Abstract
The developmental stage-specific expression of human globin proteins is characterized by a switch from the coexpression of ζ- and α-globin in the embryonic yolk sac to exclusive expression of α-globin during fetal and adult life. Recent studies with transgenic mice demonstrate that in addition to transcriptional control elements, full developmental silencing of the human ζ-globin gene requires elements encoded within the transcribed region. In the current work, we establish that these latter elements operate posttranscriptionally by reducing the relative stability of ζ-globin mRNA. Using a transgenic mouse model system, we demonstrate that human ζ-globin mRNA is unstable in adult erythroid cells relative to the highly stable human α-globin mRNA. A critical determinant of the difference between α- and ζ-globin mRNA stability is mapped by in vivo expression studies to their respective 3′ untranslated regions (3′UTRs). In vitro messenger ribonucleoprotein (mRNP) assembly assays demonstrate that the α- and ζ-globin 3′UTRs assemble a previously described mRNP stability-determining complex, the α-complex, with distinctly different affinities. The diminished efficiency of α-complex assembly on the ζ 3′UTR results from a single C→G nucleotide substitution in a crucial polypyrimidine tract contained by both the human α- and ζ-globin mRNA 3′UTRs. A potential pathway for accelerated ζ-globin mRNA decay is suggested by the observation that its 3′UTR encodes a shortened poly(A) tail. Based upon these data, we propose a model for ζ-globin gene silencing in fetal and adult erythroid cells in which posttranscriptional controls play a central role by providing for accelerated clearance of ζ-globin transcripts.
Eukaryotic genes exhibiting complex patterns of expression may be regulated through an array of controls that act both transcriptionally and posttranscriptionally. Globin genes, which are expressed in a well-characterized developmental stage-specific manner, serve as an informative model for such regulatory complexity. The α-globin gene cluster in humans and mice contains three functional genes: ζ, α2, and α1. These genes undergo a switch in expression at the transition between embryonic and fetal development, from coexpression of the α- and ζ-globin genes, to exclusive expression of the two fetal/ adult α-globin genes. This event, which occurs at gestational weeks 6 to 8 in humans and postcoital days 8.5 to 10.5 in mice, is known as ζ-globin gene silencing. The switch is remarkably efficient: although ζ-globin protein is expressed at high levels in primitive erythroblasts in the embryonic yolk sac, it cannot be detected in definitive erythroid cells from normal term fetuses or adults. Recent evidence indicates that ζ-globin gene silencing is a complex event requiring both transcriptional downregulation (23, 29, 36, 37), as well as other, less well-defined mechanisms (23). Specifically, the ζ-globin transcribed region was noted to play an important and previously unanticipated role in this process (23). In experiments utilizing transgenic mice, levels of ζ-globin mRNAs transcribed from full-length human ζ-globin (hζ-globin) transgenes were appropriately silenced during the embryonic-to-fetal transition. In contrast, chimeric ζ-globin transgenes into which the α-globin transcribed region was substituted (thereby preserving ζ-globin promoter elements) were incompletely silenced, with significant residual mRNA expressed in adult erythroid cells. While confirming the importance of promoter elements to regulation of hζ-globin gene expression (29, 36, 37), these transgenic studies also indicated that promoter elements alone are insufficient to effect its full developmental silencing.
The high-level expression of hα- and hβ-globins in adult erythrocytes is dependent on the unusually long half-lives of their fully-processed mRNAs in erythrocyte progenitors. These half-lives have been estimated to be between 16 and 72 h in a broad range of cell culture and in vivo experiments (2, 5, 18, 21, 24, 31, 32). The high stability of α- and β-globin mRNAs facilitates their accumulation in transcriptionally active erythroid precursors and ensures that they remain at high levels for continued translation in subsequent, transcriptionally silent stages of terminal erythroid differentiation. A molecular basis for the stability of hα-globin mRNA has recently been established with both cultured cells and transgenic mice (19, 43, 45, 46). The stability of α-globin mRNA in vivo is paralleled by the ability of its 3′ untranslated region (3′UTR) to assemble a messenger ribonucleoprotein (mRNP) complex (the α-complex) in vitro (42). Mutations that disrupt α-complex assembly in vitro decrease α-globin mRNA stability in vivo. The cis elements crucial to α-globin mRNA stability and α-complex assembly map to a defined pyrimidine-rich region that is highly conserved between human and mouse α-globin 3′UTRs (15, 43, 44). These data, combined with the observation that the stabilizing activity of the α-globin mRNA 3′UTR can be successfully transferred to a reporter mRNA (34), suggest that α-globin mRNA stability is determined by a mechanism that acts locally within the 3′UTR. Although the nature of this mechanism is not yet clear, recent studies indicate that mutations that destabilize α-globin mRNA by interfering with α-complex assembly promote accelerated shortening of the poly(A) tail (26). These studies demonstrate that assembly of the sequence-specific 3′UTR RNP α-complex is crucial to α-globin mRNA stability, and is consequently an important determinant of α-globin gene expression.
The studies mentioned above illustrate the importance of mRNA stability to α-globin gene expression. The possibility that these controls might also be relevant to the observed posttranscriptional component to silencing of the evolutionarily related ζ-globin gene has not been previously investigated. In the current work, we establish a transgenic mouse model system that permits direct in vivo comparison of the stabilities of hα- and hζ-globin mRNAs. We demonstrate that hζ-globin mRNA is significantly less stable than hα-globin mRNA in definitive erythroid cells, that the hζ-globin 3′UTR encompasses major structural determinants of this instability, and that the relative stabilities of the hα- and hζ-globin mRNAs in vivo parallel their capacities to assemble a specific 3′UTR RNP complex in vitro. A specific difference in the sequences of the hα- and ζ-globin mRNA 3′UTRs which contributes to their distinctly different affinities for the mRNP stability complex is identified. Finally, we demonstrate that mRNA instability encoded by the hζ-globin 3′UTR is accompanied by a shortened poly(A) tail, suggesting a mechanistic link between ζ-globin mRNA destabilization and deadenylation. Collectively, these data provide a detailed account of a posttranscriptional mechanism that in conjunction with transcriptional controls, is essential for the full developmental silencing of embryonic ζ-globin gene expression.
MATERIALS AND METHODS
Transgene construction.
The construction of plasmids containing the full-length hα- and hζ-globin genes has been previously described (23). The high-level transcription of hζ-globin mRNA in adults was facilitated in some lines by deleting a 3′ flanking region silencer element (23, 44a). The hα-globin gene is carried as the 1.5-kb PstI human genomic DNA fragment inserted into the EcoRI site of pSP72, and the hζ-globin gene is carried as the 2.3-kb EcoRI-BamHI human genomic DNA fragment, also in the EcoRI site of pSP72 (23). Chimeric α3′ζ and ζ3′α transgenes were constructed by a splice overlap extension-PCR strategy utilizing either of the two full-length genes as templates (23, 26). First-stage reaction mixtures (Tables 1 and 2) were assembled from 10 ng of template DNA, 200 pmol (each) forward and reverse primer, 4 μl of deoxynucleoside triphosphates (2 mM each), and 2 U of Vent polymerase in 100 μl of 1× reaction buffer provided by the manufacturer (New England Biolabs, Beverly, Mass.). Reaction mixtures were initially denatured at 95°C (5 min), annealed at 57°C (15 s), and extended at 73°C (25 s), and then were cycled an additional 28 times at 92°C (1 min), 57°C (15 s), and 73°C (25 s), with a terminal extension at 73°C (25 s). Second-stage reactions were identical to first-stage reactions, except that they comprised 1 μl each of the two related first-stage reactions (α3′ζ-A and -B, and ζ3′α-A and -B) along with oligomer sp72poly and either oligomer α589–607 or ζ1431–1448. Second-stage reactions were amplified as described above, except that the extension times were prolonged to 30 s. The amplified DNAs were digested with BstEII and SphI and directionally cloned into the corresponding sites of the original hα-globin plasmid to create α3′ζ, or into the hζ-globin plasmid to create ζ3′α. Ampicillin-resistant transformants were verified by restriction digest analysis and by sequencing both strands of PCR-amplified sequences. EcoRI fragments containing the 1.6-kb chimeric genes were ligated into the unique pSP72 EcoRI site adjacent to a previously inserted 6.5-kb DNA fragment containing core elements of the β-locus control region (β-LCR) (μβLCR [40]) as described previously (23). Insert orientations were verified by restriction analysis. DNA fragments containing the 8.0-kb μβLCR linked to the transgene were released by digestion with EcoRV and SalI and purified for microinjection as previously described (23). The construction of the αPζ transgene, containing the α-globin promoter linked to the ζ-globin transcribed region, has been previously described (23). The hζ mRNAs transcribed from the hα and hζ promoters of the αPζ and hζ transgenes, respectively, differ only in the sizes of their 5′UTRs (23).
