Abstract
We have developed a new method for prediction of cis-regulatory binding sites and applied it to predicting NtcA regulated genes in cyanobacteria. The algorithm rigorously utilizes concurrence information of multiple binding sites in the upstream region of a gene and that in the upstream regions of its orthologues in related genomes. A probabilistic model was developed for the evaluation of prediction reliability so that the prediction false positive rate could be well controlled. Using this method, we have predicted multiple new members of the NtcA regulons in nine sequenced cyanobacterial genomes, and showed that the false positive rates of the predictions have been reduced on an average of 40-fold compared to the conventional methods. A detailed analysis of the predictions in each genome showed that a significant portion of our predictions are consistent with previously published results about individual genes. Intriguingly, NtcA promoters are found for many genes involved in various stages of photosynthesis. Although photosynthesis is known to be tightly coordinated with nitrogen assimilation, very little is known about the underlying mechanism. We postulate for the fist time that these genes serve as the regulatory points to orchestrate these two important processes in a cyanobacterial cell.
INTRODUCTION
Cyanobacteria are among the oldest life form on Earth, which are broadly classified as Gram-negative, oxygenic phototrophs (1). These organisms inhabit a broad region of ecological niches from fresh water, soil to diverse open oceanographic areas (1). Some cyanobacteria are capable of producing commercially important natural products, including complex organic compounds with unique structure and stereochemistry (2). Often these compounds are either too expensive or too difficult to be produced in laboratories (3). Moreover, it is estimated that several species/strains of cyanobacteria, mainly in the genera of Synechococcus and Prochlorococcus living in a broad region of open ocean, contribute a significant fraction of Earth's primary production (4). Therefore, their activities have significant impacts on the global environmental changes. Because of these reasons, cyanobacteria have recently attracted broad interests. Nine cyanobacterial genomes have been sequenced as of today, and 31 are in the process of being sequenced (http://www.pdg.cnb.uam.es/cursos/Complutense2001/pages/GenomAna/GOLD0.htm).
Nitrogen is an important element for all forms of life. Numerous sources of nitrogen can be utilized by cyanobacteria. These include nitrate/nitrite, ammonium, urea, cyanate and dinitrogen through fixation (5). Nitrogen control is a phenomenon that occurs widely among microbes, and it consists of repression of the pathways for assimilation of some forms of nitrogen when more easily assimilated forms of nitrogen become available to the cell (5). Nitrogen control in cyanobacteria is mediated by NtcA, a transcriptional regulator that belongs to the CRP (cAMP receptor protein) family, which is different from the well-characterized NtrB–NtrC two-component system in enterics such as Escherichia coli and other proteobacteria (6). All known NtcA sequences from cyanobacteria are highly conserved (5), suggesting that they bind to similar binding sites. A few NtcA binding sites on DNA in some cyanobacteria have been determined using DNase footprinting and found to contain the palindromic motif GTAN8TAC (5). In addition to this motif, the promoter regions of known NtcA-activated genes also contain a −10, E.coli σ70-like box in the form of TAN3T, with the NtcA binding site replacing the −35 box that is present in the E.coli σ70-type promoters (5). NtcA-regulated genes are involved not only in the nitrogen assimilation process but also in the cell differentiation of heterocyst development in some diazotrophic species, such as Nostoc sp. PCC 7120 (PCC7120) (5). Nevertheless, NtcA-regulated genes are far from completely known even in some relatively well studied species, such as Synechocystis sp. PCC 6803(PCC6803) or Synechococcus PCC 7942 (PCC7942) (7), needless to mention some newly sequenced and less-studied species.
The availability of an increasing number of complete genome sequences has made it possible to conduct systematic analyses of NtcA-regulated genes in the cyanobacteria using comparative genomics approaches. Phylogenetic footprinting is one of the most popular approaches for identification of new cis-regulatory binding sites (8–13). In this method, the intergenic regions of orthologous genes from closely related genomes are used to identify a conserved motif based on the assumption that the regulatory elements are highly conserved in closely related species. The resulting profile is then used to scan the intergenic (or inter-operonic) regions of the whole genomes to identify additional cis-regulatory sites. Though meaningful results have been obtained through careful manual analyses of the predicted candidates, such a scanning process unavoidably results in rather high false positive rates due to random occurrences of the similar motifs (9,12,13). Thus, reducing the false positive rate of this scanning process without losing the sensitivity remains a major goal of the phylogenetic footprinting procedure. In order to achieve more accurate predictions of NtcA binding sites in cyanobacterial genomes, we first need to reduce the false positive rate significantly.
The high false positive rate of such a scanning process is mainly due to the often variable nature of cis-regulatory binding sites and their short lengths, and therefore the possibility of having similar sequences by chance is rather high for whole-genome applications. To exclude randomly occurring ‘conserved motifs’, additional information about the promoter structure is needed. We believe that at least two pieces of information can be used for this purpose. First, it is generally true that as in the case of NtcA regulated genes, the regulation of transcription in prokaryotes often requires the binding of a σ-factor of the RNA polymerase to a specific sequence box in the flanking region of a cis-regulatory element (14). Therefore, the information about the σ-factor binding box can be used to reduce the false positives of the scanning process. However, it is not a simple task to identify the σ-factor binding box using phylogenetic footprinting, since the box is only ∼6 bp long, and often has only ∼3 positions conserved. Hence, the existing motif finding algorithms generally return too many equally ‘high quality’ motifs if, for instance, 500 bp upstream intergenic regions are searched. However, in many cases, the σ-factor binding box is located in a relatively fixed downstream region of the cis-regulatory binding site. If only the limited flanking regions are searched, most motif finding algorithms can identify the true motifs in the greatly reduced search space. Second, the presence of similar motifs in the regulatory regions of the orthologous genes in other related genomes can increase the prediction accuracy as has been shown previously (8,13). However, a quantitative evaluation of this approach has not been done, which we intend to conduct in this paper.
In this paper, we have improved the phylogenetic footprinting procedure by looking for multiple binding sites in the promoter regions of genes, and have designed a new scoring function that incorporates the information of a predicted promoter of a gene and the information from promoters of its orthologues in related genomes. Furthermore, based on the observation that the cis-regulatory sites occur more frequently in the intergenic region than in the coding regions, we have introduced a probabilistic model to distinguish true binding sites from the randomly occurring ones. We have successfully applied the new method to the analyses of NtcA regulons in nine sequenced cyanobacterial genomes, and thus we have observed a number of interesting results.
MATERIAL AND METHODS
Materials
Sequences and annotation files for nine sequenced cyanobacteria genomes were downloaded from the GenBank (ftp.ncbi.nih.gov/genomes/Bacteria/). These nine cyanobacterial genomes are Gloeobacter violaceus PCC 7421(PCC7421), Nostoc sp. PCC 7120(PCC7120), Prochlorococcus marinus CCMP1375(PCC1375), Prochlorococcus marinus MED4(MED4), Prochlorococcus marinus MIT9313(MIT9313), Synechococcus elongatus PCC 6301(PCC6310), Synechococcus sp. WH8102(WH8102), Synechocystis sp. PCC 6803(PCC6803) and Thermosynechococcus elongates BF-1(thermosynechococcus). The NtcA sequences of other cyanobacteria were also downloaded from the GenBank.
Transcription unit and orthologue predictions
In order to assign each gene in a genome to a transcription unit, we used a simple rule to predict transcription units, i.e. we predicted tandem genes on the same strand with an intergenic distance less than 45 bp to be a transcription unit. A single gene that was not predicted to belong to any transcription unit was predicted to be a single gene transcription unit. We predicted two genes in two genomes to be orthologous to each other if they are a pair of reciprocal best hit in BLASTP searches with an E-value <10−20 in both directions when the two genomes are searched against each other.
Phylogenetic footprinting for NtcA binding sites and their downstream −10, E.coli σ70-like boxes
We pooled entire upstream intergenic regions (if it is longer than 800 bp, then only the immediate upstream 800 bp was pooled) of the following genes in each of the nine cyanobacterial genomes (if it encodes the gene) to identify conserved palindromic 14mers as putative NtcA binding sites for each gene using the CUBIC program (15). These genes are known to be regulated by NtcA in at least one cyanobacterium [for a review see ref. (5)], including ammonia permease amt, nitrogen global regulator ntcA, glutamine synthetase glnA, signal transduction protein PII glnB, urea transporter subunit A urtA, nitrite reductase nirA, heterocyst differentiation protein hetC, heterocyst specific ABC-transporter devB, group 2 σ70 factor rpoD-V, nitrate assimilation transcriptional activator ntcB and isocitrate dehydrogenase icd. The identified motifs with a score above a pre-selected cutoff were returned. The 31 bp downstream regions of the identified putative NtcA binding sites were pooled to identify 6 bp motifs in the form of BBN3B using the CUBIC program, where B stands for a conserved base and N3 for three variable bases.
Extraction of inter-transcription unit regions and coding sequences
Let U(g1, …, gn) be any transcription unit containing n genes g1, …, gn in a genome G. For each U(g1, …, gn) in G, we extracted its entire upstream intergenic region (if it is longer than 800 bp, then only the immediate upstream 800 bp was extracted), denoted by IU(g1,…,gn) (or simply IU) for scanning the possible cis-regulatory motifs. Meanwhile, we extracted a randomly chosen coding sequence with the same length as IU(g1,…,gn) from G, denoted as CU(g1,…,gn) (or simply CU) for scanning the randomly occurring motifs. We call both IU(g1,…,gn) and CU(g1,…,gn) are associated with U(g1, …, gn) and with each of genes g1, …, and gn as well.
