an objective lens. Images were processed using Adobe Photoshop 7.0 (Adobe Systems).
RESULTS
Isolation of a multicopy suppressor of the mid1-1 mutant.
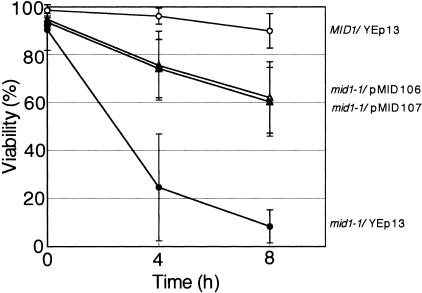
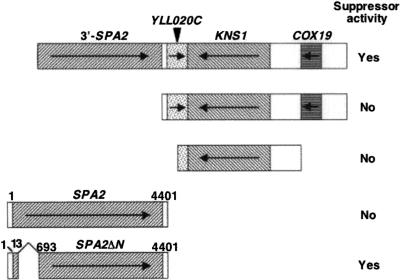
The mid1-1 mutant produces a nonfunctional Mid1 protein lacking a carboxy-terminal, cytoplasmic regulatory region (33) and dies in response to α-factor in the low-Ca2+ medium SD.Ca100 (20). To identify proteins that either regulate, are regulated by, or function in parallel with the Mid1 protein, we searched for multicopy suppressors of the mid1-1 mutant. This mutant was transformed with a yeast genomic library carried on a multicopy vector, and approximately 12,000 transformants were tested for the ability to survive in the presence of α-factor. Two plasmids, pMID106 and pMID107, derived from survivors had the ability to suppress the mid phenotype of the mid1-1 mutant, although the suppression was not complete (Fig. 1). Sequence analysis revealed that the two plasmids contained an identical 8.3-kb DNA fragment of chromosome 12 (101,612 to 109,924) (Fig. 2). This fragment contained three complete ORFs (YLL020C, KNS1, and COX19) and one incomplete SPA2 ORF that lacked the first 666 bp of the 4,398-bp ORF as well as its 5′ noncoding region.
FIG. 1.
Suppressor activity of pMID106 and pMID107. The MID1 strain H207 containing the empty vector YEp13 and the mid1-1 mutant strain H301 containing YEp13, pMID106, or pMID107 were incubated with 6 μM α-factor in SD.Ca100 medium. The viability of the cells was examined 0, 4, and 8 h after the addition of α-factor by the methylene blue liquid method. Open circles, MID1/YEp13; filled circles, mid1-1/YEp13; open triangles, mid1-1/pMID106; filled triangles, mid1-1/pMID107. Note that open and filled triangles are overlapped. Values are the means ± standard deviations (SD) of three independent experiments.
FIG. 2.
Maps of the insert of the suppressor plasmid pMID106 and the fragments tested for suppressor activity. Coding regions are shaded or hatched. Arrows indicate the direction of transcription in each gene. Fragments having suppressor activity are marked Yes and those having no activity No. Numbers above the SPA2 and SPA2ΔN genes are the positions of nucleotides from the initiation codon. The SPA2ΔN gene lacks the region spanning from nucleotides 14 to 692.
The specific gene responsible for the suppression was identified by subcloning individual or clustered ORFs from pMID106 into multicopy vectors (Fig. 2). Unexpectedly, a DNA fragment containing all three complete ORFs did not have the suppressive activity. This suggests that the incomplete SPA2 ORF, which encodes a truncated protein spanning from Asp223 to the carboxy-terminal amino acid Lys1,466, is responsible for the suppression. In other words, the mutant Spa2 protein lacking an amino-terminal region may have the multicopy suppressor activity. To test this possibility, we tried repeatedly to subclone the incomplete SPA2 ORF from pMID106 but failed for unknown reasons. Therefore, we employed a different approach as follows.
The amino-terminally truncated Spa2 lacking the SHD-I region is a multicopy suppressor of the mid1 mutant.
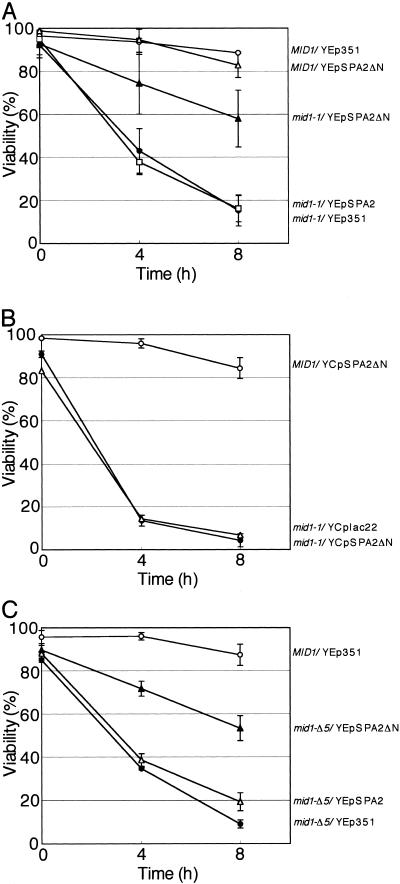
To test the above-mentioned possibility, the amino-terminal region spanning from Ser5 to Leu230, which contains the SHD-I region responsible for interaction with Ste11, Ste7, and Mkk1/2 (49), was deleted from the complete Spa2 to produce Spa2ΔN (Fig. 2). A multicopy plasmid producing this mutant protein from its own promoter was named YEpSPA2ΔN. The mid1-1 mutant was transformed with YEpSPA2ΔN, and the resulting transformant (mid1-1/YEpSPA2ΔN) was examined for cell viability after the addition of α-factor by the methylene blue liquid method. Figure 3A shows quantitatively that the viability of mid1-1/YEpSPA2ΔN cells was 58% at 8 h after the addition of α-factor and that the viability level was comparable to that of mid1-1/pMID106 and mid1-1/pMID107 cells (Fig. 1). These results are consistent with the possibility described in the former section and indicate that Spa2ΔN has the ability to partially suppress the mid phenotype. This suppression was dose dependent. When expressed from a low-copy plasmid, SPA2ΔN did not suppress the mid phenotype at all (Fig. 3B). In addition, overexpression of intact Spa2 from the multicopy plasmid YEpSPA2 did not suppress the mid phenotype at all (Fig. 3A). Thus, the deletion of the amino-terminal region of Spa2 is important for the multicopy suppressor activity.