TABLE 1.
Primers used for synthesis of chimeric globin transgenes
| Primera | Length (nt) | Orientation | Sequence (5′→3′) | Characteristic(s) |
|---|---|---|---|---|
| α589–607 | 19 | + | CTGCACAGCTCCTAAGCCA | α-Exon 3 |
| αC/ζUTR | 38 | + | TGCTGACCTCCAAATACCGTTGA/GCGCCGCCTCCGGGA | Compound α-exon 3/ζ-3′UTR |
| ζ1431–1448 | 18 | + | CCCAGCTCCTGTCCCACT | ζ-Exon 3 |
| ζC/αUTR | 39 | + | TCCTGACCGAGAAGTACCGCTAA/GCTGGAGCCTCGGTAG | Compound ζ-exon 3/α-3′UTR |
| ζUTR/αC | 38 | − | TCCCGGAGGCGGCGC/TCAACGGTATTTGGAGGTCAGCA | Compound ζ-3′UTR/α-exon 3 |
| sp72poly | 22 | − | TTAGGTGACACTATAGAACTCG | SP72 polylinker |
| αUTR/ζC | 39 | − | CTACCGAGGCTCCAGC/TTAGCGGTACTTCTCGGTCAGGA | Compound α-3′UTR/ζ-exon 3 |
Primer designations reflect their positions and/or content. For example, α589–607 extends between α-globin positions 589 and 607 (numbered from the transcription start site). Compound primers which comprise elements from both the α- and ζ-globin sequences are distinguished by a backslash, with their origin identified either from the coding region (C) or from the 3′UTR.
TABLE 2.
Composition of first- and second-stage splice overlap extension-PCRs
| Reaction | Primers | Template(s) |
|---|---|---|
| First stage | ||
| α3′ζ-A | α589–607, ζUTR/αC | α |
| α3′ζ-B | αC/ζUTR, sp72poly | ζ |
| ζ3′α-A | ζ1431–1448, αUTR/ζC | ζ |
| ζ3′α-B | ζC/αUTR, sp72poly | α |
| Second stage | ||
| α3′ζ | α589–607, sp72poly | α3′ζ-A, α3′ζ-B |
| ζ3′α | ζ1431–1448, sp72poly | ζ3′α-A, ζ3′α-B |
Generation and characterization of transgenic mice.
Mice transgenic for the hζ-globin transgene have been previously characterized (23). Mice transgenic for chimeric globin genes were generated by the Transgenic Mouse Core Facility at the University of Pennsylvania as previously described (23). Founder and F1 transgenic mice were screened by dot blot analysis of tail DNA (26) and verified by Southern analysis. Tail DNA (5 μg) from transgenic (F1 or subsequent generations) and control mice was digested with EcoRI (hα- and hα3′ζ transgenes) or BamHI (hζ- and hζ3′α transgenes), resolved on an 0.8% agarose gel in 1× TEA buffer, transferred to Zetabind (CUNO, Meriden Conn.), and probed with 32P-labeled DNA fragments complementary to regions of the hα- or hζ-globin promoter regions and mouse ζ-globin (mζ-globin) 3′ flanking region (X-region [26]). These probes detect a 1.6-kb EcoRI fragment of the hα and hα3′ζ globin transgenes or a 2.3-kb BamHI fragment of the hζ- and hζ3′α-globin transgenes, and either a 4.1-kb (EcoRI) or 1.3-kb (BamHI) fragment of genomic DNA originating from the mζ-globin 3′ flanking region. For all lines, transgene copy number was established as previously described (23).
Assay of transgenic globin mRNA stability.
Adult transgenic mice were pretreated with three intraperitoneal injections of acetyl-2-phenylhydrazine (Sigma) as described previously (26), resulting in an initial peripheral hemolysis followed by a compensatory erythropoietic response characterized by an increase in both the marrow erythroid/myeloid ratio and the peripheral reticulocyte count. RNA was purified from unfractionated marrow cells and peripheral reticulocytes as described previously (26). Internally 32P-labeled antisense-oriented RNA probes for RNase protection assays were transcribed in vitro from template DNA by using SP6 RNA polymerase (SP6 Maxiscript kit; Ambion). RNA samples were desiccated and resuspended in 20 μl of Berk buffer {80% formamide, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.4), 400 mM NaCl, 1 mM EDTA} supplemented with both transgene-specific and murine α-globin probes, heat denatured for 10 min at 80°C, and incubated overnight at 52°C. Digestion buffer (200 μl of 10 mM Tris [pH 7.5], 300 mM NaCl, 5 mM EDTA, 20 mg of RNase A per ml, 1 mg of RNase T1 per ml) was added, and the samples were digested at room temperature for 25 min. To terminate the reaction, 17 μl of a 4:1 mixture of 10% sodium dodecyl sulfate–10-mg/ml proteinase K was added, and the samples were incubated at 37°C for an additional 20 min. The samples were extracted with phenol-chloroform-isoamyl alcohol, ethanol precipitated, and analyzed on an 8 M urea–6% acrylamide gel. This method permits calculation of normalized stabilities for mRNAs which are highly reproducible (26). The specific activities of individual probes was adjusted to facilitate visual comparison of band intensities on single autoradiographs; consequently, observed band intensities do not reflect actual transgene expression levels.
Gel mobility shift analysis.
hα- and hζ-globin RNA 3′UTR probes were transcribed in vitro from PCR-generated cDNA templates according to the manufacturer’s instructions (T7 Maxiscript kit). Murine erythroleukemia (MEL) cells (2.5 × 108) washed twice in ice-cold phosphate-buffered saline were resuspended in 3.5 ml of buffer A (10 mM KCl, 1.5 mM MgCl, 10 mM Tris HCl [pH 7.4], 0.5 mM dithiothreitol) supplemented with pepstatin (0.2 μg/ml), leupeptin (0.2 μg/ml), and aprotinin (10 μg/ml). Cells were lysed by 1 pass through a 23-gauge needle and four additional passes through a 25-gauge needle. KCl (1 M, 0.49 ml) was added, and the mixture was centrifuged in an HS-4 rotor (Beckman) at 3,200 rpm for 10 min at 4°C. The supernatant was transferred into 5-ml Beckman tubes and spun at 32,500 rpm for 60 min at 4°C in an SW41 Ti rotor. Glycerol (0.1 volume) was added to the supernatant, which was subsequently stored in 50-μl aliquots at −80°C. Quick-thawed aliquots were preincubated with 1 μl of RNasin (5 Prime→3 Prime, Boulder, Colo.). and 1 μl of β-mercaptoethanol for 30 min at room temperature. 32P-labeled probe (5 × 104 cpm/ml) and specific cold competitor (100 to 200 μg) were added to a total volume of 15 μl, and the incubation continued for an additional 30 min. The reaction was terminated by addition of RNase T1 (20 U/reaction) for 10 min at room temperature. Heparin (1 μl of a 50-mg/ml stock) and loading dye were added, and the reaction was resolved on a 60:1 acrylamide–bis-acrylamide gel prerun at 110 V for 1 h in 1× TBE. α-Complex assembly on α42, α42G, and ζ42 DNA oligomers was assessed by a similar method which omitted digestion with RNase (14). Pilot experiments (not shown) indicated that the α-complex assembles with equal affinity on sequence-identical RNA and DNA oligomers.
Estimation of apparent Kd.
Cytoplasmic extract (5 μg) was incubated for 30 min with 32P-labeled α- or ζ-globin 3′UTR RNA probe in concentrations ranging from 0.05 to 10 nM (α 3′UTR) or 0.1 to 24 nM (ζ 3′UTR), and the resulting RNP complexes were resolved on a 5% native polyacrylamide gel. Quantitation of bound and free RNA probe was performed by PhosphorImager densitometry (Molecular Dynamics) with ImageQuant software. The values of apparent equilibrium dissociation constant (Kd) were calculated by linear regression in a double-reciprocal plot for the one-binding-site hyperbola (10). Each curve comprises data points from two independent experiments.
RNase H mapping.