Scanning genomic sequences and the scoring functions
Each extracted sequence t (IU or CU) from each genome was scanned first by the above-constructed profile of the NtcA binding sites. For each t, we return the motif with the highest score defined by the following formula (1). Then up to 31 bp downstream region of each identified putative NtcA binding site is scanned for a −10 like box with the highest score defined by the formula (1), using the corresponding profile.
The score of a motif found in a sequence segment t by scanning with a profile M is defined as
| 1 |
| 2 |
| 3 |
where l is the length of the motifs of M, h any substring of t with length l, h(i) the base at position i of h, p(i, b) the relative frequency of base b at position i in M, q(b) the relative frequency of base b occurring in the background, and n the number of motifs in M. A pseudo-count 1 is added to the frequency of each base at each position in the profile when computing p(i,b). The coefficient a is for normalization so that Ii is in the region [0,1].
When multiple profiles M1, …, Mz are used for scanning, the score of concurrence of multiple putative binding sites in the sequence t is defined as
| 4 |
We now define a new score for concurrence of multiple binding sites in a sequence associated with a transcription unit by also considering the presence of similar motifs in the sequences associated with its orthologous genes in other genomes. Let t be the extracted sequence (IU or CU) associated with a transcription unit U(g1, …, gn) in genome T. If gi has orthologues in mi closely related genomes G1, …, Gmi, let ok(gi) be the same type of extracted sequence (IU or CU) associated with the orthologue of gi in genome Gk. Then the score of concurrence of the multiple binding sites in t is redefined as
| 5 |
where di,j,k, is the Hamming distance between the motif found by profile Mj in t, and the corresponding motif found in ok(gi), and lj is the length of the motifs of profile Mj.
Statistical significance of cis-regulatory binding site prediction
When we consider all the extracted IU's and CU's in a genome, the scores of binding sites found in IU's and CU's, i.e. S(IU) and S(CU), respectively, are random variables. Let p[S(t) > s] be the probability that the extracted sequence t (IU or CU) has putative binding sites with a score S(t) > s as defined by (1), (4) or (5). To compute p[S(CU) > s] for a genome, we generated 100 CU's associated with each transcription unit U(g1, …, gn) in each genome and computed S(CU) for each CU to avoid possible biased sampling. We then used the following log odds ratio (LOR) to estimate the confidence of predictions,
| 6 |
Since p[S(CU) > sc] is the probability of type I error for testing the null hypothesis that IU does not contain a motif when S(IU) is greater than a cutoff sc, we used it to estimate the false positive rate of our predictions. The algorithm was implemented using Perl scripts, which are freely available upon request.
Estimation of prediction sensitivity
Since the entire set of NtcA binding site are not known in any cyanobacterial genome, it is difficult to calculate the sensitivity of the predictions directly. We estimated the prediction sensitivity of a cutoff score for each genome by computing the portions of the sites in the whole training sets from the nine genomes, which can be recovered by the cutoff as if they all appeared in that genome.
RESULTS AND DISCUSSION
The DNA binding domains of NtcA in cyanobacteria are conserved
As shown in Figure 1A, the DNA binding domain of the available NtcA sequences from 17 cyanobacteria is predicted to form a helix–turn–helix motif like that of the CRP of E.coli (α-helices E and F) (16). The amino acid sequences of these helix–turn–helix motifs are identical except that Ala at position 4 in the consensus sequence is replaced by Ser in MIT9313 and CCMP1375, and Val at position 16 in the consensus sequence is replaced by Ile in WH7803, MIT8313, WH8102, CCMP1375 and MED4. Arg at position 13 in the consensus sequence is conserved in all sequences, in which CRP is in direct contact with the nucleotides in the cis-regulatory binding sites (17). Thus it is highly likely that NtcA will recognize similar DNA sequences in different cyanobacteria. Figure 1B shows the phylogenetic relationship of the whole NtcA protein sequences in the nine cyanobacterial genomes in our analyses and that of CRP of E.coli. Clearly, WH8102 and the three Prochlorococcus strains CCMP1379, MED4 and MIT9313 form a group, and the rest five genomes form another group on this tree, which is similar to their taxonomic tree based on 16S rDNA sequences [data not shown, also see ref. (18)].
Figure 1.
(A) Multiple sequence alignments of the DNA binding domains of the known NtcA sequences of 17 cyanobacteria and that of the CRP of E.coli. The domain corresponds to the E and F α-helices in the crystal structure of the CRP of E.coli. Abbreviations: Synechococcus sp. WH 7803(WH7803), Prochlorococcus marinus sp. MIT 9313(MIT9313), Synechococcus sp. WH 8102(WH8102), Prochlorococcus marinus sp. CCMP1375(CCMP1375), Prochlorococcus marinus subsp. pastoris (Proch_pastoris), Prochlorococcus marinus sp. CCMP1986(MED4), Crocosphaera watsonii WH 8501(WH8501), Cyanothece sp. ATCC 51142(Cyanothece), Synechocystis sp. PCC 6803(PCC6803), Nostoc sp. PCC 7120(PCC 7120), Synechococcus sp. PCC 7002(PCC7002), Trichodesmium sp. IMS101(Trichodesmium), Plectonema boryanum (Plectonema), Synechococcus elongatus PCC 6301(PCC6301), Synechococcus sp. PCC 7942(PCC7942), Thermosynechococcus elongatus BP-1 (Thermosynechococcus), Gloeobacter violaceus PCC 7421(PCC7421), E.coli CRP (E.coliCRP). (B) Phylogenetic relationships of NtcAs of the nine genomes in our analyses. The unrooted tree was generated by the neighbor-joining method based on the multiple amino acid sequence alignments by the ClustalX program using the default settings. Scale bar, substitutions per position. (C) and (D) Logo representations of the NtcA binding sites and −10 like boxes in the training sets. Logos are generated by the Weblogo server (http://weblogo.berkeley.edu/logo.cgi).
The profiles of NtcA binding sites and −10, E.coli σ70 consensus-like boxes
We pooled the upstream regions of the orthologues in each of the nine cyanobacterial genomes of the 11 genes (see Materials and Methods) to identify putative NtcA binding sites for each of the genes. We identified 51 putative NtcA binding sites from a total of 65 pooled upstream sequences (Supplementary Table s1). Among these 51 sites, we have correctly found 10 of 11 known NtcA binding sites in PCC6803 and PCC7120 (Supplementary Table s1), suggesting that at least most of the NtcA binding sites we found are likely to be correct. We failed to identify the known NtcA binding site for the icd of PCC6803, as this site diverges from the canonical NtcA binding sites (GTAN8TAC) in the second triplet (see below). These 51 sites constitute the training set A1 for the NtcA binding site (Supplementary Table s1). The -31 bp downstream regions of these predicted NtcA binding sites are further searched for 6 bp, −10, E.coli σ70-like boxes (−10 like boxes) and the identified sites form the training set B1 for the −10 like box (Supplementary Table s1). The 10 known −10 like boxes downstream the respective known NtcA sites in PCC6803 and PCC7120 are also correctly identified (Supplementary Table s1), suggesting that at least most of the −10 like boxes we found are likely to be correct. In addition, we have collected 12 experimentally verified NtcA binding sites and their downstream −10 like boxes from seven other cyanobacteria (5) (Supplementary Table s2). We also included in this list the sites for icd in PCC6803 that we failed to find by phylogenetic footprinting (Supplementary Table s2). They constitute the training sets A2 and B2 for NtcA binding sites and −10 like boxes, respectively. We combined A1 and A2 to construct the profile of NtcA binding sites, and B1 and B2 to construct the profile of −10 like boxes. The logo representations of the profiles of the combined NtcA binding sites and −10 like boxes are shown in Figure 1C and 1D, respectively.
The conventional scanning method results in high false positive rates
As has been previously noted (11,13), scanning the intergenic (or inter-operonic) regions (we shall call such regions inter-transcription unit regions) of a whole genome using the profile of a transcription regulator often results in many false positive binding sites. However, the seriousness of this problem has not been quantitatively evaluated to date owing to the fact that it is often difficult to be sure that an uncovered high-scoring motif is really a false positive. Here we address this problem by taking advantage of the observation that all the known NtcA binding sites are located in the inter-transcription unit regions, and none has been found in the coding regions (5). Therefore, we assume that at least the majority of the high-scoring putative NtcA binding sites found in the coding regions are false positives. We then ask the question as how likely a high-scoring spurious motif is returned by scanning with a profile of binding sites.