FIG. 3.
Suppressor activity of YEpSPA2ΔN and YEpSPA2. (A) The MID1 strain bearing YEp351 or YEpSPA2ΔN and the mid1-1 mutant bearing a multicopy plasmid (either YEpSPA2ΔN, YEpSPA2, or YEp351) were incubated with 6 μM α-factor in SD.Ca100 medium and examined for viability as described in the legend to Fig. 1. Open circles, MID1/YEp351; filled circles, mid1-1/YEp351; open triangles, MID1/YEpSPA2ΔN; filled triangles, mid1-1/YEpSPA2ΔN; open squares, mid1-1/YEpSPA2. (B) The MID1 strain bearing the low-copy plasmid YCpSPA2ΔN and the mid1-1 mutant bearing either YCpSPA2ΔN or empty vector YCplac22 were incubated and examined for viability as described above. Open circles, MID1/YCpSPA2ΔN; filled circles, mid1-1/YCpSPA2ΔN; open triangles, mid1-1/YCplac22. (C) The MID1 strain bearing YEp351 and the mid1-Δ5 mutant bearinga multicopy plasmid (either YEpSPA2ΔN, YEpSPA2, or YEp351) were incubated and examined for viability as described above. Open circles, MID1/YEp351; filled circles, mid1-Δ5/YEp351; open triangles, mid1-Δ5/YEpSPA2; filled triangles, mid1-Δ5/YEpSPA2ΔN. Values are the means ± SD of three independent experiments.
Spa2ΔN is overexpressed from YEpSPA2ΔN.
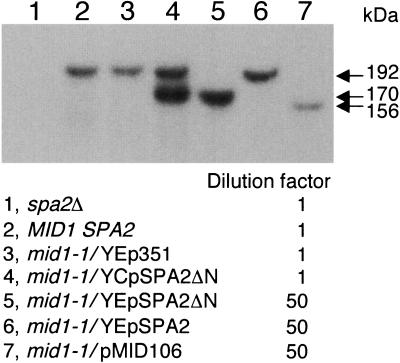
To examine whether the Spa2ΔN protein is overexpressed in mid1-1/YEpSPA2ΔN cells or missing in these cells because of amino-terminal truncation that might result in instability of the protein, we performed immunoblot analysis. As shown in Fig. 4, the amount of Spa2ΔN in mid1-1/YEpSPA2ΔN cells (Fig. 4, lane 5) was more than 50 times as much as that of Spa2 in control cells (Fig. 4, lanes 2 and 3). In addition, the amount of Spa2 in mid1-1/YEpSPA2 cells (Fig. 4, lane 6) and that of the truncated Spa2 protein in mid1-1/pMID106 cells (Fig. 4, lane 7) were nearly 50 times as much as that of Spa2 in the control cells (Fig. 4, lanes 2 and 3). Furthermore, Spa2ΔN was not significantly overproduced in mid1-1/YCpSPA2ΔN cells (Fig. 4, lane 4). These results indicate that Spa2ΔN is definitely overproduced from the multicopy plasmid YEpSPA2ΔN. Therefore, the amount of Spa2ΔN correlates well with the viability data shown in Fig. 1 and 3.
FIG. 4.
Estimation of the amount of overexpressed Spa2ΔN. Yeast strains used were H207-SD (spa2Δ), H207 (MID1 SPA2), and H301 (mid1-1). Each strain with or without an appropriate plasmid (see the panel) was grown to the exponential phase, and total cell extracts were prepared by homogenization with glass beads. The protein concentration of all samples was adjusted to 2 mg/ml, and the extracts of mid1-1/YEpSPA2ΔN (lane 5), mid1-1/YEpSPA2 (lane 6), and mid1-1/pMID106 (lane 7) were diluted 50-fold in the extracts of spa2Δ cells. Samples (20 μg protein each) were then subjected to 4 to 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with antibodies against the Spa2-trpE protein. Note that the wild-type Spa2 protein with the apparent molecular mass of 192 kDa, which has been expressed from the chromosomal SPA2 gene, is not detectable in lanes 5 and 7 because of the 50-fold dilution. Arrows represent the apparent molecular mass estimated with molecular weight markers. A representative of three independent experiments that gave essentially the same results is shown.
Direct interaction of Spa2ΔN with mutant Mid1 is unnecessary in multicopy suppression.
If Spa2ΔN requires interaction with the mutant Mid1 protein to suppress the mid1-1 mutation, the plasmid YEpSPA2ΔN must fail to suppress the mid phenotype of cells lacking the entire Mid1 protein. To examine this possibility, YEpSPA2ΔN was introduced into cells bearing another mid1 mutant allele, mid1-Δ5, which produces no Mid1 polypeptide because of the introduction of a stop codon just after the initiation codon of the MID1 gene (53), and the resulting transformant mid1-Δ5/YEpSPA2ΔN was examined for cell viability as described above. Figure 3C shows that the viability of mid1-Δ5/YEpSPA2ΔN was essentially the same as that of mid1-1/YEpSPA2ΔN. YEpSPA2ΔN did not affect the viability of the MID1 strain (Fig. 3A). These results indicate that SPA2ΔN is not an allele-specific suppressor and that the Spa2ΔN protein does not interact with mutant Mid1 proteins to partially suppress the mid1 mutations.