Purified RNA (0.1 to 1.0 μg) and specific oligomer (300 ng) were heat denatured and renatured in a 10-μl reaction mixture (20 mM Tris [pH 7.5], 1 mM EDTA, 50 mM NaCl). Details of analysis of the α-globin mRNA poly(A) tail have been previously described (26); a 21-nucleotide (nt) oligomer which anneals 15 nt 5′ to the TGA termination site was used for analysis of the ζ-globin mRNA poly(A) tail (5′TCAGGACAGAGGATACGACCG). The samples were supplemented with 10 μl of a mixture of 20 mM Tris (pH 7.5), 20 mM MgCl2, 100 mM NaCl, 2 mM dithiothreitol, 60 mg of bovine serum albumin per ml, and 0.1 U of RNase H and incubated at 30°C for 60 min. The reaction was terminated by the addition of 130 ml of stop mix (5 μg of tRNA, 20 mM EDTA, 300 mM NaOAc), and the products were phenol extracted, ethanol precipitated, and resuspended in 10 μl of 95% formamide–5 mM EDTA loading dye. The products were resolved on a 6% acrylamide–8 M gel and then were electrotransferred to a Nytran membrane (Schleicher and Schuell, Keene, N.H.) as directed by the manufacturer (Hoefer Scientific, San Francisco, Calif.). The membrane was hybridized with random-primer 32P-labeled probe corresponding to a region immediately 3′ to the oligomer annealing site and then washed according to a standard Northern transfer protocol (26). The probe for the ζ-globin 3′ fragment was generated with forward and reverse primers that anneal within its 3′UTR (5′CAGGACAGGCTGCGGC3′ and 5′ATTGGTTTATTGGCGC3′, respectively) as previously described (26).
RESULTS
In vivo determination of hα- and hζ-globin mRNA stabilities.
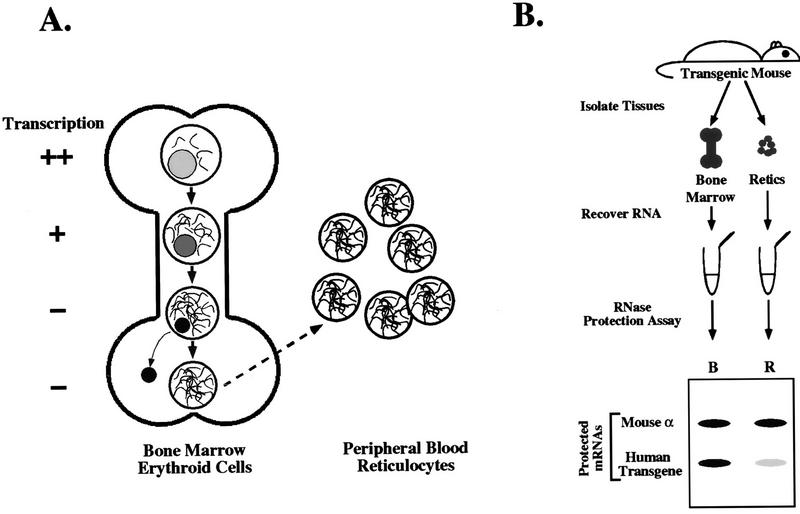
An assay was designed to compare the relative stabilities of transgenic α- and ζ-globin mRNAs in the intact mouse. This assay exploits the non-steady-state conditions resulting from the generalized transcriptional silencing that occurs midway through terminal erythroid differentiation in all mammalian species. Transcriptionally active erythroid progenitors in the bone marrow of adult animals mature into transcriptionally silent intermediates and finally into anucleate reticulocytes that migrate into the peripheral circulation (Fig. 1A). Thus, bone marrow erythroid cells, which are mostly posttranscriptional, and peripheral blood reticulocytes, which are entirely posttranscriptional, represent early and late time points, respectively, in a physiologic transcriptional chase experiment. The uncertain length of the transcriptionally silent interval precludes calculation of absolute mRNA half-lives. Therefore, the stabilities of a variety of transgenic globin mRNAs are determined by assessing the extent to which their levels decline in the interval between these two points in time relative to the level of a standard control globin mRNA. In each tissue, transgenic globin mRNA levels are normalized to the level of stable, endogenous mα-globin mRNA (Fig. 1B). The relative stability of a transgenic mRNA in terminally differentiating erythroid cells is calculated as the ratio of its normalized level in reticulocytes to that in the marrow (the “normalized stability”). As defined, the normalized stability of a transgenic globin mRNA would be 1.0 if it were as stable as mα-globin mRNA, while lower values would indicate relative instability of the transgenic mRNA (26).
FIG. 1.
In vivo determination of globin mRNA stabilities in transgenic mice. (A) Bone marrow erythroid cells and circulating peripheral reticulocytes represent two sequential stages in terminal erythroid differentiation. The sequence of events during terminal differentiation of erythroid cells in the bone marrow (left) and peripheral circulation (right) is illustrated. In the bone marrow, transcriptionally active erythroid progenitors (top) undergo global transcriptional silencing, nuclear condensation, and nuclear extrusion (middle), resulting in transcriptionally silent, anucleate marrow reticulocytes (bottom). Marrow reticulocytes, which contain substantial levels of actively translating globin mRNA, are subsequently released into the bloodstream as peripheral reticulocytes. Reticulocyte mRNA degrades as the cell matures into an erythrocyte over the ensuing 2 to 3 days (not illustrated). (B) Assessment of relative human globin mRNA stabilities in transgenic mice. Bone marrow hematopoietic cells and peripheral blood reticulocytes were harvested from individual transgenic mice previously rendered anemic by treatment with phenylhydrazine. Total RNA was purified from each tissue, and the levels of transgenic (α- or ζ-) globin mRNA and control endogenous mα-globin mRNA were determined by RNase protection with corresponding 32P-labeled antisense RNA probes. Protected probe fragments were resolved on a denaturing acrylamide-urea gel, and band densities were quantitated by PhosphorImager analysis. All assays were carried out under conditions of probe excess. B, bone marrow; R, peripheral blood reticulocytes (Retics).
hζ-Globin mRNA is less stable than hα-globin mRNA in adult-stage erythroid cells.
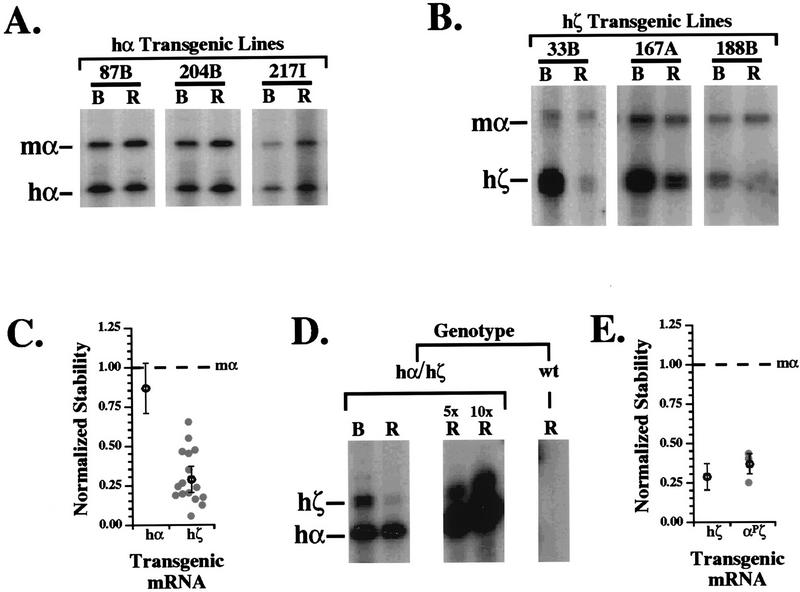
Mouse lines containing hα- and hζ-globin transgenes have been previously generated and characterized for transgene copy number and overall level of transgene expression by standard assays (23, 26). The stabilities of hα- and hζ-globin mRNAs were determined relative to that of α-globin mRNA in individual transgenic mice by the RNase protection assay (RPA [Fig. 1B]). Representative analyses of mice from each of three independent lines transgenic for the hα-globin gene demonstrate that hα-globin mRNA is as stable as endogenous mα-globin mRNA in terminally differentiating adult erythroid cells (Fig. 2A). In contrast, analyses of mice from each of three independent lines transgenic for the hζ-globin gene demonstrate a rapid loss of the hζ-globin mRNA relative to endogenous mα-globin mRNA (Fig. 2B). The normalized stability of hζ-globin mRNA was determined in two or more mice from each of 16 independent transgenic lines; these data are plotted in Fig. 2C. The mean stability of 0.28 for the hζ-globin mRNAs is significantly lower than the mean stability of 0.85 for hα-globin mRNA (P < 0.005).