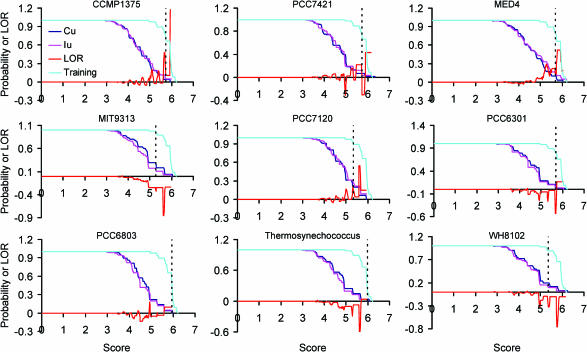
To do this, we have computed the LOR (see Materials and Methods) for the distribution of the scores of putative NtcA binding sites found in the extracted inter-transcription unit regions (IU) over that of sites found in the randomly selected coding regions (CU) (see Materials and Methods). Intuitively, if a scanning process and its associated scoring function capture the important features of the binding sites, then the probability to find a high-scoring binding site should be much higher in the IU's than in the CU's, because the former should have more chance of having binding sites than the latter. However, as shown in Figure 2, in the worse cases as for the genomes MIT9313, PCC6301, thermosynechococcus and WH8102, the probabilities of high-scoring putative binding sites to occur in the IU's are lower than in the CU's. Even for the best cases as for the genomes CCMP1375, PCC7421, MED4, PCC7120 and PCC6803 where the probabilities of high-scoring putative motifs to occur in the IU's are higher than in the CU's, the values of LOR are generally small and often highly oscillating, indicating that this conventional scanning procedure as well as the associated scoring function (1) (see Materials and Methods) does not capture the essence of the real binding sites. Furthermore, if a cutoff of the scores is chosen for each genome such that all the members from that genome in the training set are included, rather large portions (3.63–22.61% in our genomes) of the IU's will be predicted to contain putative NtcA binding sites as shown in the column 5 of Table 1. This means that 56–1200 NtcA sites are predicted with a score as high as the real ones for each genome (column 6 of Table 1). The associated P-values for these predictions estimated by our probabilistic model (6) are also rather insignificant in most cases, ranging from 0.0355 to 0.2961 with an average 0.1183 (column 4 of Table 1), suggesting again that a large portion of these predicted NtcA binding sites (56–1200) are false positives. To evaluate how likely weak real binding sites can be uncovered by these cutoffs for each genome, we estimated the prediction sensitivities at these cutoffs by computing the portion of binding sites in the training sets from all the nine genomes (51 sites in Supplementary Table s1 +1 site in Supplementary Table s2 from PCC6803) that can be predicted by the score cutoff for each genome as if all these sites appeared in this genome (see Materials and Methods). As shown in Figure 2 and column 7 of Table 1, the sensitivities are rather unstable and generally low, ranging from 21.28 to 95.75% with an average 71.87%, even with such rather large numbers of predictions (56–1200 sites). If we reduce the false positive rates by choosing a higher cutoff so that the P-value is <0.01, then only rather low sensitivities could be obtained, ranging from 0 to 46.81% with an average 17.97% (column 8 of Table 1). These results unequivocally demonstrate that the conventional method for scanning the inter-transcription unit regions with a profile of a transcriptional factor binding sites may result in rather high false positives, and at the same time, many true binding sites might be missed.
Figure 2.
The probability [p(S > s)] of the scores of the 52 NtcA binding sites in the training sets from the nine cynaobacteria (cyan), and those of the scores of putative NtcA binding sites found in the IU's (pink) and UU's (blue) and their log odds ratio (red) when only the profile of NtcA binding sites is used for scanning. The doted vertical line in each panel indicates the largest score cutoff for the genome to include all the binding sites from that genome in the training sets.
Table 1.
Summary of the scanning results using only the profile of NtcA binding sites
| Genome | No. of transcription units | Cutoffa | P-value at cutoff | Percentile at cutoff | No. of sites predicted at cutoff | Sensitivity at cutoff | Sensitivity at P < 0.01 |
|---|---|---|---|---|---|---|---|
| CCMP1375 | 1285 | 5.72 | 0.0375 | 0.0475 | 61 | 0.7447 | 0.4681 |
| PCC7421 | 3042 | 5.72 | 0.0726 | 0.0700 | 213 | 0.7447 | 0.0000 |
| MED4 | 1097 | 5.68 | 0.0960 | 0.1285 | 141 | 0.7447 | 0.1489 |
| MIT9313 | 1592 | 5.26 | 0.2961 | 0.1470 | 234 | 0.9575 | 0.1489 |
| PCC7120 | 4509 | 5.34 | 0.1968 | 0.2661 | 1200 | 0.8936 | 0.4681 |
| PCC6301 | 1805 | 5.66 | 0.1050 | 0.0310 | 56 | 0.8085 | 0.0851 |
| PCC6803 | 2540 | 5.94 | 0.0355 | 0.0441 | 112 | 0.4681 | 0.2128 |
| Thermosyn | 1600 | 5.98 | 0.0334 | 0.0363 | 58 | 0.2128 | 0.0851 |
| WH8102 | 1480 | 5.34 | 0.1916 | 0.1155 | 171 | 0.8936 | 0.0000 |
| Average | 2106 | 5.63 | 0.1183 | 0.0985 | 250 | 0.7187 | 0.1797 |
aScore cutoff to include the sites in the training sets from the genome.
Information integration greatly improves binding site prediction
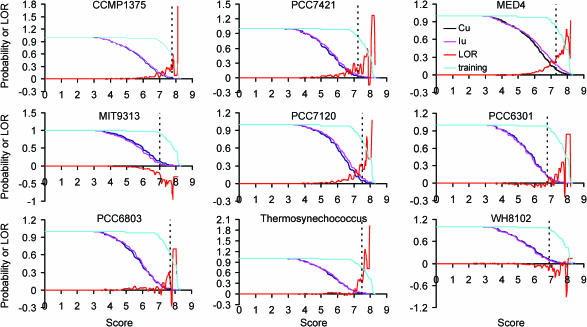
Based on the observation that a conserved −10, E.coli σ70-like box (−10 like box) in the form of TAN3T appears in the downstream of all known NtcA binding sites, our first attempt in improving the prediction accuracy (both the specificity and sensitivity) of NtcA binding sites was to incorporate the information of a putative −10 like box in the downstream of the putative NtcA binding site. To do so, we have computed a combined score as defined by (4) (see Materials and Methods) for each pair of the putative NtcA site and its down stream −10 σ70-like box found in each extracted genomic sequences (IU or CU). As shown in Figure 3, the LORs of the scores are greatly improved for all the genomes except for PCC6301, suggesting that the combined scores are more likely to be able to discriminate the real binding sites in the IU's from the randomly occurring spurious ones in the CU's. In agreement with this conclusion, the P-values at the cutoffs of scores to include the members from the respective genomes in the training set decrease (on an average of 2-fold) for all genomes except for PCC6301 compared to the results when scanning with only the profile of NtcA binding sites (Table 4), indicating that the prediction specificities are greatly improved for most of the genomes. Meanwhile, the sensitivities (79.20% on average) at these cutoffs also increase except for CCMP1375, MIT9313 and PCC7120 (column 7 of Table 2), compared to the results (71.87% on average) when scanning with only the profile of NtcA binding sites (column 7 of Table 1). More importantly, the sensitivities at cutoffs with P-values <0.01, have greatly increased for all nine genomes (60.04% on average, column 8 in Table 2), compared to the results (17.97% on average) when scanning with only the profile of NtcA binding sites (column 8 of Table 1). These results indicate that scanning for binding sites with two profiles greatly improves the prediction accuracy in most of the cases.
Figure 3.
The probability [p(S > s)] of the scores of the 52 NtcA promoters in the training sets from the nine cynaobacteria (cyan), and those of the scores of putative NtcA promoters found in the IU's (pink) and CU's (blue) and their log odds ratio (red) when the profiles of NtcA binding sites and −10 like boxes are used for scanning. The doted vertical line in each panel indicates the largest score cutoff for the genome to include all the binding sites from that genome in the training sets.
Table 4.
Improvement of P-values through information integration
| Genome | s/s + pa | s + p/s + p + ob | s/s + p + oc |
|---|---|---|---|
| CCMP1375 | 1.943 | 14.846 | 28.846 |
| PCC7421 | 2.022 | 44.875 | 90.750 |
| MED4 | 1.075 | 7.633 | 8.205 |
| MIT9313 | 2.257 | 6.798 | 15.342 |
| PCC7120 | 4.364 | 5.638 | 24.600 |
| PCC6301 | 0.791 | 17.013 | 13.462 |
| PCC6803 | 2.247 | 31.600 | 71.000 |
| Thermosyn | 2.141 | 39.000 | 83.500 |
| WH8102 | 1.378 | 15.796 | 21.773 |
| Average | 2.024 | 20.355 | 39.720 |
as/p + s: ratio of the P-value at the score cutoff when only NtcA profile is used over that when profiles of NtcA sites and −10 boxes are used.
bs + p/p + s + o: ratio of P-value at the score cutoff when the profiles of NtcA sites and −10 boxes are used over that when the profiles of NtcA sites and −10 boxes as well as appearance of similar sites for the orthologues are used.
cs/s + p + o; the ratio of P-value at the score cutoff when only the profile of NtcA sites is used over that when the profiles of NtcA sites and −10 boxes as well as appearance of similar sites for the orthologues are used.
Table 2.
Summary of the scanning results using the profiles of NtcA binding sites and −10 like boxes
| Genome | No. of transcription units | Cutoffa | P-value at cutoff | Percentile at cutoff | No. of sites predicted at cutoff | Sensitivity at cutoff | Sensitivity at P < 0.01 |
|---|---|---|---|---|---|---|---|
| CCMP1375 | 1285 | 7.74 | 0.0193 | 0.0381 | 49 | 0.5958 | 0.5319 |
| PCC7421 | 3042 | 7.22 | 0.0359 | 0.0533 | 162 | 0.8511 | 0.6809 |
| MED4 | 1097 | 7.30 | 0.0893 | 0.1641 | 180 | 0.8085 | 0.8085 |
| MIT9313 | 1592 | 7.02 | 0.1312 | 0.0766 | 122 | 0.8936 | 0.5106 |
| PCC7120 | 4509 | 7.53 | 0.0451 | 0.0783 | 353 | 0.6809 | 0.5745 |
| PCC6301 | 1805 | 6.76 | 0.1327 | 0.1230 | 222 | 0.9575 | 0.6809 |
| PCC6803 | 2540 | 7.60 | 0.0158 | 0.0315 | 80 | 0.6809 | 0.5106 |
| Thermosyn | 1600 | 7.48 | 0.0156 | 0.0344 | 55 | 0.7234 | 0.6809 |
| WH8102 | 1480 | 6.89 | 0.1390 | 0.0791 | 117 | 0.9362 | 0.4255 |
| Average | 2106 | 7.28 | 0.0693 | 0.0754 | 149 | 0.7920 | 0.6004 |
aScore cutoff to include the sites in the training sets from the genome.