Low Ca2+ uptake activity of mid1-1 cells is not remedied by SPA2ΔN.
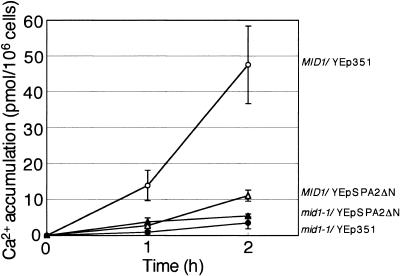
To investigate whether the partial suppression of the mid1 mutations by SPA2ΔN is due to an increase in Ca2+ uptake activity by Spa2ΔN, Ca2+ accumulation in MID1 and mid1-1 cells expressing Spa2ΔN was measured after the addition of α-factor. Figure 5 shows that Spa2ΔN did not increase Ca2+ accumulation in mid1-1 cells. This indicates that a possible increase in Ca2+ uptake does not account for the suppression of the mid1 mutations by Spa2ΔN. Interestingly, Spa2ΔN markedly decreased Ca2+ accumulation in MID1 cells. The implication of this result will be discussed below.
FIG. 5.
Ca2+ accumulation in mid1-1 and MID1 cells bearing YEpSPA2ΔN. Cells were incubated in SD.Ca100 medium and received 6 μM α-factor, after which Ca2+ accumulation was measured. Open circles, MID1/YEp351; filled circles, mid1-1/YEp351; open triangles, MID1/YEpSPA2ΔN; filled triangles, mid1-1/YEpSPA2ΔN. Values are the means ± SD of three independent experiments.
Cell death associated with the mid1 mutation depends on SPA2.
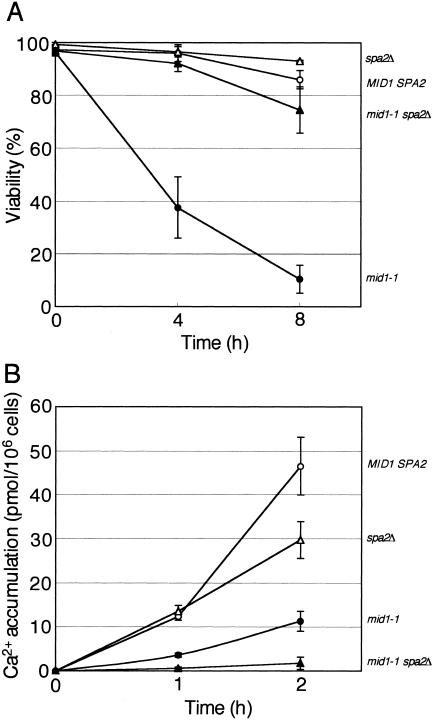
To investigate the phenotypes of mutant cells lacking the entire SPA2 gene (spa2Δ cells) and double mutant mid1-1 spa2Δ cells, these cells were subjected to the viability and Ca2+ accumulation assays. Figure 6A shows that both spa2Δ cells and mid1-1 spa2Δ cells did not die even 8 h after the addition of α-factor, like MID1 SPA2 cells. This result indicates that SPA2 is epistatic to MID1. Therefore, it is likely that Mid1 becomes functional after Spa2 has functioned.
FIG. 6.
SPA2 is epistatic to MID1 and required for Ca2+ accumulation. The viability (A) and Ca2+ accumulation (B) of the MID1 SPA2 strain, the mid1-1 and spa2Δ single mutants, and the mid1-1 spa2Δ double mutants after exposure to α-factor in SD.Ca100 medium are shown. Viability and Ca2+ accumulation were examined as described in the legends to Fig. 1 and 4, respectively. Open circles, MID1 SPA2 cells; filled circles, mid1-1 cells; open triangles, spa2Δ cells; filled triangles, mid1-1 spa2Δ cells. Values are the means ± SD of three independent experiments.
Ca2+ accumulation significantly decreased in spa2Δ cells (Fig. 6B). Furthermore, Ca2+ accumulation in mid1-1 spa2Δ cells was lower than that in mid1-1 cells and spa2Δ cells. Thus, the mid1-1 mutation and the spa2Δ mutation have an additive effect on the decrease in Ca2+ accumulation.
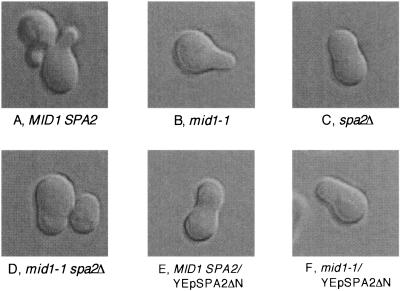
Cells overexpressing Spa2ΔN are unable to develop a well-polarized mating projection.
It has been shown that when exposed to α-factor, spa2Δ cells change into incomplete shmoos that do not have a well-polarized mating projection (13, 49). We thus examined the morphology of mid1-1 or MID1 cells lacking SPA2 or expressing SPA2ΔN after exposure to α-factor for 2 h. As shown in Fig. 7A and B, MID1 and mid1-1 cells differentiated into shmoos having a well-polarized mating projection. In contrast, 7C shows that spa2Δ cells developed a peanut shell-like morphology without a mating projection as reported previously (13). Interestingly, the morphology of mid1-1 spa2Δ cells was essentially the same as that of spa2Δ cells (Fig. 7D), indicating that SPA2 is epistatic to MID1. This is consistent with the result of the viability assay shown in Fig. 6A.
FIG. 7.