FIG. 2.
hζ-globin mRNA is less stable than hα-globin mRNA in adult murine erythroid cells. (A) hα-Globin mRNA is stable in adult erythroid cells. Levels of transgenic α-globin mRNA and endogenous mα-globin mRNA were determined by RPA of bone marrow erythrocytes (B) and peripheral blood reticulocytes (R) from individual mice transgenic for the hα-globin gene. Autoradiographs of three representative lines are shown. The positions of the protected hα- and mα-globin mRNA fragments are indicated. Transgenic line designations are shown at the top of respective lanes. In this experiment and others, the specific activities of individual probes were adjusted to ensure that the intensities of the experimental and control bands were both within the linear range of autoradiographic detection. (B) hζ-Globin mRNA is unstable in adult erythroid cells. RPA analyses of hζ-globin mRNA stability in mice from three representative transgenic lines is shown. The positions of the protected hζ- and mα-globin mRNA fragments are indicated. (C) ζ-Globin mRNA is less stable than α-globin mRNA in terminally differentiating adult erythroid cells. The normalized stability values of hζ-globin mRNA determined in each of 16 independent transgenic lines ( ) are plotted. A minimum of two mice from each transgenic line were studied. The normalized stability of endogenous mα-globin mRNA (defined as 1.0) is indicated by a dashed horizontal line. The mean ± 2 standard errors for hζ-globin mRNA is indicated by an open circle with error bars. The mean ± 2 standard errors for hζ-globin mRNA is reproduced from a similar analysis of 10 independent α-globin transgenic lines (26). The difference between the mean stabilities of hα- and hζ-globin mRNA is significant at P < 0.005. (D) Direct comparison of hα- and hζ-globin mRNA stabilities in a mouse coexpressing both transgenes. A mouse was generated which was transgenic for both the human α- and ζ-globin genes, and the levels of expressed hα- and hζ-globin mRNA in its bone marrow (B) and peripheral blood reticulocytes (R) was determined by RPA. An autoradiograph from the study is illustrated, with the positions of the protected hα- and hζ-globin mRNA fragments indicated to the left. A linear increase in the amount of probe protected by 5- and 10-fold amounts of reticulocyte mRNA confirmed that the analysis was performed under conditions of probe excess. Parallel assay of reticulocyte mRNA from a wild-type (wt) mouse illustrates the specificity of the probes for their transgenic hα- and hζ-globin mRNA targets. The above results were confirmed by the study of an additional doubly hemizygous mouse (not shown). (E) Apparent differences in hα- and hζ-globin mRNA stabilities do not reflect differences in promoter function. The stability of an hζ-globin mRNA transcribed from an hα-globin promoter was studied in transgenic mice with the described stability assay. The normalized stability values of hζ-globin mRNA in αPζ mice was determined for mice from each of three independent transgenic lines ( ); the mean ± 2 standard errors is indicated by an open circle with error bars. This value does not differ significantly from the mean stability ± 2 standard errors for hζ-globin mRNA transcribed from the ζ-globin promoter, which has been reproduced from Fig. 2C. The normalized stability of endogenous mα-globin mRNA (defined as 1.0) is indicated by a dashed horizontal line.
We next directly compared the stabilities of the hα- and hζ-globin mRNAs in mice that carried both the hα- and hζ-globin transgenes. These doubly hemizygous mice were generated by mating hα- and hζ-globin transgenic mice and identifying offspring that carried both transgenes by analysis of tail DNA (not shown). The level of hζ-globin mRNA falls relative to hα-globin mRNA in the interval between the bone marrow and reticulocyte stages of terminal differentiation (Fig. 2D). This direct demonstration of the relative instability of hζ-globin mRNA relative to hα-globin mRNA is in full agreement with their normalized stabilities generated from simple transgenic lines (Fig. 2C).
To address the possibility that asynchronous transcriptional silencing of the hα and hζ transgenes contributed to the relative decline in hζ mRNA levels, mouse lines were generated from chimeric transgenes in which the hα promoter was substituted for the hζ promoter in the full-length hζ transgene (αPζ transgene [23]). hζ mRNAs transcribed from the hα promoter in these mice appeared to be as unstable as hζ mRNAs transcribed from the native hζ promoter (Fig. 2E). Hence, although the identity of the promoter affected overall levels of expression, it did not significantly affect the decline in hζ-globin levels during terminal erythroid differentiation. These data support the conclusion that the observed decline in ζ-globin mRNA levels in the interval between the marrow and reticulocyte stages of terminal differentiation is due to a posttranscriptional mechanism(s) affecting ζ-globin mRNA stability.
Differences in the stabilities of hα- and hζ-globin mRNAs are encoded by elements within their 3′UTRs.
We have previously linked the stability of hα-globin mRNA to the structure of its 3′UTR (26, 43, 45, 46). To test whether the relative instability of hζ-globin mRNA is related to structural divergence in this region, we created chimeric hα- and hζ-globin transgenes with exchanged 3′UTRs (Fig. 3A). The chimeric α3′ζ gene encodes an mRNA comprising the hα-globin 5′UTR and coding region and the hζ-globin 3′UTR. The reciprocal transgene, ζ3′α, encodes an mRNA comprising the hζ-globin 5′UTR and coding region, and the hα-globin 3′UTR. As in the case of the hα- and hζ-globin transgenes, high-level expression of the chimeric transgenes was achieved by linking each to core elements of the β-LCR (23, 41). Transgenic lines were established from each of the chimeric globin genes. In each case, transgene copy numbers (between one and five per mouse genome in all but one line [data not shown]) were similar to those previously determined for hα- and hζ-globin transgenic lines (23).
FIG. 3.
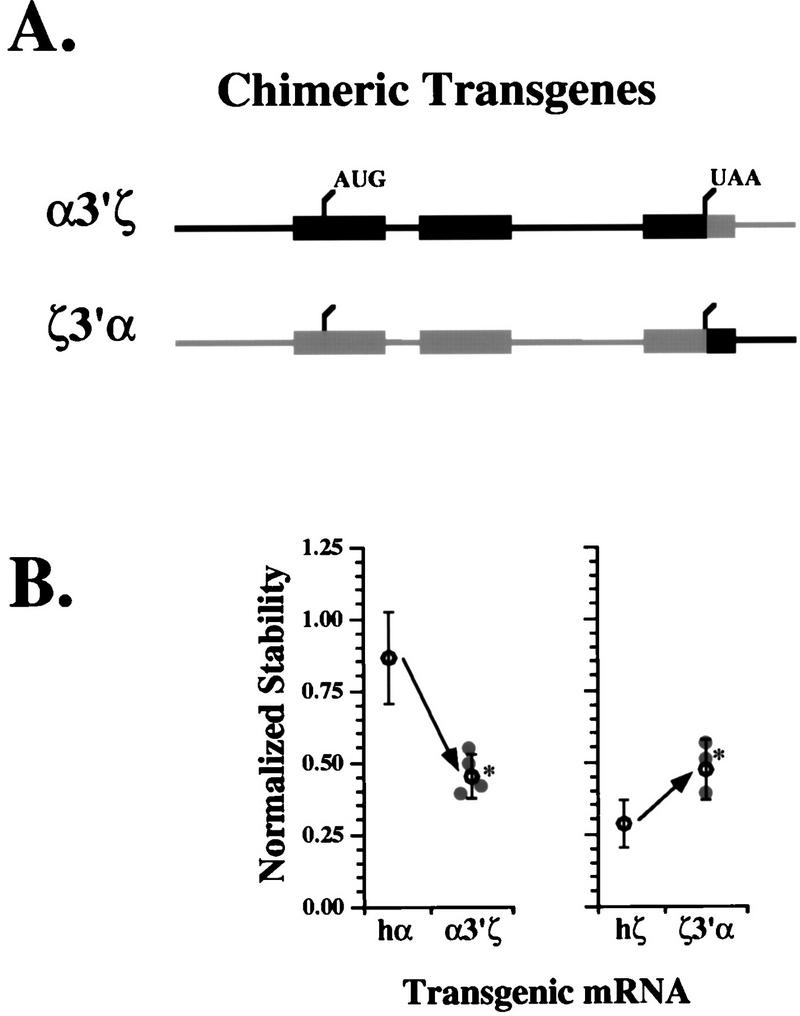
Elements within the ζ-globin 3′UTR mediate mRNA destabilization. (A) Construction of chimeric hα- and hζ-globin transgenes with exchanged 3′UTRs. Two chimeric transgenes were constructed from segments of hα- and hζ-globin genes (black and gray, respectively). The α3′ζ gene comprises the full length α-globin gene in which the ζ-globin 3′UTR and 3′ flanking region have been substituted. The ζ3′α transgene contains the full-length hζ-globin gene in which the α-globin 3′UTR and 3′ flanking region have been substituted. The positions of the normal translation initiation and termination sites are indicated. (B) Distinct stability phenotypes are determined by hα- and hζ-globin mRNA 3′UTRs. Transgenic mouse lines were generated from each chimeric globin transgene illustrated in panel A, and the stabilities of their encoded mRNAs were determined by RPA (Fig. 1). The normalized stability value for transgenic mRNAs from each individual line is plotted ( ). The mean ± 2 standard errors from all α3′ζ lines is shown to the left, along with the average stability of transgenic hα-globin mRNA reproduced from Fig. 2C. The mean ± 2 standard from all ζ3′α lines is shown to the right, along with the average stability of transgenic human ζ-globin mRNAs reproduced from Fig. 2C. Average stabilities which differ significantly from either hα- or hζ-globin mRNA at the P < 0.05 level are indicated by asterisks (∗).