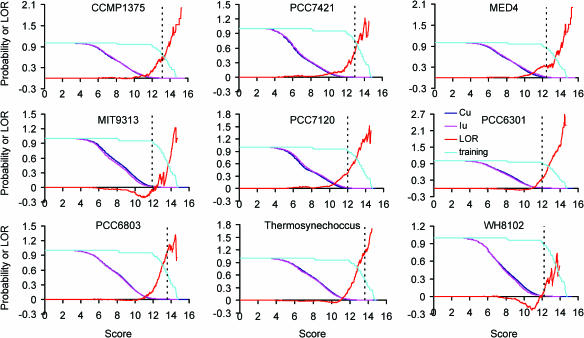
In order to further improve the prediction accuracy, we looked for the presence of the similar putative NtcA binding sites as well as −10 σ70-like boxes in the regulatory regions of the orthologous genes across multiple genomes, and a score that combines all these sources of information as defined in (5) (see Materials and Methods) was computed for each putative promoter in the IU's and CU's. As shown in Figure 4, the LOR's for all nine genomes increases monotonically beyond a certain value of the scores, indicating that this new score function can well differentiate between the putative binding sites found in the regulatory regions and those found in coding regions. Since most of the high-scoring sites, if not all, found in the coding regions are supposed to occur by accident, the much higher probability of occurrence of the high-scoring sites found in the regulatory regions must have been under great selection pressure according to the evolutionary theory, and thus, they are biologically meaningful. In this case, they are more likely to be of regulatory function. As summarized in the Tables 3 and 4, the P-values (column 4 of Table 3) (0.0004–0.0193, average 0.0066) at the cutoffs to include the members of the binding sites from the respective genomes in the training sets decreases on an average of 20.355 (5.638–44.875) and 39.7197 (8.205–90.750) fold when compared to the results in Tables 2 and 1 where only two and one source of information about the binding sites in the regulatory regions were utilized, respectively. The sensitivities at these cutoffs to uncover the members from all the genomes in the training sets decrease slightly in PCC7421, PCC6301, PCC6803, thermosynechococcus and WH8102 (column 7 of Table 3) when compared to the results in column 7 of Table 2 where information about the binding sites in the regulatory regions of orthologues is not utilized, an indication of possible overtraining in these genomes. However, this problem can be corrected by choosing a relatively lower score cutoff so that a certain statistical significance level is still guaranteed. For instance, the sensitivities at score cutoffs with P-value < 0.01 greatly increase (column 8 of Table 3) (82.99–95.75%, average 90.78%) for all genomes when compared to those in Table 2 (42.55–80.85%, average 60.04%) where information about the binding sites in the regulatory regions of orthologues is not utilized. All these results strongly suggest that our new scoring function can greatly improve the accuracy of binding site predictions.
Figure 4.
The probability [p(S > s)] of the scores of the 52 promoters in the training sets from the nine cynaobacteria (cyan), and those of the scores of putative promoters found in the IU's (pink) and IC's (blue) and their log odds ratio (red) when both the profiles of NtcA binding sites and −10 like boxes are used for scanning, and the presence of similar sites in the regulatory regions of orthologues in other genomes are also considered. The doted vertical line in each panel shows the largest score cutoff for the genome to include all the binding sites from that genome in the training sets except for PCC6801 and PCC7120, where the largest cutoff is chosen so that only icd of PCC8603, and hetC and icd of PCC7120 are excluded, respectively (see text for details).
Table 3.
Summary of the scanning results using multi-source of information
| Genome | No. of transcription units | Cutoffa | P-value at cutoff | Percentile at cutoff | No. of sites predicted at cutoff | Sensitivity at cutoff | Sensitivity at P < 0.01 | Percentile at P < 0.01 | No. of sites predicted at P < 0.01 |
|---|---|---|---|---|---|---|---|---|---|
| CCMP1375 | 1285 | 13.14 | 0.0013 | 0.0047 | 6 | 0.7021 | 0.8936 | 0.0195 | 25 |
| PCC7421 | 3042 | 12.98 | 0.0008 | 0.0033 | 10 | 0.7021 | 0.9575 | 0.0151 | 46 |
| MED4 | 1097 | 12.46 | 0.0117 | 0.0246 | 27 | 0.8723 | 0.8299 | 0.0191 | 21 |
| MIT9313 | 1592 | 11.88 | 0.0193 | 0.0176 | 28 | 0.9574 | 0.8723 | 0.0101 | 16 |
| PCC7120 | 4509 | 12.02 | 0.0080 | 0.0189 | 85 | 0.9149 | 0.9575 | 0.0235 | 106 |
| PCC6301 | 1805 | 12.04 | 0.0078 | 0.0194 | 35 | 0.9149 | 0.9575 | 0.0222 | 40 |
| PCC6803 | 2540 | 13.56 | 0.0005 | 0.0047 | 12 | 0.5319 | 0.8936 | 0.0169 | 43 |
| Thermosyn | 1600 | 13.60 | 0.0004 | 0.0056 | 9 | 0.4893 | 0.8936 | 0.0212 | 34 |
| WH8102 | 1480 | 12.20 | 0.0088 | 0.0115 | 17 | 0.8723 | 0.8936 | 0.0115 | 17 |
| Average | 2106 | 12.53 | 0.0066 | 0.0122 | 25 | 0.7754 | 0.9078 | 0.0175 | 39 |
In order to predict new members of the NtcA regulons in the nine sequenced cyanobacterial genomes, one can choose for each genome a cutoff of the combined score S(t) as defined in (5) (see Materials and Methods) so that a predefined statistical significance level can be achieved. In general, the lower a P-value is chosen, the higher confidence one has with the predictions. Specifically, we consider the predictions with a P-value < 0.01 as highly statistically significant, and those with a P-value < 0.05 as statistically significant. Shown in the Table 3 (column 10) are the numbers of NtcA promoters predicted in each genome at score cutoffs with P-value < 0.01. At these cutoffs, the binding sites from the respective genomes in the training set are recovered for all the genomes except for MED4, MIT9313, PCC6803 and PCC7120, where promoters for glnB (pmm1463, P-value < 0.0117), amt1 (pmt1853, P-value < 0.0193), icd(slr1289), and hetC (alr2817, P-value < 0.4078) and icd (alr1827, P-value < 0.1356) in the training set are not recovered, respectively. The predicted NtcA promoters at a score cutoff with P-value < 0.01 for each genome are shown in Supplementary Tables s3–s11 as Supplementary Data. The complete prediction results for each genome can be found at http://csbl.bmb.uab.edu/~zhx/nitrogen/ntca/. As can be seen from these results, genes bearing a high scoring putative NtcA promoter (e.g. P-value < 0.05) always have at least an orthologue in other genomes.
Regulation by NtcA of nitrogen assimilation related genes
It is well known that different species of cyanobacteria can utilize various sources of nitrogen as a result of acclimation to their ecological niches (5). In general, ammonium is the preferred nitrogen source for cyanobacterial cells. In an ammonium-replete environment, the expression of genes for the assimilation of other sources of nitrogen is suppressed, a phenomenon called nitrogen control (5). In ammonium-limited environments, however, genes involved in the uptake and metabolism of alternative sources of nitrogen will be induced through the binding of NtcA to the cis-regulatory elements in the form of GTAN8TAC in the promoter regions of the genes. A downstream −10 σ70-like box in the form TAN3T/A is also required for the action of NtcA (5). All forms of assimilated nitrogen are eventually converted into ammonium by relevant enzymes. Ammonium is then incorporated in the carbon backbones through the glutamine synthetase-glutamate synthase (GS-GOGAT) cycle (5) in which glutamine synthetase GlnA is the major player to catalyze the formation of glutamine from ammonium and 2-oxoglutarate produced by NADP+-isocitrate dehydrogenase (the product of icd gene) (19). Since cyanobacteria do not encode a 2-oxoglutarate dehydrogenase, the only fate of 2-oxoglutarate is to be converted into glutamate/glutamine (5). Hence, 2-oxoglutarate is widely believed to be an indicator of C/N balance in a cyanobacterial cell (5). A high level of 2-oxoglutarate signals a high C/N ratio in a cyanobacterial cell (20). It has been shown that 2-oxoglutarate is the effector molecule for NtcA activation (21–24). The activities of NtcA and some other nitrogen assimilation related molecules are also subject to modulations of the signal transduction protein PII encoded by the glnB gene at transcriptional (7) and/or posttranslational levels (25). The activities of PII are controlled by both its phosphorylation state in a seryl residue and levels of 2-oxoglutrate and ATP in the cell (21,24,26). These genes are known to be regulated by NtcA in some species of cyanobacteria [for a schematic summary see ref. (22)]. However, the regulations by NtcA of nitrogen assimilation related genes are far from being understood in all cyanobacterial species/strains, in particular, in some newly sequenced genomes used in our analyses.