Morphology of α-factor-treated cells. Cells of various strains were incubated in SD.Ca100 medium containing 6 μM α-factor for 2 h, after which the cells were photographed under a differential interference contrast microscope (DIC). Typical examples are shown. A, MID1 SPA2 cells; B, a mid1-1 cell; C, a spa2Δ cell; D, mid1-1 spa2Δ cells; E, a MID1 SPA2 cell bearing YEpSPA2ΔN; F, a mid1-1 cell bearing YEpSPA2ΔN.
MID1 and mid1-1 cells overexpressing SPA2ΔN (Fig. 7E and F) as well as those containing the original suppressing plasmid pMID106 (data not shown) also changed into cells without a mating projection. This result suggests that the Spa2ΔN protein interferes with the function of the normal Spa2 protein.
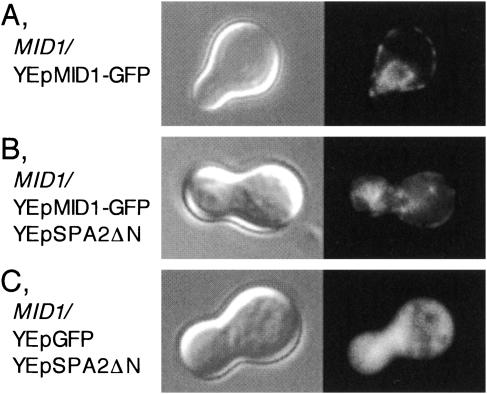
Spa2ΔN overexpression partially interferes with the targeting of Mid1.
As described above, Spa2ΔN overexpression resulted in a marked decrease in Ca2+ accumulation in MID1 cells (Fig. 5). The decrease may be due to Mid1 mislocalization caused by Spa2ΔN overexpression. We examined this possibility by observing the distribution of Mid1-GFP in Spa2ΔN-overexpressing cells incubated with α-factor for 2 h. The Mid1-GFP protein used in this study was fully active because this protein expressed from plasmids complemented the lethality and low Ca2+ uptake activity of the mid1 mutant (C. Ozeki-Miyawaki, Y. Moriya, H. Tatsumi, H. Iida, and M. Sokabe, submitted for publication). Figure 8A shows that in control cells, Mid1-GFP was present in the plasma membrane and the endoplasmic reticulum membrane as reported previously (56). By contrast, in Spa2ΔN-overexpressing cells a considerable amount of Mid1-GFP was accumulated in the cytoplasm, although a part of Mid1-GFP was duly present in the plasma membrane (Fig. 8B). GFP was distributed throughout the cytoplasm (Fig. 8C). These results indicate that Spa2ΔN overexpression results in a partial retardation of the membrane targeting of Mid1.
FIG. 8.
Localization of the Mid1-GFP fusion protein in Spa2ΔN-overexpressing cells. Exponentially growing MID1 cells transformed with various plasmids were incubated with 6 μM α-factor for 2 h, harvested, and observed by DIC (left panels) or fluorescence microscopy (right panels). (A) A cell transformed with YEpMID1-GFP. (B) A cell transformed with YEpMID1-GFP and YEpSPA2ΔN. (C) A cell transformed with YEpGFP and YEpSPA2ΔN. Typical examples of the cells are shown. Essentially the same results were obtained in at least two additional experiments, independently.
Spa2ΔN overexpression results in mislocalization of normal Spa2.
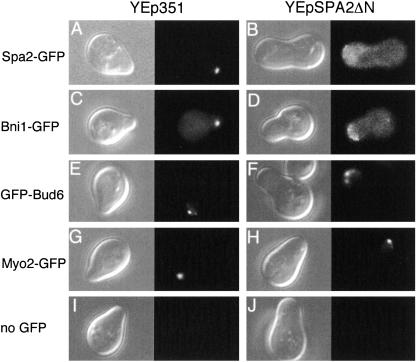
Spa2 is localized at the tip of the mating projection of α-factor-treated cells (13). Spa2ΔN may interfere with the localization of wild-type Spa2 when overexpressed and thus restrict Spa2 function. To examine this possibility, we analyzed the effect of Spa2ΔN overexpression on the localization of Spa2-GFP produced from the chromosomal SPA2-GFP gene (2) by fluorescence microscopy. Cells expressing Spa2-GFP with or without YEpSPA2ΔN were grown to the exponentially growing phase and incubated with α-factor for 2 h. As shown in Fig. 9A and B, in control cells Spa2-GFP was present at the tip of the mating projection as reported previously (13), whereas in Spa2ΔN-overexpressing cells it was distributed throughout the cytoplasm with a slight accumulation around the edge of the cells. This result clearly indicates that Spa2ΔN interferes with the localization of intact Spa2-GFP.
FIG. 9.
Localization of polarisome proteins in Spa2ΔN-overexpressing cells. Exponentially growing cells expressing Spa2-GFP, Bni1-GFP, GFP-Bud6, Myo2-GFP, or no GFP fusion protein were transformed with the empty vector YEp351 or YEpSPA2ΔN, incubated with 6 μM α-factor for 2 h, harvested, and subjected to microscopic analysis as described in the legend to Fig. 8. The left panel for each transformant shows DIC analysis; the right panel for each transformant shows fluorescence microscopy. (A and B) A Spa2-GFP-expressing cell (strain YKT570) with YEp351 and YEpSPA2ΔN, respectively. (C and D) A Bni1-GFP-expressing cell (YKT455) with YEp351 and YEpSPA2ΔN, respectively. (E and F) A GFP-Bud6-expressing cell (YKM14) with YEp351 and YEpSPA2ΔN, respectively. (G and H) A Myo2-GFP-expressing cell (YKT512) with YEp351 and YEpSPA2ΔN, respectively. (I and J) A cell expressing no GFP fusion protein (YKT38) with YEp351 and YEpSPA2ΔN, respectively. Typical examples of the cells are shown. Essentially the same results were obtained in at least two additional experiments, independently.