The normalized stabilities of each of the three chimeric mRNAs were established by using multiple mice from each of several independent transgenic lines, employing the standard methods described above. The stability of hα-globin mRNA is halved upon substitution of a ζ-globin 3′UTR (α3′ζ [Fig. 3B, left]). In comparison, the stability of hζ-globin mRNA nearly doubles upon substitution of an α-globin 3′UTR (ζ3′α [Fig. 3B, right]). The stabilities of the α3′ζ and ζ3′α mRNAs differ significantly (P < 0.05) from the stabilities of the parental hα- and hζ-globin mRNAs, respectively. Notably, neither 3′UTR exchange results in full stabilization (or destabilization) of the chimeric mRNA (see Discussion). These data confirm the importance of the 3′UTR to hα-globin mRNA stability, demonstrate that this activity can be transferred to a heterologous mRNA, and indicate that the absence of a functionally equivalent element in the hζ-globin 3′UTR is responsible, in part, for the relative instability of ζ-globin mRNA in adult erythroid cells.
The ζ-globin mRNA 3′UTR assembles a stability-determining α-complex with reduced affinity.
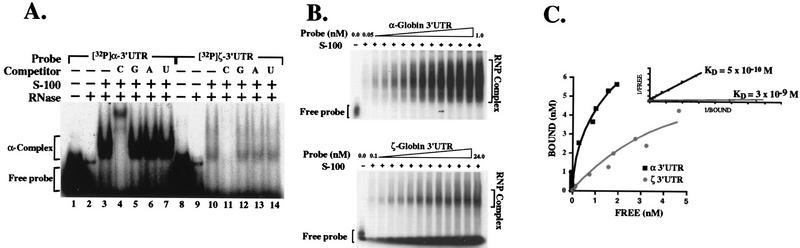
The stability of hα-globin mRNA in vivo has been linked to the assembly of an mRNP complex (the α-complex) on a pyrimidine-rich tract within its 3′UTR (15, 19, 43). The α-complex has a characteristic mobility on nondenaturing polyacrylamide gels, and its assembly is specifically competed by poly(C). When incubated under standard conditions in cytosolic extract from MEL cells, the hζ-globin 3′UTR assembles an mRNP complex with the same migration (Fig. 4A, lanes 3 and 10) and competition profile (lanes 11 to 14) as the authentic α-complex (lanes 4 to 7). These data suggest that the hζ-globin mRNA 3′UTR assembles a complex that is identical to or closely related to the authentic α-complex.
FIG. 4.
hα- and hζ-globin mRNA 3′UTRs differ in their abilities to assemble an RNP α-complex. (A) The α- and ζ-globin mRNAs assemble similar RNP complexes on their respective 3′UTRs. 32P-labeled hα- and hζ-globin mRNA 3′UTRs were incubated in cytosolic (S-100) extract prepared from MEL cells, and RNA not protected by RNP complexes was subsequently digested by addition of RNase T1. Protected RNP complexes were resolved on a nondenaturing 5% polyacrylamide gel. Assays were carried out in the absence (lanes 3 and 10) or presence (lanes 4 to 7 and 11 to 14) of excess homoribopolymer competitors [poly(C), poly(G), poly(A), or poly(U)]. Control lanes (1, 2, 8, and 9) verify the integrity of the probes and the efficiency of RNase digestion. The positions of the α-complex and free RNA probe are indicated to the left of the autoradiograph. (B) Determination of the Kd of the RNP complexes which assemble on the α- and ζ-globin mRNA 3′UTRs. Defined quantities of 32P-labeled hα or hζ 3′UTR transcripts were incubated in MEL cell cytosolic extract (5 μg) and resolved on nondenaturing 5% polyacrylamide gels. For each sample, the quantity of probe incorporated into α-complexes (bracketed) was determined by PhosphorImager analysis. The position of free hα and hζ 3′UTR probes are indicated. hα-3′UTRs are nearly completely incorporated into α-complexes when present in concentrations of <1 nM, consistent with the extremely low Kd of this interaction. Free probe can be visualized upon prolonged autoradiograph exposure and in samples which contain probe in concentrations of >2.5 nM (not shown). (C) The α-complex assembles on the α- and ζ-globin mRNA 3′UTRs with distinct affinities. The affinity of α-complex interaction with the α- and ζ-globin 3′UTRs (black and gray, respectively) was estimated as the apparent Kd, calculated from linear regression analysis of a double-reciprocal plot (inset). Curves comprise points from results illustrated in panel B and from additional independent experiments (not shown).
The comparatively weak intensity of its mRNP signal suggested that complex assembly on the ζ3′UTR was inefficient relative to assembly of the same complex on the α 3′UTR (Fig. 4A). To quantitate this difference, the apparent Kd for α-complex assembly on the hα- and hζ-globin mRNA 3′UTRs was determined. Defined quantities of hα- and hζ-globin 3′UTR probes were incubated in MEL cell cytosolic extract and resolved on a nondenaturing polyacrylamide gel, and the intensity of the α-complex was quantitated by PhosphorImager (Fig. 4B). A double-reciprocal plot of complex-bound (gel-shifted) versus free 3′UTR probes indicated Kds of 5 × 10−10 M for the hα 3′UTR and 3 × 10−9 M for the hζ 3′UTR (Fig. 4C). The sixfold-higher Kd for the hζ 3′UTR was supported by cross-competition studies of 32P-labeled hα- and hζ-globin mRNA 3′UTRs which demonstrated that the ζ 3′UTR was a less efficient competitor for RNP complex assembly than the hα 3′UTR (data not shown). Hence, the α-globin 3′UTR contains a target sequence which assembles an α-complex sixfold more efficiently than a corresponding sequence contained within the ζ-globin 3′UTR.
A C→G substitution in the major polypyrimidine track within the ζ-globin 3′UTR reduces the efficiency of α-complex assembly.