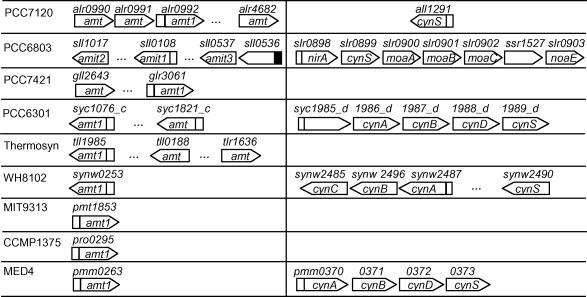
As shown in Figure 5, strong NtcA promoters are found without exception for the ammonium transporters of the amt family, at least a copy of which is encoded in all the nine cyanobacterial genomes analyzed, suggesting that ammonium uptake is tightly under NtcA regulation in these genomes. Strong binding sites are also found for the second copy of amt in PCC6803 and PCC6301 but not in PCC7120 and thermosynecococcus. These NtcA promoters might be responsible for the Amt1 induction under low ammonia concentrations as it has been shown in PCC7942 (27).
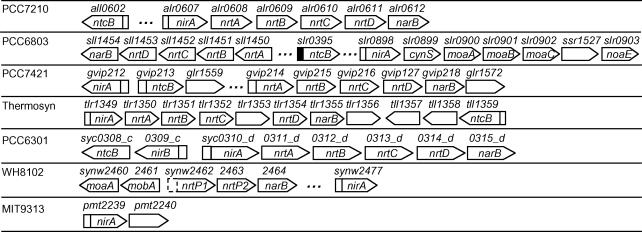
Figure 5.
Putative NtcA promoters found for gene clusters involved in ammonia and cyanate uptakes. A open or solid box upstream of a gene (arrowed box) represents a putative NtcA promoter identified at P-value <0.01 or 0.05, respectively. A ‘…’ indicates that there is more than one gene in between its two flanking genes.
Nitrate and nitrite are the most common alternative nitrogen sources for cyanobacteria. However, it seems that not all cyanobacteria analyzed in this study can utilize these forms of nitrogen since the genomes of CCMP1375 and MED4 do not seem to have any known genes responsible for nitrate/nitrite uptake and their subsequent reduction into ammonium. This might be related to their ammonium-rich ecological niches in deeper seawater (4,18). On the other hand, three types of nitrate/nitrite transporters are exclusively encoded in the other seven cyanobacterial genomes, namely, the ABC type transporters nrtABCD in PCC7120, PCC7421, PCC6803, PCC6301 and thermosynecococcus, the major facilitator type nrtP in WH8102 and the furmate/nitrite transport type in MIT9313 (pmt2240). These transporters are often clustered with the related genes, i.e. the nitrate reductase narB is often located downstream of the transporters, and nitrite reductase nirA and regulator ntcB upstream of the transporters (Figure 6). Strong NtcA promoters are found in the regulatory regions of nirA in these seven genomes, suggesting that they and their downstream transporters are under tight control of NtcA (Figure 6). It has been demonstrated that the positive effect of nitrite on the expression of NirA and NrtABCD-NarB is mediated by the LysR family protein NtcB (28–31). The gene nirA is splited from the nrtABCD-narB operon in PCC421, however, an NtcA promoter is found for the latter. Furthermore, an NtcA binding site (GTGacaaccgcTAC) and −10 σ70-like box (TGagtA) is found in the upstream of the nrtP transporters synw2462–2463 in WH8102 (Figure 6), though its score is not high due to the lack of an orthologue in the genomes in our analyses. It will be interesting to investigate whether these two genes and the downstream narB are under the regulation of this putative NtcA promoter (Figure 6). Interestingly, NtcB is only encoded in the genomes where NrtABCD transporter is also encoded such as in PCC7120, PCC7421, PCC6803, PCC6301 and thermosynechococcus, but not in the genomes where a different type of transporter is utilized, such as NrtP in WH8102 and furmate/nitrite transporter in MIT9313 (Figure 6). Strong NtcA promoters are found in the regulatory regions of NtcB in PCC7120, PCC7421, PCC6803, PCC6801, PCC6301 (forming an operon with nirB) and thermosynechococcus, suggesting that they are all regulated by NtcA as suggested previously in PCC7120 (28,29,31) (Figure 6).
Figure 6.
Putative NtcA promoters found for gene clusters involved in nitrate/nitrite uptake. A open or solid box upstream of a gene (arrowed box) represents a putative NtcA promoter identified at P-value <0.01 or 0.05, respectively. A dashed box represents a strong putative NtcA promoter, but it is not identified at these two statistical significant levels. A ‘…’ indicates that there is more than one gene in between its two flanking genes.
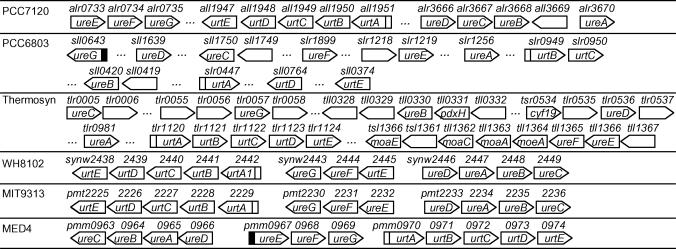
Urea is another common form of nitrogen source for some cyanobacteria living in oligotrophic oceans where the concentration of urea is at a level of 0.1–1 µM (32). High affinity ABC type urea transporters UrtABCDE are encoded in PCC7120, PCC6803, MIT9313, MED4, WH8102 and thermosynechococcus. The subunits of this transporter are located in the same operon except for PCC6803 in which they are scattered along the chromosome (Supplementary Table s7). High-scoring promoters are found without exception for urtA in these genomes (Supplementary Table s7), suggesting that urtA and its downstream genes urtBCDE that might form an operon with urtA are under NtcA regulation as it has been demonstrated in PCC7120 (33). Scattered urtA and urtB in PCC6803 are also likely to be regulated by NtcA since high-scoring promoters are found in their respective regulatory regions (Figure 7). Moreover, the three subunits of urease UreABC (urea amidohydrolase) and accessory proteins involved in urease assembly (UreDEFG) are encoded in these six genomes. They form two divergently transcribed operons in the forms of ureDABC and ureEFG next to the urt genes in MED4, MIT9313 and WH8102 (Figure 7). In contrast, the ure genes are scattered along the chromosome in at least three transcription units in PCC7120, PCC6803 and thermosynechocossus (Figure 7). No high-scoring NtcA promoters are found for ure genes except for the ureG in PCC6830 and ureEFG in MED4, suggesting that ure genes are not subjected to NtcA regulation at least in most of the genomes analyzed. This is consistent with the finding that the transcription of urease in PCC7120 is not subjected to NtcA regulation, instead, it is constitutively expressed (33).
Figure 7.
Putative NtcA promoters found for gene clusters involved in urea uptake. A open or solid box upstream of a gene (arrowed box) represents a putative NtcA promoter identified at P-value < 0.01 or 0.05, respectively. A ‘…’ indicates that there is more than one gene in between its two flanking genes.
Cyanase which catalyzes the decomposition of cyanate(NCO−) into CO2 and ammonium are encoded in the genomes of PCC7120, PCC6803, MED4, PCC6301 and WH8102 (Figure 5). In the last three genomes, the cyanase gene cynS is clustered with the ABC type cyanate transporter cynABC/D in the same orientation, and they probably form an operon in PCC6801 and MED4. Thus these three genomes are likely to use cyanate as a nitrogen source as it has been demonstrated in PCC9742 (34) and PCC6301 (35). However, no known cyanate transporters are encoded in the genomes of PCC7120 and PCC6803. Thus it remains unknown as how cyanate is taken up by their cells. Alternatively, no cyanate transporter is encoded in these two genomes, and the encoded CynS is mainly for the degradation of the endogenously produced cyanate (34). High-scoring NtcA promoters are predicted for the cynABC/D/S operons in MED4, PCC6301 and WH8102 and cynS in PCC7120 and PCC6803 (forming a gene cluster with nirA and moa genes) (Figure 5), suggesting that they are under NtcA regulation as it has been demonstrated in PCC9742 (34). Interestingly, the cyn genes are not found in CCMP1375 and MIT9313 that live in the same oligotrophic oceans as MED4 and WH8102 do. The major difference in their ecological niches is that the former two organisms live in nutrient-rich deeper layers of seawater, respectively, whereas the latter two are found in the nutrient-limited upper layer of seawater. This might suggest that the different living environments have caused the former two to lose the genes for cyanate utilization or the latter two to gain the relevant genes during the evolution to cope with environments with limited nitrogen sources.
Some cyanobacterial species are diazotrophic, and a large number of them conduct nitrogen fixation in aerobic conditions. In the case of PCC7120, the only genome capable of nitrogen fixation in our analyses, a specialized cell form called a heterocyst is developed for this purpose. It has been shown that nitrogen fixation (nif) and related genes are not expressed in cultures supplemented with combined nitrogen (36), suggesting that some of these genes might be under NtcA control. Notwithstanding low scoring in general due to the lack of orthologues in the other genomes in our analyses, putative NtcA promoters are found for several nitrogen fixation related genes (Table 5), including experimentally verified ones for devBCA, xisA and hetC genes (Table 5), which are all known to be involved in the development of heterocysts (5). We expect that some of the other predictions are correct, and thus they are likely to be regulated by NtcA. In addition, it has been shown that differentiation into heterocyst from a vegetative cell can be induced by derivation of nitrogen sources, and that the differentiation is strictly dependent on NtcA (22). Although the positive regulator of the heterocyst development, HetR in PCC7120, is impaired in an NtcA mutant (37), it is unlikely to be regulated directly by NtcA, since no NtcA prompter is found for this gene in our analyses. Intriguingly, however, we have identified high-scoring NtcA promoters for 13 putative transcription regulators in PCC7120 (Supplementary Table s9), suggesting that a complex transcription regulatory network is under the control of NtcA, which mediates the indirect regulation effects of NtcA, as for the case of HetR. Clearly, inclusion into our analyses in the future of at least another genome capable of nitrogen fixation and development of heterocysts will undoubtedly improve the quality of the prediction and uncover more genes involved in the nitrogen fixation and/or the heterocyst development.