Spa2 associates with Bni1 and Bud6 to make the polarisome (23, 49). Spa2ΔN may also interfere with the localization of Bni1 and Bud6. To examine this possibility, the same experiments as those for Spa2-GFP were performed on Bni1-GFP- or GFP-Bud6-expressing cells, in which Bni1-GFP was produced from the chromosomal BNI1-GFP gene (36) and GFP-Bud6 from the GFP-BUD6 gene under the control of the ACT1 promoter on a low-copy plasmid (1). Bni1-GFP underwent a mislocalization similar to Spa2-GFP in Spa2ΔN-overexpressing cells (Fig. 9C and D), but GFP-Bud6 was less affected by Spa2ΔN overexpression than the former two proteins (Fig. 9E and F).
Myo2 is an unconventional type V myosin required for polarized delivery of secretory vesicles and localized at the projection tip but is not a polarisome protein (24, 30). We employed Myo2-GFP as a negative control and found that the localization of Myo2-GFP was not affected by Spa2ΔN overexpression (Fig. 9G and H).
YEpSPA2ΔN also suppresses the mid phenotype of the cch1 and cnb1 null mutants.
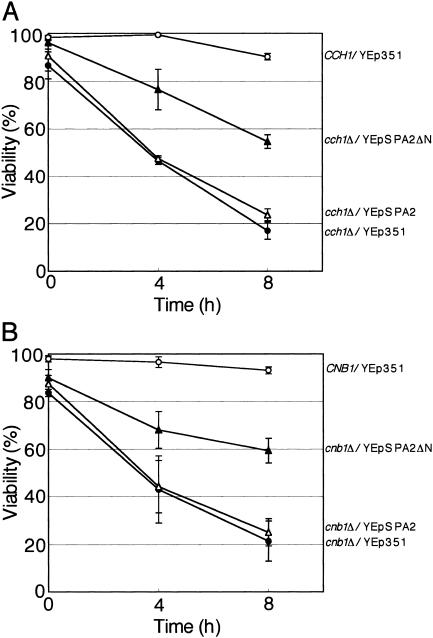
Because Mid1 constitutes HACS with Cch1 as mentioned above, the mid phenotype of cells lacking Cch1 may be suppressed by YEpSPA2ΔN. We examined this prediction and found that this is the case (Fig. 10A).
FIG. 10.
Suppression of the cch1Δ and cnb1Δ mutations by YEpSPA2ΔN. (A) The CCH1 strain bearing YEp351 and the cch1 null mutant (cch1Δ) bearing a multicopy plasmid (either YEpSPA2ΔN, YEpSPA2, or YEp351) were incubated with 6 μM α-factor in SD.Ca100 medium and examined for viability as described in the legend to Fig. 1. Open circles, CCH1/YEp351; filled circles, cch1Δ/YEp351; open triangles, cch1Δ/YEpSPA2; filled triangles, cch1Δ/YEpSPA2ΔN. (B) The CNB1 strain bearing YEp351 and the cnb1Δ mutant bearing a multicopy plasmid (either YEpSPA2ΔN, YEpSPA2, or YEp351) were incubated and examined for viability as described above. Open circles, CNB1/YEp351; filled circles, cnb1Δ/YEp351; open triangles, cnb1Δ/YEpSPA2; filled triangles, cnb1Δ/YEpSPA2ΔN. Note that the wild-type strains CCH1, CNB1, and MID1 are identical to each other. Values are the means ± SD of three independent experiments.
Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is suggested to function downstream of Mid1 and Cch1 (4, 34, 37, 55), and cells defective in calcineurin die in response to α-factor or endoplasmic reticulum stress agents, such as tunicamycin, like mid1 cells (4, 37, 55). It is therefore possible that the mid phenotype of cnb1Δ cells lacking the regulatory subunit of calcineurin and thereby having no functional calcineurin would be suppressed by Spa2ΔN. Figure 10B shows that YEpSPA2ΔN suppressed the mid phenotype of cnb1Δ cells, supporting the above possibility.
DISCUSSION
We have shown that the Spa2ΔN protein lacking a region from Ser5 to Leu230 acts as a multicopy suppressor of the mid phenotype of the mid1-1 mutant. The mid1-1 mutation has been identified to be a nonsense mutation and thereby produces a mutant Mid1 protein lacking the carboxy-terminal region (33). Because overexpression of the Spa2ΔN protein also suppresses the mid phenotype of the null mutant bearing the mid1-Δ5 allele resulting in no production of the Mid1 polypeptide (53), it was suggested that the suppressive effect is not due to a direct binding of the Spa2ΔN protein to the mutant Mid1 protein. We have also shown by immunoblot analysis that the Spa2ΔN protein is actually overproduced from the multicopy plasmid YEpSPA2ΔN with at least a 50-fold increase (Fig. 4).
The relationship between Spa2 and Mid1 on the mid phenotype pathway.
In general, a multicopy suppressor complements a mutation by one of the following mechanisms. (i) The suppressor product binds to and remedies the mutated gene product. (ii) The suppressor product has essentially the same function as the mutated gene product. In the case of this study, it could be another Ca2+-permeable channel or an activator. (iii) The suppressor product acts downstream of the mutated gene product in a common signaling pathway. (iv) The suppressor product acts before the execution phase of the mutated gene product and interferes with a step that is prerequisite for the functioning of the mutated gene product when overproduced, and eventually the mutant cells do not enter into a regular pathway and then survive. Our genetic analysis has suggested the fourth mechanism for the Spa2ΔN protein (see below).