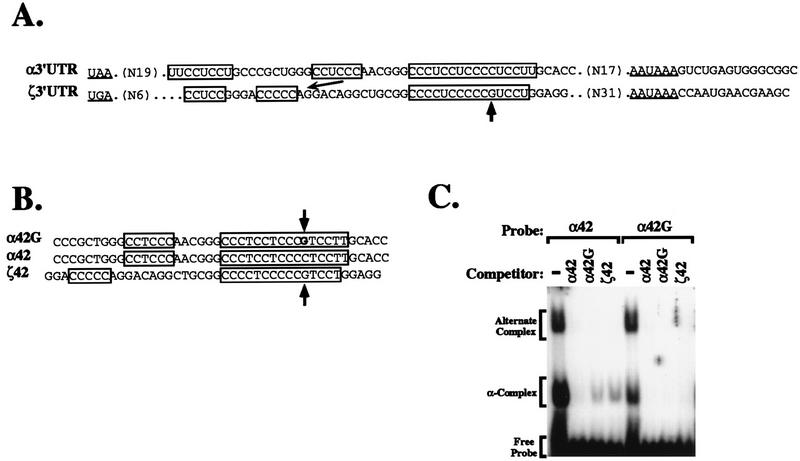
An analysis of the structural basis for the difference in α-complex assembly and mRNA stability was initiated by inspection of the α- and ζ-globin 3′UTR nucleotide sequences (Fig. 5A). The ζ-globin 3′UTR was aligned to maximize homology to three pyrimidine-rich elements of the α-globin 3′UTR, which have been directly implicated in both α-globin mRNA stability and α-complex assembly (14, 43, 45). All three of the pyrimidine-rich tracts within the α-globin 3′UTR are conserved within the ζ-globin 3′UTR. However, the central tract is shifted 5′ a distance of 7 nt in the ζ-globin 3′UTR relative to the α-globin 3′UTR, and the longest of the three polypyrimidine tracts within the ζ-globin 3′UTR is interrupted by a single purine (G) transversion. Either (or both) of these structural differences might account for the decreased affinity of the α-complex for the ζ-globin 3′UTR and for the consequent decrease in ζ-globin mRNA stability. Evolutionary comparisons and structural mapping studies suggest, however, that the major polypyrimidine tract is the most important feature determining α-globin mRNA stability and α-complex assembly (14, 44). Thus, the effect of the single-base transversion on α-complex assembly was studied. The α-complex readily assembled on a 42-nt oligomer (α42) corresponding to the segment of the native α-3′UTR previously demonstrated to be fully sufficient for α-complex assembly (14). A single C→G mutation in the major polypyrimidine track of α42 (α42G) markedly reduced α-complex assembly (Fig. 5C). Moreover, α42G and a 42-nt oligomer corresponding to the native ζ3′UTR (ζ42), which contains a similarly positioned G, both compete poorly for α-complex assembly relative to native α42. These data indicate that a naturally occurring G which interrupts the major polypyrimidine tract in the native ζ-globin mRNA 3′UTR decreases its capacity to assembly a stabilizing α-complex.
FIG. 5.
Assembly of an mRNP stability complex on the α-globin 3′UTR is inhibited by a C→G transversion within the major polypyrimidine tract. (A) Alignment of hα- and hζ-3′UTR minimal sequences. The hα- and hζ-globin mRNA 3′UTRs are aligned to maximize homology; the lengths of 3′UTR sequences on either end of the aligned segments are indicated parenthetically. The termination codons (UAA or UGA) and poly(A) hexanucleotides (AAUAAA) are underlined. The polypyrimidine tracts implicated in α-globin mRNA stability are boxed, as are corresponding polypyrimidine-rich regions of the ζ-globin mRNA 3′UTR. The 5′ displacement of the central ζ-globin polypyrimidine tract, relative to the related α-globin sequence, is emphasized by a narrow arrow; interruption of the major polypyrimidine tract by a purine (G) is indicated by a thick vertical arrow. (B) Oligomers used to assess the importance of an uninterrupted pyrimidine tract to high-efficiency α-complex assembly. α42 corresponds to the 42-nt region of the α-globin mRNA 3′UTR protected from RNase digestion by a fully assembled α-complex. α42G differs from the wild-type α42 by the substitution of a G within the 3′ terminal of the major polypyrimidine tract. ζ42 is the wild-type ζ-globin 3′UTR sequence corresponding to the α42 segment. The substituted G in α42G is in boldface, and both it and the native G which interrupts the major polypyrimidine tract in the ζ-3′UTR sequence (ζ42) are indicated by arrows. (C) Interruption of the major polypyrimidine tract of the α-3′UTR by a purine G inhibits α-complex assembly. 32P-labeled α42 and α42G were incubated in MEL cell S-100 extract in the absence (−) or presence of unlabeled competitors present in 100-fold excess, and resolved on a native polyacrylamide gel as described in the legend to Fig. 4. The positions of the α-complex, the previously defined alternate complex (43), and free probe are indicated to the left of the autoradiograph.
Destabilization of hζ-globin mRNA is linked to its accelerated deadenylation.
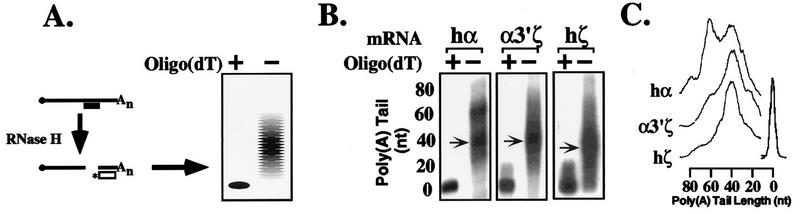
The observation that destabilization of the hζ-globin mRNA in vivo parallels inefficient assembly of the α-complex on its 3′UTR in vitro suggests that the two processes are related. To begin to characterize the molecular steps that accompany accelerated hζ-globin mRNA decay, we assessed the length of poly(A) tails on the hα-, hζ-, and hα3′ζ-globin mRNAs (Fig. 3A). RNA was isolated from reticulocytes of mice carrying each of the three transgenes (Fig. 6A). The transgenic RNAs were subjected to site-specific RNase H cleavage, resolved on a denaturing polyacrylamide gel, and electroblotted to a nylon membrane, and the 3′-terminal fragments containing the poly(A) tails were visualized with ζ or α 3′UTR sequence-specific probes. The poly(A) tail on the hα-globin 3′UTR demonstrated discrete peaks of activity that occurred at intervals of approximately 20 nt (Fig. 6B). The major peak corresponded to a poly(A) tail length of approximately 60 nt (A60), while large proportions of shorter (A40) and longer (A80) poly(A) tails were also observed. In contrast, both mRNAs with ζ-3′UTRs (the ζ and α3′ζ mRNAs) contained poly(A) tails almost entirely composed of the shorter A40 variety. Hence, two characteristics of ζ-globin mRNA—its relative instability and a short poly(A) tail—are encoded by elements contained within the 105-nt ζ 3′UTR.
FIG. 6.
The hζ-globin mRNA 3′UTR contains a short poly(A) tail. (A) Measurement of poly(A) tail length (An) on transgenic globin mRNAs. Site-specific RNase H cleavage of globin mRNAs was targeted by an antisense DNA oligomer (horizontal bar) complementary to a sequence located approximately 100 nt 5′ to its poly(A) addition site. The RNA fragments were resolved on a denaturing acrylamide gel and electrotransferred to a nylon membrane. The 3′-terminal fragment, which contains the poly(A) tail, was visualized by hybridization with a site-specific 32P-labeled 3′UTR DNA probe, the position of which is indicated (∗□). The length of the fully deadenylated 3′-terminal RNA fragment was established by addition of excess oligo(dT) to the hybridization reaction prior to the RNase H digestion. (B) The hζ and α3′ζ mRNAs have shortened poly(A) tails. The poly(A) tail lengths of hα-, hζ-, and chimeric α3′ζ-globin mRNAs from transgenic reticulocytes were determined with the RNase H assay. The positions of the fully deadenylated 3′-terminal fragments are aligned, and the positions equivalent to a poly(A) tail length of 40 residues (determined by DNA size markers electrophoresed in parallel for each study) are indicated by an arrow. The results for each mRNA were confirmed in experiments with reticulocytes from mice representing two or more independent transgenic lines. (C) Densitometric analyses of the poly(A) tails from panel B. Signal intensities from hα-, hζ-, and chimeric α3′ζ-globin poly(A) tails from panel B were quantitated by scanning densitometry, and the adjusted scans were aligned to a standard scale to facilitate direct comparison.
DISCUSSION
The induction of α-globin gene expression and reciprocal silencing of ζ-globin gene expression in erythroid cells between the 6th and 8th weeks of human gestation define the end of the embryonic and the initiation of the fetal stages of erythroid development. This process is paralleled in the β-globin gene cluster by a switch from embryonic ɛ- to fetal γ-globin gene expression (reviewed by Russell and Liebhaber [33]). It is hypothesized that coordination of globin gene switching between the two clusters provides a mechanism through which hemoglobin heterotetramers with specific O2 affinities can be expressed during different developmental stages; such an arrangement might facilitate transplacental diffusion of O2 from maternal to embryonic or fetal erythrocytes. A potential hazard of such an arrangement is that failure to fully silence expression of embryonic globin genes might result in the inappropriate expression of embryonic hemoglobin heterotetramers with high O2 affinities in fetal and adult erythrocytes (14). This situation might be detrimental to the fetus by altering sensitive transplacental O2 gradients and to adults by triggering a secondary polycythemia (11). These possibilities emphasize the potential importance of full ζ-globin gene silencing at the appropriate gestational age.