Table 5.
Putative NtcA promoters found in nitrogen fixation related genes in PCC7120
| Rank | Transcription unit | Name | Putative NtcA sitea | Downstream of NtcA binding site and −10 like boxb | NtcA site position | Score |
|---|---|---|---|---|---|---|
| 84 | alr3710 alr3711 | devB devC | GTAcagtctgtTAC | CTTTACCTGAAACAGATGAATGTAGAATTTA | −738 | 12.048 |
| 260 | alr1442 | xisA | GTAagcaacgcAAC | CTTGAGAACCTTGTACGAAAGGTAGAGTACC | −741 | 11.3296 |
| 182 | alr1911 | nifJ | GTGctgaaaaaAAC | TTGAATTTGATGTAACCTCACCCCTAGCGGG | −284 | 11.5923 |
| 1888 | alr2817 | hetC | GTAacatgagaTAC | ACAATAGCATTTATATTTGCTTTAGTATCTC | −607 | 8.12595 |
| 2018 | alr1407 asr1408 asr1409 | nifV1 nifZ nifT | GTTctctgtcaTAC | AGATATATTGTTATTGTTTAGCTAATATTTA | −191 | 7.85222 |
| 2262 | alr2968 | nifV2 | GTAgtactaaaTAC | TGTGGCTTAAATAATTGAGTGTTTTGTCGCA | −164 | 7.44696 |
| 2553 | alr1430 | fdxH | GTAcatcagttAAC | AGTTAACCGAATCAAGTCTTCCTGTAAACGG | −149 | 7.09269 |
| 2765 | alr0874 | nifH2 | GTCctagaagcCAC | TCTAAAACTCCGACTTCATTTTAGGAAACTA | −155 | 6.83681 |
| 2784 | all1437 all1438 | nifN nifE | GTTatctaattGAC | CGTATTTCTTGCAAAGGTCTGTTTATGAAAA | −239 | 6.82542 |
aBold, experimentally verified binding sites.
bBold, −10 like boxes.
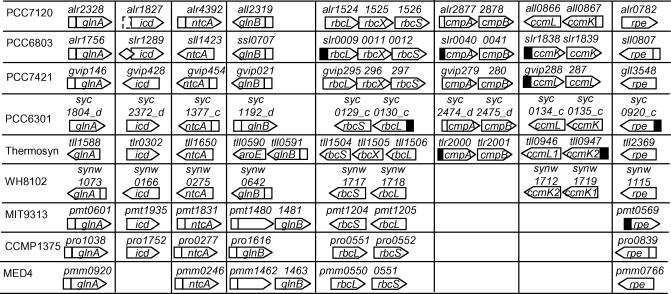
Interestingly, high-scoring NtcA promoters are found for glnA and glnB in all genomes analyzed except for the glnA in PCC6301 and thermosynechococcus (Figure 8), suggesting that both of these two genes are under the regulation of NtcA in these genomes as it has already been demonstrated in PCC9742, PCC6803 and PCC7120 (5). The only exceptions are glnA genes in PCC6301 and thermosynechococcus. High-scoring NtcA promoters are also found for ntcA itself in PCC7421, PCC7120, CCMP13785, MED4, MIT9313 and PCC6301, but not in PCC6803, WH8102 and thermosynechococcus (Figure 8), suggesting that ntcA is auto-regulated in the first six organisms but might not auto-regulate in the last three. The implication of this difference is not clear, but may be related to the environments they inhabit. Although an NtcA binding site has been identified in the regulatory region of icd in PCC6803 (38), we failed to predict it because this site deviates from the canonical GTAN8TAC with a G replacing the highly conserved A in the second triplet (Supplementary Table s2). However, a canonical NtcA site is found for the icd in PCC7120 (Supplementary Table s1), though deviant sites for the icd genes in the other genomes might have been missed by our prediction (Figure 8).
Figure 8.
Putative NtcA promoters found for genes involved in nitrogen incorporation into carbon skeleton and carbon fixation. A open or solid box upstream of a gene (arrowed box) represents a putative NtcA promoter identified at P-value < 0.01 or 0.05, respectively. A dashed box represents a strong putative NtcA promoter, but it is not identified at these two statistical significant levels. A diamond box represents an experimentally verified NtcA promoter, but it is not identified by our algorithm. A ‘…’ indicates that there is more than one gene in between its two flanking genes.
Cross-talk between nitrogen assimilation, photosynthesis and other biochemical processes
As shown in Supplementary Tables s3–s11, we have found high-scoring NtcA promoters for numerous genes whose relationships to the nitrogen assimilation process are largely unknown. Nevertheless, occurrences of strong NtcA promoters in their regulatory regions strongly suggest that these genes are likely to be involved in the nitrogen assimilation-related local or global responses. A large portion of these genes encode highly conserved hypothetical proteins, making them good candidates for further experimental investigation to explore their possible functions in the nitrogen assimilation related biological processes, especially for those genes that encode putative transcription factors. On the other hand, we have also found strong NtcA promoters for genes whose functions or the functions of their orthologues in other cyanobacterial species are known. Surprisingly, a subset of these genes are involved in photosynthesis and carbon fixation processes.
As shown in Figures 8 and 9, strong NtcA promoters are found in the nine genomes for a variety of genes involved in various stages in the photosynthesis and carbon fixation processes from light harvesting in the antenna complex to electron transfers in the photosystems II and I (Figure 9), and from CO2/HCO3− uptake and concentration to key reactions in the Calvin cycle (Figure 8). These findings strongly suggest that these photosynthetic genes are likely to be somehow regulated by NtcA. In fact, it has been shown that nitrogen assimilation and photosynthesis are highly orchestrated processes (39). On one hand, nitrogen assimilation is linked to photosynthesis. The uptake of nitrogen containing compounds is powered by ATP generated by photophosphorylation. Reduced ferrodoxin from photosynthesis acts an electron donor to nitrate and nitrite reductases, and the reducing power is necessary for the action of GOGAT. Furthermore, the expression of NtcA in PCC6801 is actually regulated by the redox state of the cell (40). As mentioned before, NtcA is activated by the C/N balance indictor 2-oxoglurate, the end product of the dark-reaction of photosynthesis (38). A higher level of 2-oxoglurate (high C/N ratio) might be an indication of relatively strong photosynthesis activity compared to the ongoing level of nitrogen assimilation. In this sense, 2-oxoglurate serves as a messenger from photosynthesis to nitrogen assimilation to speed up the latter, so that the activities of the two systems are coordinated. On the other hand, the intensity of photosynthesis depends on the availability of nitrogen in the environment as has been shown in the field studies (41)—lower levels of nitrogen assimilation are associated with lower levels of photosynthesis, and vise visa. It is also well known that nitrogen-deprivation depresses photosynthesis by inducing degradation of photosynthetic apparatus, a process known as chlorosis (42). Although chlorosis is a complex multi-stage global response (42), it has been recently shown that nitrogen starvation-induced chlorosis is an adaptation of a cell to long term survival (42,43), in which the photosynthesis is kept at a low level (44) to match the low availability of nitrogen. Interestingly, upon nitrogen becoming available, photosynthesis resumes rapidly (42,44). These facts strongly suggest that the photosynthesis process is able to sense the availability of nitrogen to the cell, and accordingly coordinate its activity with the current nitrogen level. Intriguingly, nitrogen starvation-induced chlorosis in PCC7942 is strictly dependent on NtcA, and an NtcA mutant fails to reinitiate photosynthesis when nitrogen becomes available (43), suggesting that NtcA plays an important role in coordinating the activities of the nitrogen assimilation and photosynthesis processes. However, how NtcA is related to such coordination is largely unknown. We postulate here for the first time that the photosynthesis-related genes and probably some others that bear NtcA promoters might serve as regulatory points to coordinate these two important processes. Although the details of such coordination are currently unclear and require experimental validation, there are several lines of evidence that support this hypothesis and suggest some possible scenarios of the coordination. First, it is recently reported that the expression of a few monitored photosynthetic genes allophycocyanin apc, phycocyanin cpc and thioredoxin trxM are down-regulated, whereas porins somA and somB are up-regulated in the early stage of the chlorosis inducted by nitrogen starvation (5). However, apc/cpc, trxM and other photosynthesis-related genes are maintained at a somewhat higher level while the expression of most other genes are dormant in the late stage of chlorosis induced by nitrogen starvation (44). In agreement with this, NtcA promoters are found for the orthologues/homologues of these genes in a number of cyanobacterial genomes analyzed (Figure 9). The active expression of these photosynthesis-related genes during the later stage of chlorosis might be mediated by NtcA, since they are necessary for the re-initiation of photosynthesis upon the availability of nitrogen, while an ntcA mutant fails to reinitiate photosynthesis when nitrogen becomes available. (43,44). Second, Bird and Wyman (45) have recently reported that the large subunit of the key enzymes ribulose bisphosphate carboxylase RbcL in the Calvin cycle is up-regulated in PCC8103 growing on nitrogen-deprived medium, a condition that leads to the activation of NtcA. Interestingly, rbcLS operon is reported to be repressed under the same condition in PCC9721 (46) and PCC9742 (7). Regulation of rbcLS operon by NtcA through an NtcA promoter has been experimentally verified (46), and strong NtcA promoters are found for rbcL in PCC6803 and PCC6301 in our analysis (Figure 8). The different responses of the rbcLS operons to nitrogen deprivation among different species might be due to their different regulatory machineries for NtcA regulated genes, as it has been demonstrated for glnA of PCC8103 where glnA is up-regulated by ammonium (45). In contrast, in both PCC6803 and PCC9742, glnA is substantially down-regulated by ammonia (47). Such differential responses have been explained as a mechanism of coordinating nitrogen assimilation and carbon fixation (45).