The Ca2+ accumulation assay precludes the first and second mechanisms. If either one of these mechanisms is involved, a low level of Ca2+ accumulation in the mid1-1 mutant should be remedied by Spa2ΔN expressed from the multicopy plasmid YEpSPA2ΔN. However, the plasmid did not increase Ca2+ accumulation in the mid1-1 mutant (Fig. 5). The observation that YEpSPA2ΔN was able to complement the mid phenotype of the mid1-Δ5 null mutant also precludes the first mechanism, as mentioned above.
The third mechanism would not be supported by multicopy suppression experiments. If the Spa2 protein acts downstream of Mid1, overproduction of the wild-type Spa2 protein from multicopy plasmid YEpSPA2 might suppress the low viability of the mid1-1 mutant. However, this is not the case (Fig. 3).
Analyses of the mid1-1 spa2Δ double mutant preclude the third mechanism and support the fourth mechanism. If Spa2 acts downstream of Mid1, then the phenotype of the double mutant should be the same as that of the mid1-1 single mutant. On the other hand, if Spa2 acts before the execution phase of Mid1, the phenotype of the double mutant should be the same as that of the spa2Δ single mutant. The viability and morphology of the mid1-1 spa2Δ mutant after exposure to mating pheromone were essentially the same as those of the spa2Δ single mutant (Fig. 6A and 7C and D), being consistent with the latter possibility, namely, the fourth mechanism. Furthermore, the fourth mechanism is also supported by the observation that the morphology of mid1-1/YEpSPA2ΔN cells is the same as that of spa2Δ cells, suggesting that the Spa2ΔN protein interferes with the normal Spa2 protein and thereby prevents cells entering the mating process, in which the function of Mid1 is required. This suggestion is supported by direct observation of the localization of Spa2-GFP in Spa2ΔN-overexpressing cells (Fig. 9). Note that, in the fourth mechanism, Mid1 is not necessarily a downstream component of Spa2 in a single signaling pathway. Spa2 would affect many cellular components through stimulation of polarized growth, and Mid1 would be one of them.
It is still possible to speculate that Spa2 acts on one pathway (viability) but not the other (Ca2+ influx), because YEpSPA2ΔN does not suppress the low Ca2+ accumulation activity of the mid1 mutant (Fig. 5) even though it suppresses the cell death phenotype (Fig. 3A). However, it should be noted that YEpSPA2ΔN markedly lowers the Ca2+ accumulation of MID1 cells (see MID1/YEpSPA2ΔN in Fig. 5). This indicates that YEpSPA2ΔN inhibits the Ca2+ uptake activity of the intact Mid1, and thus it is reasonable that YEpSPA2ΔN does not remedy the low Ca2+ accumulation of the mid1 mutant. Therefore, the Ca2+ accumulation assay does not support the above speculation.
Another speculation might be possible: Mid1 negatively regulates Spa2 function sometime in the mating process, probably through the influx of Ca2+, and Spa2 function leads to cell death in the mid1 mutants. Loss of Spa2 function caused by either YEpSPA2ΔN or the spa2Δ mutation, therefore, rescues the mid1 mutants from death. This speculation can explain all of the results on cell viability presented in Fig. 1, 3A and C, and 6A. However, it cannot explain why YEpSPA2ΔN and the spa2Δ mutation result in a decrease in Ca2+ accumulation (Fig. 5 and 6B) unless one of the functions of Spa2 is postulated to activate Mid1 on the basis of the fourth mechanism.
A role for Spa2 in Mid1 function.
The Ca2+ accumulation assay has provided a novel insight into a mechanism of Ca2+ uptake. The observation that the level of Ca2+ accumulation was significantly lower in spa2Δ mutant cells than in SPA2 cells (Fig. 6B) suggests that the Spa2 protein is required for Ca2+ uptake to some extent. Because the level in the mid1-1 spa2Δ mutant is lower than that in the mid1-1 mutant, the Spa2 protein may also have the ability to regulate a Ca2+-permeable channel other than the Ca2+ channel composed of Mid1. Muller et al. reported that Spa2 is required for Ca2+ uptake through a low-affinity Ca2+ influx system (LACS) that probably involves the Fig1 protein but not Mid1 and Cch1 (39, 40). LACS is reported to be stimulated by mating pheromone in rich media, such as YPD, while a HACS composed of Mid1 and Cch1 is activated by mating pheromone in low-Ca2+ media, such as SD.Ca100.
Fluorescence microscopy has indicated that although there is a considerable accumulation of Mid1-GFP in the cytoplasm of Spa2ΔN-overexpressing cells, a part of Mid1-GFP is successfully localized to the plasma membrane (Fig. 8). We speculate that the properly targeted Mid1 might be nonfunctional in Spa2ΔN-overexpressing cells because Ca2+ accumulation in the cells is markedly decreased (Fig. 5).
Fluorescence microscopy has also indicated that Spa2ΔN overexpression interferes with the localization of the normal Spa2-GFP protein to the projection tip (Fig. 9). Furthermore, Spa2ΔN overexpression has the same effect on one of the polarisome proteins, Bni1-GFP, while it has a small effect on the localization of another polarisome protein, GFP-Bud6, and no effect on a nonpolarisome protein, Myo2-GFP. The results are consistent with the observations that Bni1 localization is markedly dependent on Spa2 (42) and that Bud6 is partially associated with Spa2 and not required for Spa2 localization (49). Therefore, we suggest that Spa2ΔN interferes with the polarisome proteins according to their ability to interact with Spa2.