The contribution of transcriptional controls to globin gene switching has been intensively studied, and the specific importance of this mechanism to ζ-globin silencing has been clearly documented (23, 29, 36, 37). However, independent lines of evidence from experiments utilizing primary human erythroid tissue suggest that additional mechanisms which act posttranscriptionally on both α- and ζ-globin gene products are also crucial to this process (1, 49). Nuclear run-on experiments utilizing progenitor erythroid cells from normal adult bone marrow yield surprisingly high levels of ζ-globin transcripts (49); however, ζ-globin mRNA can be detected at only trace levels in near fully differentiated adult reticulocytes (1). This apparent paradox is resolved if one postulates a posttranscriptional defect in ζ-globin mRNA accumulation due to instability of the ζ-globin mRNA. This hypothesis is supported by our recent studies of transgenic mice, which demonstrate that hζ-globin transcriptional control elements are only partially effective in ζ-globin gene silencing and that differences within the α- and ζ-globin transcribed regions are important determinants of their expression (23). Collectively, these data support a model in which posttranscriptional events—specifically, instability of ζ-globin mRNA—play a necessary role in the full silencing of ζ-globin gene expression.
The general importance of mRNA stability to the control of eukaryotic gene expression is well established. The half-lives of eukaryotic mRNAs vary more than 1,000-fold, from as short as several minutes to as long as several days (30). The short half-lives of labile mRNAs such as c-myc, c-fos, and other cytokine and lymphokine mRNAs, permit changes in transcription to be rapidly reflected in the level of cytoplasmic mRNA (6, 30). In comparison, “bulk” proteins, such as collagens (13, 25) or crystallins (22), are typically encoded by highly stable mRNAs, providing the cell with a substantial savings in the energy required for their transcription, processing, and transport. In some cases, mRNA stability is dynamic, as occurs with transferrin receptor (TfR) mRNA, whose half-life varies in response to physiologically relevant changes in cellular iron levels (20). Hence, mRNA stability plays an important regulatory role in the expression of a wide variety of genes central to cellular differentiation and function.
In the current work, we test the hypothesis that posttranscriptional controls of ζ-globin gene expression act through destabilization of ζ-globin mRNA in fetal and adult erythroid cells. A mouse model system was established in which the stabilities of transgenic globin mRNAs could be measured in vivo (Fig. 1). We have previously verified the utility of the transgenic approach for studies of globin mRNA stability by comparing the stability of hαwt mRNA to an hα-globin mRNA with a known destabilizing mutation (αCS) (26). This system closely models the physiology of normal human erythropoiesis and precisely models the compensatory erythropoietic response to a variety of anemias, including those (as typified by the thalassemias) that result from ineffective erythropoiesis and accelerated peripheral hemolysis. The current approach also permits long-term monitoring of highly stable globin mRNAs under non-steady-state conditions that occur in cells undergoing rapid changes in morphology, function, and gene expression. The “physiologic transcriptional chase” has obvious advantages over the use of global, nonspecific inhibitors of transcription, which may impart artifactual effects in studies of mRNA stability (27, 39, 48). Moreover, cell culture-based model systems which permit interval assessment of mRNA levels following a transcriptional pulse (such as the fos promoter response to serum induction) are poorly suited for the study of highly stable mRNAs, because it is difficult to maintain cell viability during the prohibitively long serum-free preparatory interval necessary to establish sufficiently low background levels of the test mRNA (45). Studies summarized in Fig. 2 demonstrated that hζ-globin mRNA is significantly less stable than hα-globin mRNA in erythroid cells from transgenic mice. This effect complements, but is independent of, transcriptional downregulation of the ζ-globin gene occurring at the embryonic-to-fetal transition. The mRNA sequences which determine the difference in stability map to the α- and ζ-globin mRNA 3′UTRs (Fig. 3). These results confirm the previously established importance of the 3′UTR to α-globin mRNA stability (45, 46) and demonstrate that the ζ-globin mRNA 3′UTR lacks an equivalent stability-determining function. These results also confirm in a whole animal system earlier observations in cultured cells that the stabilizing function accompanies physical transfer of the α-globin 3′UTR (34).
Although exchange of the hα- and hζ-globin mRNA 3′UTRs significantly alters the stability of the resultant chimeric mRNAs in the anticipated direction, this effect is incomplete. Compared to the stabilities of the native hα- and hζ-globin mRNAs, the intermediate values for the chimeric mRNAs suggest that additional, functionally distinct regions outside the 3′UTR of either (or both) the α- and ζ-globin mRNAs may also contribute to their stability. A similar arrangement of spatially and functionally distinct elements mediates accelerated decay of several short-lived mRNAs, including those of c-myc, c-fos, and beta interferon (17, 40, 47, 48), and may mediate high-level stability of the human β-globin mRNA in adult erythroid cells (34). The evolutionary forces that favor a multiplicity of stability-determining features remain obscure, although it seems likely that such an arrangement functions in part to ensure the stability of α-globin mRNA, which is crucial to the generation of a fully functional erythrocyte.
An unexplained aspect of the current data is the variation in the normalized stabilities of transgenic mRNAs among lines generated from identical transgene constructs. This effect is best appreciated by the scatter in the stabilities of hζ-globin mRNA illustrated in Fig. 2; a similar scatter has also been observed in measurements of the stability of transgenic hα-globin mRNAs (26). The stability of a transgenic mRNA determined in multiple mice from the same line is highly reproducible (∼12% variation), as are serial determinations from an individual mouse. This high reproducibility suggests that biologic factors contribute to the scatter in mRNA stabilities observed among independent lines. An analysis of possible variables that could result in line-to-line variation in mRNA stability failed to demonstrate linkage to transgene structure, copy number, level of expression, or the sex of the adult mouse. Additional experiments (Fig. 2E) indicated that the stability of transcribed ζ-globin mRNAs is independent of promoter identity. This raises the possibility that the observed differences might reflect local influences at each transgene insertion site. Such random integration site-dependent effects might explain some of the variation that has previously been observed in studies of transgenic α-globin gene expression (38). Although of potential interest, the observed line-to-line variation in mRNA stability does not detract from the highly significant, threefold difference in the stability of the human α- and ζ-globin mRNAs in the large number of independent transgenic lines studied.
A molecular basis for the difference in the stabilities of the α- and ζ-globin mRNAs was suggested by RNA gel mobility shift analysis (Fig. 4). The high stability of α-globin mRNA is believed to result from assembly of an RNP complex (the α complex) on the α-globin 3′UTR (14, 19, 43). It was thus surprising to find that despite its relatively low stability, the ζ-globin mRNA 3′UTR assembles an mRNP complex which, as judged by its gel migration and its susceptibility to competition by poly(C), appears similar, if not identical, to an authentic α-complex. However, the affinity of the α-complex for the ζ-globin 3′UTR was sixfold lower than its affinity for the α-globin 3′UTR (Fig. 4), thus linking the relative instability of ζ-globin mRNA to the relatively high Kd of α-complex formation on its 3′UTR. The structural basis for this difference in the efficiency of α-complex assembly was suggested by alignment of the α- and ζ-globin mRNA 3′UTR sequences (Fig. 5A). Both 3′UTRs contain a series of three pyrimidine-rich tracts which have been largely conserved in the 350 to 400 million years since separation of the hα- and hζ-globin genes (12). These pyrimidine-rich tracts appear to be essential for assembly of an α-complex on the α-globin 3′UTR (14, 43, 44). We have shown that interruption of the major pyrimidine-rich tract by a single purine negatively affects the efficiency of α-complex assembly (Fig. 5). Still, the fact that these pyrimidine tracts have been largely conserved within the context of otherwise poorly conserved 3′UTRs suggests that they may serve an additional unrecognized function(s). For example, the ζ 3′UTR might play a limited role in enhancing ζ-globin mRNA stability in primitive (embryonic) erythroid cells. To date, our attempts to test this possibility have been frustrated by technical barriers that limit direct measurement of the stability of globin mRNAs in this tissue.