Figure 9.
Putative NtcA promoters found for genes involved in photosynthesis. A open or solid box upstream of a gene (arrowed box) represents a putative NtcA promoter identified at P-value < 0.01 or 0.05, respectively. Each numbered column represents a functional group: 1, allophycocyanin complex; 2, phycoerythrin or phycocyanin complex; 3, chlorophyll a/b binding proteins; 4, photosystem II proteins; 5, photosystem I proteins; 6, light repressed protein; 7, cytochrome oxidases; 8, NADH dehydrogenases; 9, thioredoxin; 10, iron stress-induced protein or flavodoxin; 11, ferredoxin-NADP reductase; 12, ferredoxins; 13, prorins.
Different regulatory machineries for NtcA regulated genes can also happen in the same species, as it has been demonstrated that though both nitrogen deprivation and alternative sources of nitrogen other than ammonia activate NtcA (5), their effects on the NtcA regulated genes can be remarkably different (45). For instance, nitrogen deprivation up-regulates rbcL and glnA in WH8103 while nitrate or nitrite down-regulates rbcL and glnA compared to cells growing on a medium containing ammonia (45). Thus, the activation of NtcA can lead to either activation or depression of a gene through the same or different NtcA promoters dependent on the level of NctA and probably other relevant molecules such as NtcB and P II protein. The latter is required for the expression of some NtcA regulated genes in cells growing on a medium with nitrate as nitrogen source (7). In this sense, it is expected that the photosynthetic genes that bear an NtcA promoter would respond differently in the different stages of nitrogen starvation, and among species. This might explain the observations that a large number of the genes are up- or down-regulated during nitrogen starvation or when nitrogen becomes available to nitrogen-starving cells (7,42–44), both of which are likely to lead to NtcA activation (5).
Another line of evidence that supports our hypothesis comes from the interesting distributions of the number of genes and specific genes that bear an NtcA promoter among the different ecotypes and species living in various ecological niches. For instance, there is only one photosynthesis gene petH (encoding the ferredoxin-NADP+ reductase in photosytem I) bearing an NtcA promoter in PCC7421 (Figure 9), living in calcareous rocks, with 4439 genes encoded in its genome. In contrast, there are 3–13 photosynthesis genes bearing NtcA promoters in the other eight cyanobacterial genomes living in either freshwater or seawater (Figure 9), most of which have much smaller genomes (Table 1). The reason for this discrepancy is not clear but may be related to their living environments. Since light is greatly attenuated in water (4), in order to coordinate the nitrogen assimilation and photosynthesis processes, more precise control of photosynthesis might be needed for species living in water than for PCC7421 living in calcareous rocks where light is more intense. Another interesting observation is that NtcA promoters are found for the chlorophyll a/b-binding light harvesting genes (pcb) in low-light adapted Prochlorococcus marine strains CCMP1375 (pcbG, pcbA and pcbD) and MIT9313 (pcbA and pcbB), but not for pcb genes in high-light adapted closely related MED4 (Figure 9). In contrast, NtcA promoters are found for the photosystem II psb genes in MED4, WH8102, thermo synechococcus, PCC6803 and PCC7120, but not for their orthologues in CCMP1375, MIT9313 and PCC6301. These differences in the NtcA-regulated genes among the strains/species or ecotype might be again the result of acclimation to their ecological niches. For instance, the low-light adapted strains for CCMP1375 and MIT9313, the rate-limiting step in photosynthesis might be light harvesting as has been suggested by multiple copies of pbc genes encoded in these genomes (4,18) and their super antenna complex structures to increase the power of light harvesting (48,49). Therefore, the NtcA regulation at this point might achieve optimal results in low-light adapted organisms, whereas for the high-light adapted species/ecotypes such as MED4, WH8102, thermosynechococcus, PCC6803 and PCC7120, the rate-limiting step in photosynthesis might be in photosystem II, so the NtcA regulation on the genes in this system might achieve optimal results in these organisms. Putative NtcA promoters are found more frequently for some other photosynthetic genes in the nine genomes analyzed, such as flavodoxin isiB/isiA, thioredoxin trxA/M, light-repressed protein lrt, subunits of NADH dehydrogenase ndh, ferredoxin petF and allophycocyanin apc (Figure 9), suggesting that these genes might play more common roles in coordinating the nitrogen assimilation and photosynthesis processes.
In addition, all the RNA polymerase σ-factors found to date in cyanobacteria belong to the σ70 family (50,51), and multiple genes of which are found in all the genomes analyzed. Interestingly, strong NtcA promoters are found for some σ-factor genes in all genomes analyzed except for MED4. In some genomes such as PCC6301, PCCd6803, PCC7120, MIT9313, CCMP1375 and thermosynechococcus, strong NtcA promoters are found for more than one member of the σ70-factor family. These σ-factors belong to the groups 1, 2 or 3 of the σ70-factor family as shown in Figure s1 as Supplementary Data. In agreement with these findings, it has been demonstrated that the group 2 σ70-factor, rpoD-2V/sigE (sll1689) in PCC6803 is regulated by NtcA, and that it is involved in the survival response under nitrogen starvation (50). Hence, we postulate that its orthologues and homolgues bearing an NtcA promoter in this and the other cyanobacteria are also highly likely to be regulated by NtcA as does the RpoD-2V/SigE(Sll1689) in PCC6803, but different groups of the σ70-factor family might mediate different types of response. It will be interesting to investigate experimentally the functions of these σ-factors in the nitrogen stress-induced global responses.
Lastly, cross-talk between nitrogen assimilation and other pathways is also possible because high-scoring promoters are found for genes in these pathways. For instance, there is possible cross-talk between the nitrogen assimilation pathway and the cell wall peptidoglycan murein biosynthesis pathway, which involves UDP-N-acetylmuramate-alanine ligase murB and murC that bear high-scoring NtcA promoters in CCMP1375 and MED4 (Supplementary Table s3 and s5). Products of mur genes catalyze the addition of l-alanine to the nucleotide precursor UDP-N-acetylmuramoyl during murein biosynthesis. Since a portion of nitrogen assimilated is used to synthesize l-alanine that is a major component of murein, biosynthesis of murein might be under the regulation of NtcA to cope the availability of nitrogen to the cell. All these pathways together with the nitrogen assimilation pathway would constitute an entire interaction network responsible for nitrogen assimilation-related global responses that involve a large number of genes (7).