In summary, a possible but not conclusive explanation for the findings in this study is as follows. Spa2 acts to form the polarized mating projection where the plasma membrane is stretched because of the reconstruction of the cell wall at the mating projection, and then Mid1 is activated by membrane stretch. The Spa2ΔN protein interferes with the function of the normal Spa2 protein and eventually inhibits the formation of the mating projection; thereby, Mid1 function is abrogated because of a lack of membrane stretch. If this speculation is correct, the mid phenotype of the cch1 mutant should also be suppressed by YEpSPA2ΔN because Cch1 and Mid1 cooperate in mating pheromone-treated cells (10, 39, 43), and this was found to be the case (Fig. 10A). Therefore, one of the functions of Spa2 is most likely to activate the Mid1/Cch1 Ca2+ channel by forwarding polarized growth.
Implication for other organisms.
Stretch-activated Ca2+-permeable channels are suggested to be important for polarized morphogenesis in various fungi and plant cells. For example, at growing hyphal tips of the oomycete Saprolegnia ferax, a tip-high gradient of stretch-activated Ca2+-permeable channels is formed (12), and the peripheral F-actin network is suggested to establish or maintain the tip-high gradient (29). At the tips of growing Lilium longiflorum pollen tubes, Ca2+ oscillations are generated, and the source of the oscillation is suggested to be Ca2+ influx through Ca2+ channels mechanically coupled to membrane stretch (18, 35). Because the molecular nature and activation mechanism of these stretch-activated Ca2+-permeable channels are still unknown, the present study on Mid1 and Spa2 should add new insight into the molecular mechanism of polarized morphogenesis.
Acknowledgments
We thank Hideki Taguchi for his encouragement; Yoshimi Takai for yeast strains and plasmids; Kazuma Tanaka and Konomi Fujimura-Kamada for yeast strains; Yoshikazu Ohya for a yeast genomic library; David C. Amberg, Robert A. Arkowitz, Tokichi Miyakawa, Chikako Ozeki-Miyawaki, and Akio Sugino for plasmids; Michael Snyder for anti-Spa2-trpE antibodies; Takashi Maruoka for his encouragement and discussion; members of our laboratory for their discussion and technical advice; and Yumiko Higashi for her secretarial assistance.
This work was supported by CREST, JST (to H.I.), a Grant-in-Aid for Scientific Research on Priority Areas (no. 15031212 to H.I.), and a Grant-in-Aid for Scientific Research B (no. 16370072 to H.I.) from the Ministry of Education, Culture, Sports, Science and Technology in Japan.
REFERENCES
- 1.Amberg, D. C., J. E. Zahner, J. W. Mulholland, J. R. Pringle, and D. Botstein. 1997. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell 8:729-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkowwitz, R. A., and N. Lowe. 1997. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J. Cell Biol. 138:17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmen, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. A compendium of methods from current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bonilla, M., K. K. Nastase, and K. W. Cunningham. 2002. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casamayor, A., and M. Snyder. 2002. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5:179-186. [DOI] [PubMed] [Google Scholar]
- 6.Chan, R. K., and C. A. Otte. 1982. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courchesne, W. E., and S. Ozturk. 2003. Amiodarone induces a caffeine-inhibited, MID1-dependent rise in free cytoplasmic calcium in Saccharomyces cerevisiae. Mol. Microbiol. 47:223-234. [DOI] [PubMed] [Google Scholar]
- 8.Delley, P.-A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Difco Laboratories. 1984. Difco manual, 10th ed., p. 1135-1141. Difco Laboratories, Detroit, Mich.
- 10.Fischer, M., N. Schnell, J. Chattaway, P. Davies, G. Dixon, and D. Sanders. 1997. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419:259-262. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara, T., K. Tanaka, A. Mino, M. Kikyo, K. Takahashi, K. Shimizu, and Y. Takai. 1998. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at bud site and regulation of actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1221-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrill, A., S. L. Jackson, R. R. Lew, and I. B. Heath. 1993. Ion channel activity and tip growth: tip-localized stretch-activated channels generate an essential Ca2+ gradient in the oomycete Saprolegnia ferax. Eur. J. Cell Biol. 60:358-365. [PubMed] [Google Scholar]
- 13.Gehrung, S., and M. Snyder. 1990. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 111:1451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, S. S., V.-K. Ton, V. Beaudry, S. Rulli, K. Cunningham, and R. Rao. 2003. Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J. Biol. Chem. 278:28831-28839. [DOI] [PubMed] [Google Scholar]
- 16.Hepler, P. K., L. Vidali, and A. Y. Cheung. 2001. Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 7:159-187. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vector with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 18.Holdaway-Clarke, T. L., J. A. Fejio, G. R. Hackett, J. G. Kunkel, and P. K. Hepler. 1997. Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9:1999-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iida, H., Y. Yagawa, and Y. Anraku. 1990. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway: a study of [Ca2+]i in single Saccharomyces cerevisiae cells with imaging of fura-2. J. Biol. Chem. 265:13391-13399. [PubMed] [Google Scholar]
- 20.Iida, H., H. Nakamura, T. Ono, M. S. Okumura, and Y. Anraku. 1994. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol. Cell. Biol. 14:8259-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iida, K., T. Tada, and H. Iida. 2004. Molecular cloning in yeast by in vivo homologous recombination of the yeast putative α1 subunit of the voltage-gated calcium channel. FEBS Lett. 576:291-296. [DOI] [PubMed] [Google Scholar]
- 22.Inoue, H., H. Nojima, and H. Okayama. 1990. High frequency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 23.Jaquinoud, M., and M. Peter. 2000. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p). Mol. Cell. Biol. 20:6244-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, G. C., J. A. Prendergast, and R. A. Singer. 1991. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113:539-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanzaki, M., M. Nagasawa, I. Kojima, C. Sato, K. Naruse, M. Sokabe, and H. Iida. 1999. Molecular identification of a eukaryotic, stretch-activated nonselective cation channel. Science 285:882-886. [DOI] [PubMed] [Google Scholar]
- 26.Kanzaki, M., M. Nagasawa, I. Kojima, C. Sato, K. Naruse, M. Sokabe, and H. Iida. 2000. Molecular identification of a eukaryotic, stretch-activated nonselective cation channel. Science 288:1347. [DOI] [PubMed] [Google Scholar]
- 27.Knoblich, J. A. 2001. Asymmetric cell division during animal development. Nat. Rev. Mol. Cell Biol. 2:11-20. [DOI] [PubMed] [Google Scholar]
- 28.Lebere, E., D. Y. Thomas, and M. Whiteway. 1997. Pheromone signaling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7:59-66. [DOI] [PubMed] [Google Scholar]
- 29.Levina, N. N., R. R. Lew, and I. B. Heath. 1994. Cytoskeletal regulation of ion channel distribution in the tip-growing organism Saprolegnia ferax. J. Cell Sci. 107:127-134. [DOI] [PubMed] [Google Scholar]
- 30.Lillie, S. H., and S. S. Brown. 1994. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 125:825-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locke, E. G., M. Bonilla, L. Liang, Y. Takita, and K. W. Cunningham. 2000. A homolog of voltage-gated Ca2+ channels stimulated by depletion of secretory Ca2+ in yeast. Mol. Cell. Biol. 20:6686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKay, V. L., S. K. Welch, M. Y. Insley, T. R. Manney, J. Holly, G. C. Saari, and M. L. Parker. 1988. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc. Natl. Acad. Sci. USA 85:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maruoka, T., Y. Nagasoe, S. Inoue, Y. Mori, J. Goto, M. Ikeda, and H. Iida. 2002. Essential hydrophilic carboxy-terminal regions including cysteine residues of the yeast stretch-activated calcium-permeable channel Mid1. J. Biol. Chem. 277:11645-11652. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto, T. K., A. J. Ellsmore, S. G. Cessna, P. S. Low, J. M. Pardo, R. A. Bressan, and P. M. Hasegawa. 2002. An osmotically induced cytosolic Ca2+ transient activates calcineurin signaling to mediate ion homeostasis and salt tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 277:33075-33080. [DOI] [PubMed] [Google Scholar]
- 35.Messerli, M. A., G. Danuser, and K. R. Robinson. 1999. Pulsatile fluxes of H+, K+, and Ca2+ lag growth pulses of Lilium longiforum pollen tubes. J. Cell Sci. 112:1497-1509. [DOI] [PubMed] [Google Scholar]
- 36.Misu, K., K. Fujimura-Kamada, T. Ueda, A. Nakano, H. Katoh, and K. Tanaka. 2003. Cdc50p, a conserved endosomal membrane protein, controls polarized growth in Saccharomyces cerevisiae. Mol. Biol. Cell 14:730-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser, M. J., J. R. Geiser, and T. N. Davis. 1996. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16:4824-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mount, R. C., B. E. Jordan, and C. Hadfield. 1996. Transformation of lithium-treated yeast cells and the selection of auxotrophic and dominant markers. Methods Mol. Biol. 53:139-145. [DOI] [PubMed] [Google Scholar]
- 39.Muller, E. M., E. G. Locke, and K. W. Cunningham. 2001. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 159:1527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, E. M., N. A. Mackin, S. E. Erdman, and K. W. Cunningham. 2003. Fig1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J. Biol. Chem. 278:38461-38469. [DOI] [PubMed] [Google Scholar]
- 41.Ohsumi, Y., and Y. Anraku. 1985. Specific induction of Ca2+ transport activity in MATa cells of Saccharomyces cerevisiae by a mating pheromone, α-factor. J. Biol. Chem. 260:10482-10486. [PubMed] [Google Scholar]
- 42.Ozaki-Kuroda, K., Y. Yamamoto, H. Nohara, M. Kinoshita, T. Fujiwara, K. Irie, and Y. Takai. 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paidhungat, M., and S. Garrett. 1997. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol. Cell. Biol. 17:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad, K. R., and P. M. Rosoff. 1992. Characterization of the energy-dependent, mating factor-activated Ca2+ influx in Saccharomyces cerevisiae. Cell Calcium 13:615-626. [DOI] [PubMed] [Google Scholar]
- 45.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Sheu, Y., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder, M. 1989. The SPA2 protein of yeast localizes to sites of cell growth. J. Cell Biol. 108:1419-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sprague, G. F., Jr., and I. Herskowitz. 1981. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene, BAR1. J. Mol. Biol. 153:305-321. [DOI] [PubMed] [Google Scholar]
- 52.Sprague, G. F., Jr., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Tada, T., M. Ohmori, and H. Iida. 2003. Molecular dissection of the hydrophobic segments H3 and H4 of the yeast Ca2+ channel component Mid1. J. Biol. Chem. 278:9647-9654. [DOI] [PubMed] [Google Scholar]
- 54.Tokes-Fuzesi, M., D. M. Bedwell, I. Repa, K. Sipos, B. Sumegi, A. Rab, and A. Miseta. 2002. Hexose phosphorylation and the putative calcium channel component Mid1p are required for the hexose-induced transient elevation of cytosolic calcium response in Saccharomyces cerevisiae. Mol. Microbiol. 44:1299-1308. [DOI] [PubMed] [Google Scholar]
- 55.Withee, J. L., J. Mulholland, R. Jeng, and M. S. Cyert. 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshimura, H., T. Tada, and H. Iida. 2004. Subcellular localization and oligomeric structure of the yeast putative stretch-activated Ca2+ channel component Mid1. Exp. Cell Res. 293:185-195. [DOI] [PubMed] [Google Scholar]
- 57.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2 (MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 15:83-91. [PMC free article] [PubMed] [Google Scholar]