CONCLUSION
Owing to the often under-representation nature of the profile of cis-regulatory sites and to their high variability and relative short length, scanning the whole genome using such a profile to uncover all possible binding sites inevitably results in a rather high false positive rate. We have unequivocally demonstrated that combining other sources of information such as the profile of another binding site and/or the information of similar binding sites in the regulatory regions of orthologues in related genomes can greatly improve the prediction accuracy. In the case of NtcA promoter prediction, the false positive rate of the scanning process can be reduced from 8.2- to 90.9-fold compared to the prediction results when only a single profile is used, while the same level of prediction sensitivity is maintained. Furthermore, using our statistical model, the prediction sensitivity or specificity can be well-controlled. Although some cis-regulatory binding sites/promoters for genes unique to a genome might be missed by this method under a stringent statistical significant level, as we have shown for hetC and xisA in PCC7120 and nrtP in WH8102, due to the lack of orthologues in the other genomes in the analyses, this problem will become less significant as more and more genomes are sequenced, providing higher coverage for genes appearing in multiple genomes. Applying this new algorithm to the prediction of NtcA binding sites in nine cyanobacterial genomes has led to numerous interesting observations about the NtcA regulons in this important group of microorganisms. Most intriguingly, high scoring NtcA promoters are found for many genes involved in the various stages of photosynthesis process. We postulate for the first time that these genes serve as the regulatory points to orchestrate the nitrogen assimilation and photosynthesis processes in a cyanobacterial cell.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Supplementary Material
Acknowledgments
This research was supported in part by the US Department of Energy's Genomes to Life program (http://doegenomestolife.org/) under project, ‘Carbon Sequestration in Synechococcus sp.: From Molecular Machines to Hierarchical Modeling’ (www.genomes2life.org), Distinguished Cancer Scholar grant from Georgia Cancer Coalition and by National Science Foundation (NSF/DBI-0354771, NSF/ITR-IIS-0407204). We would like to thank Dr Brian Palenik for helpful discussion during the course of this work, Dr Mary Ann Moran for her critical reading of this manuscript and anonymous reviewers for their insightful suggestions that greatly improve this paper. Funding to pay the Open Access publication charges for this article was provided by NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant D.A., editor. Molecular Biology of Cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 2–25. [Google Scholar]
- 2.Luesch H., Harrigan G.G., Goetz G., Horgen F.D. The cyanobacterial origin of potent anticancer agents originally isolated from sea hares. Curr. Med. Chem. 2002;9:1791–1806. doi: 10.2174/0929867023369051. [DOI] [PubMed] [Google Scholar]
- 3.Burja A.M., Banaigs B., Abou-Mansour E., Grant Burgess J., Wright P.C. Marine cyanobacteria–a prolific source of natural products. Tetrahedron. 2001;57:9347–9377. [Google Scholar]
- 4.Ting C.S., Rocap G., King J., Chisholm S.W. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 2002;10:134–142. doi: 10.1016/s0966-842x(02)02319-3. [DOI] [PubMed] [Google Scholar]
- 5.Herrero A., Muro-Pastor A.M., Flores E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 7.Fadi Aldehni M., Sauer J., Spielhaupter C., Schmid R., Forchhammer K. Signal transduction protein P(II) is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942. J. Bacteriol. 2003;185:2582–2591. doi: 10.1128/JB.185.8.2582-2591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelfand M.S., Novichkov P.S., Novichkova E.S., Mironov A.A. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 2000;1:357–371. doi: 10.1093/bib/1.4.357. [DOI] [PubMed] [Google Scholar]
- 9.Mironov A.A., Koonin E.V., Roytberg M.A., Gelfand M.S. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchette M., Tompa M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 2002;12:739–748. doi: 10.1101/gr.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGuire A.M., Hughes J.D., Church G.M. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 2000;10:744–757. doi: 10.1101/gr.10.6.744. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Tan K., Stormo G.D. Computational identification of the Spo0A-phosphate regulon that is essential for the cellular differentiation and development in Gram-positive spore-forming bacteria. Nucleic Acids Res. 2003;31:6891–6903. doi: 10.1093/nar/gkg879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan K., Moreno-Hagelsieb G., Collado-Vides J., Stormo G.D. A comparative genomics approach to prediction of new members of regulons. Genome Res. 2001;11:566–584. doi: 10.1101/gr.149301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber T.M., Gross C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 15.Olman V., Xu D., Xu Y. CUBIC: identification of regulatory binding sites through data clustering. J. Bioinform. Comput. Biol. 2003;1:21–40. doi: 10.1142/s0219720003000162. [DOI] [PubMed] [Google Scholar]
- 16.Vega-Palas M.A., Flores E., Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 17.Schultz S.C., Shields G.C., Steitz T.A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 18.Rocap G., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 19.Muro-Pastor M.I., Florencio F.J. NADP(+)-isocitrate dehydrogenase from the cyanobacterium Anabaena sp. strain PCC 7120: purification and characterization of the enzyme and cloning, sequencing, and disruption of the icd gene. J. Bacteriol. 1994;176:2718–2726. doi: 10.1128/jb.176.9.2718-2726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muro-Pastor M.I., Reyes J.C., Florencio F.J. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 2001;276:38320–38328. doi: 10.1074/jbc.M105297200. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez-Bermudez M.F., Herrero A., Flores E. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 2002;512:71–74. doi: 10.1016/s0014-5793(02)02219-6. [DOI] [PubMed] [Google Scholar]
- 22.Flores E., Herrero A. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans. 2005;33:164–167. doi: 10.1042/BST0330164. [DOI] [PubMed] [Google Scholar]
- 23.Tanigawa R., Shirokane M., Maeda Si S., Omata T., Tanaka K., Takahashi H. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl Acad. Sci. USA. 2002;99:4251–4255. doi: 10.1073/pnas.072587199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez-Bermudez M.F., Herrero A., Flores E. Carbon supply and 2-oxoglutarate effects on expression of nitrate reductase and nitrogen-regulated genes in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 2003;221:155–159. doi: 10.1016/S0378-1097(03)00208-8. [DOI] [PubMed] [Google Scholar]
- 25.Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 2004;28:319–333. doi: 10.1016/j.femsre.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Lee H.M., Flores E., Forchhammer K., Herrero A., Tandeau De Marsac N. Phosphorylation of the signal transducer PII protein and an additional effector are required for the PII-mediated regulation of nitrate and nitrite uptake in the Cyanobacterium Synechococcus sp. PCC 7942. Eur. J. Biochem. 2000;267:591–600. doi: 10.1046/j.1432-1327.2000.01043.x. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez-Bermudez M.F., Paz-Yepes J., Herrero A., Flores E. The NtcA-activated amt1 gene encodes a permease required for uptake of low concentrations of ammonium in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology. 2002;148:861–869. doi: 10.1099/00221287-148-3-861. [DOI] [PubMed] [Google Scholar]
- 28.Aichi M., Takatani N., Omata T. Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2001;183:5840–5847. doi: 10.1128/JB.183.20.5840-5847.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frias J.E., Flores E., Herrero A. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol. Microbiol. 2000;38:613–625. doi: 10.1046/j.1365-2958.2000.02156.x. [DOI] [PubMed] [Google Scholar]
- 30.Frias J.E., Herrero A., Flores E. Open reading frame all0601 from Anabaena sp. strain PCC 7120 represents a novel gene, cnaT, required for expression of the nitrate assimilation nir operon. J. Bacteriol. 2003;185:5037–5044. doi: 10.1128/JB.185.17.5037-5044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S., Kawaguchi Y., Ohe T.A., Omata T. cis-acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1998;180:4080–4088. doi: 10.1128/jb.180.16.4080-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier J.L., Brahamsha B., Palenik B. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology. 1999;145:447–459. doi: 10.1099/13500872-145-2-447. [DOI] [PubMed] [Google Scholar]
- 33.Valladares A., Montesinos M.L., Herrero A., Flores E. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 2002;43:703–715. doi: 10.1046/j.1365-2958.2002.02778.x. [DOI] [PubMed] [Google Scholar]
- 34.Harano Y., Suzuki I., Maeda S., Kaneko T., Tabata S., Omata T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J. Bacteriol. 1997;179:5744–5750. doi: 10.1128/jb.179.18.5744-5750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson P.M., Sung Y.C., Fuchs J.A. The cyanase operon and cyanate metabolism. FEMS Microbiol. Rev. 1990;7:247–252. doi: 10.1111/j.1574-6968.1990.tb04920.x. [DOI] [PubMed] [Google Scholar]
- 36.Huang T.C., Lin R.F., Chu M.K., Chen H.M. Organization and expression of nitrogen-fixation genes in the aerobic nitrogen-fixing unicellular cyanobacterium Synechococcus sp. strain RF-1. Microbiology. 1999;145:743–753. doi: 10.1099/13500872-145-3-743. [DOI] [PubMed] [Google Scholar]
- 37.Frias J.E., Flores E., Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 38.Muro-Pastor M.I., Reyes J.C., Florencio F.J. The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J. Bacteriol. 1996;178:4070–4076. doi: 10.1128/jb.178.14.4070-4076.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsac N.T.d., Lee H.M., Hisbergues M., Castets A.M., Bédu S. Control of nitrogen and carbon metabolism in cyanobacteria. J Appl Phycol. 2001;13:287–292. [Google Scholar]
- 40.Alfonso M., Perewoska I., Kirilovsky D. Redox control of ntcA gene expression in Synechocystis sp. PCC 6803. Nitrogen availability and electron transport regulate the levels of the NtcA protein. Plant Physiol. 2001;125:969–981. doi: 10.1104/pp.125.2.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morel F.M., Price N.M. The biogeochemical cycles of trace metals in the oceans. Science. 2003;300:944–947. doi: 10.1126/science.1083545. [DOI] [PubMed] [Google Scholar]
- 42.Gorl M., Sauer J., Baier T., Forchhammer K. Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology. 1998;144:2449–2458. doi: 10.1099/00221287-144-9-2449. [DOI] [PubMed] [Google Scholar]
- 43.Sauer J., Gorl M., Forchhammer K. Nitrogen starvation in Synechococcus PCC 7942: involvement of glutamine synthetase and NtcA in phycobiliprotein degradation and survival. Arch. Microbiol. 1999;172:247–255. doi: 10.1007/s002030050767. [DOI] [PubMed] [Google Scholar]
- 44.Sauer J., Schreiber U., Schmid R., Volker U., Forchhammer K. Nitrogen starvation-induced chlorosis in Synechococcus PCC 7942. Low-level photosynthesis as a mechanism of long-term survival. Plant Physiol. 2001;126:233–243. doi: 10.1104/pp.126.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bird C., Wyman M. Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression. Appl. Environ. Microbiol. 2003;69:7009–7018. doi: 10.1128/AEM.69.12.7009-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramasubramanian T.S., Wei T.F., Golden J.W. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J. Bacteriol. 1994;176:1214–1223. doi: 10.1128/jb.176.5.1214-1223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyes J.C., Muro-Pastor M.I., Florencio F.J. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J. Bacteriol. 1997;179:2678–2689. doi: 10.1128/jb.179.8.2678-2689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bibby T.S., Mary I., Nield J., Partensky F., Barber J. Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature. 2003;424:1051–1054. doi: 10.1038/nature01933. [DOI] [PubMed] [Google Scholar]
- 49.Bibby T.S., Nield J., Partensky F., Barber J. Oxyphotobacteria. Antenna ring around photosystem I. Nature. 2001;413:590. doi: 10.1038/35098153. [DOI] [PubMed] [Google Scholar]
- 50.Muro-Pastor A.M., Herrero A., Flores E. Nitrogen-regulated group 2 sigma factor from Synechocystis sp. strain PCC 6803 involved in survival under nitrogen stress. J. Bacteriol. 2001;183:1090–1095. doi: 10.1128/JB.183.3.1090-1095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imamura S., Yoshihara S., Nakano S., Shiozaki N., Yamada A., Tanaka K., Takahashi H., Asayama M., Shirai M. Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J. Mol. Biol. 2003;325:857–872. doi: 10.1016/s0022-2836(02)01242-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.