An analysis of the poly(A) tails of the α- and ζ-globin mRNAs indicated a potential mechanism through which ζ-globin mRNA decay is accelerated (Fig. 6). Although mRNA degradation may proceed via a number of overlapping pathways in both yeast and higher eukaryotes, a common pathway involves initial deadenylation followed by cleavage of the 5′ 7mG(5′) ppp(5′)N mRNA cap (8, 9, 16, 28, 40). The demonstration that functional characteristics, such as mRNA stability, and structural attributes, such as poly(A) tail length, both accompany physical transfer of the ζ-globin 3′UTR to the α-globin mRNA emphasizes the importance of this region to normal ζ-globin gene expression. The shortened poly(A) tails may result from inefficient polyadenylation of the nascent mRNA in the nucleus and/or from accelerated shortening of a normally polyadenylated mRNA in the cytoplasm. Attempts to resolve this issue by analysis of poly(A) tail length in early-stage (marrow) erythroid progenitors have been frustrated by the limited amount of tissue and the low-level expression of the hζ transgene. It seems likely, however, that accelerated deadenylation related to degradation occurs in the cytoplasm, because we have observed that unusually short poly(A) tail lengths accompany mRNAs destabilized by either of two mechanisms: the loss of cis elements in the ζ-globin mRNA 3′UTR (this study) or through physical disruption of the α-complex by ribosomal read-through into the 3′UTR (26). A causal link between accelerated mRNA decay and a short poly(A) tail has been demonstrated in yeast, but has not yet been firmly established in higher eukaryotes (8, 9).
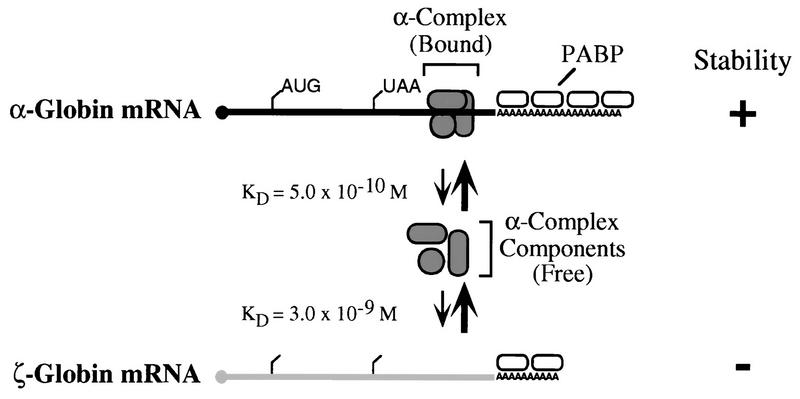
The data from the current study support a model for ζ-globin gene silencing which incorporates both transcriptional downregulation and posttranscriptional destabilization of ζ-globin mRNA (Fig. 7). In this model, high-affinity cis elements within the α-globin mRNA 3′UTR assemble α-complexes efficiently, resulting in stabilization of α-globin mRNA. In contrast, the accelerated deadenylation and decay of human ζ-globin mRNA in adult erythroid cells reflect relatively inefficient assembly of a stabilizing α-complex on low-affinity cis elements within its 3′UTR. This relatively inefficient assembly of the α-complex is likely to be of only minimal importance to ζ-globin expression in circulating primitive (embryonic) erythroblasts, because these cells contain low levels of competitor α-globin mRNA. Moreover, ζ-globin mRNA is continually replenished in circulating primitive erythroblasts, which maintain a transcriptionally active nucleus. In contrast, the relative instability of ζ-globin mRNA is a crucial determinant of full ζ-globin gene silencing in definitive (fetal or adult) erythrocytes. Reciprocal induction of the α-globin genes and downregulation of ζ-globin gene transcription result in a large increase in the ratio of α to ζ mRNAs. According to the model, the α-globin mRNAs assemble stabilizing α-complexes on their high-affinity 3′UTRs from limiting quantities of essential components. ζ-globin mRNAs, which compete poorly for these limiting factors, are left unprotected and are subject to accelerated decay. These molecular events are paralleled by a cellular switch from transcriptionally active, nucleated primitive erythrocytes to transcriptionally silent, anucleate definitive erythrocytes. The shortened half-life of ζ-globin mRNA results in an exponential fall in its levels relative to α-globin mRNA in the 3- to 5-day transcriptionally silent (but translationally active) period of terminal differentiation. Hence, the temporal confluence of these two events—the molecular and cellular switches—results in a situation in which direct molecular communication or “cross-talk” between the α- and ζ-globin 3′UTRs assumes a prominent role in ζ-globin gene silencing. This model would predict that naturally occurring mutations which downregulate α-globin gene transcription would result in increased levels of ζ-globin mRNA. In fact, this pattern is observed in individuals with some forms of deletional α-thalassemia who express especially low levels of α-globin mRNA (7, 42). A detailed knowledge of these mechanisms would permit formulation of therapeutic strategies designed to alter the balance between α- and ζ-globin gene expression through modulation of these posttranscriptional controls. In addition, the identification of additional mRNAs whose functions depend on assembly of RNP complexes which share components with the α-complex (15) will suggest a wider role for direct mRNA-mRNA interaction in the control of gene expression.
FIG. 7.
Model in which instability of ζ-globin mRNA reflects its reduced affinity for the α-complex and is initiated through accelerated decay of the poly(A) tail. In terminally differentiating adult erythroid cells, limiting quantities of one or more components of the α-complex (gray objects) assemble preferentially on high-affinity sites within the α-globin mRNA 3′UTR. Lower-affinity sites in the ζ-globin 3′UTR compete poorly for α-complex assembly, subjecting the unprotected ζ-globin mRNA to accelerated degradation. In this model, assembly of the poly(A) tail by PABP multimers results in discontinuous deadenylation, observed experimentally as phasing of poly(A) tail lengths (26). The α- and ζ-globin mRNA sequences are indicated in black and gray, respectively; the mRNA cap (•) and translation initiation and termination sites (tick marks) are noted, as are PABP monomers (open ovals).
ACKNOWLEDGMENTS
We thank Alice Lee and Faith Fox for expert technical assistance and Nancy Cooke and William Lee for critical reading of the manuscript.
This work was supported in part by NIH grants HL-K11-02623 (J.E.R.), HL-38632 (S.A.L.), and CA-72765 (S.A.L.). S.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Albitar M, Peschle C, Liebhaber S A. Theta, zeta, and epsilon globin messenger RNAs are expressed in adults. Blood. 1989;74:629–637. [PubMed] [Google Scholar]
- 2.Aviv H, Volloch Z, Bastos R, Levy S. Biosynthesis and stability of globin mRNA in erythroleukemic Friend cells. Cell. 1976;8:495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- 3.Baer B W, Kornberg R D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer B W, Kornberg R D. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastos R N, Aviv H. Theoretical analysis of a model for globin messenger RNA accumulation during erythropoiesis. J Mol Biol. 1977;110:205–218. doi: 10.1016/s0022-2836(77)80069-7. [DOI] [PubMed] [Google Scholar]
- 6.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 7.Chui D H K, Wong S C, Chung S-W, Patterson M, Bhargava S, Poon M-C. Embryonic ζ-globin chains in adults: a marker for α-thalassemia-1 haplotype due to a >17.5 kb deletion. N Engl J Med. 1986;314:76–79. doi: 10.1056/NEJM198601093140203. [DOI] [PubMed] [Google Scholar]
- 8.Decker C J, Parker R. Mechanisms of mRNA degradation in eucaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 9.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 10.Feldman H A. Mathematical theory of complex ligand-binding systems at equilibrium: some methods for parameter fitting. Anal Biochem. 1972;48:317–338. doi: 10.1016/0003-2697(72)90084-x. [DOI] [PubMed] [Google Scholar]
- 11.Gau G T, Fairbanks V F, Maldonado J E, Bassingthwaighte J B, Tancredi R G. Cardiac dysfunction in a patient with hemoglobin Malmo treated with repeated transfusions. Clin Res. 1974;22:276A. [Google Scholar]
- 12.Goodman M, Weiss M L, Czelusniak J. Molecular evolution above the species level: branching pattern, rates, and mechanisms. Syst Zool. 1982;31:376. [Google Scholar]
- 13.Hamalainen L, Oikarinen J, Kivirikko K. Synthesis and degradation of type 1 procollagen mRNA in cultured human skin fibroblasts and the effects of cortisol. J Biol Chem. 1985;260:720–726. [PubMed] [Google Scholar]
- 14.Hofmann O, Mould R, Brittain Y. Allosteric modulation of oxygen binding to the three human embryonic hemoglobins. Biochem J. 1995;306:367–370. doi: 10.1042/bj3060367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holcik M, Liebhaber S A. Four highly stable eukaryotic mRNAs assemble 3′UTR RNA-protein complexes sharing cis- and trans-components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 17.Jones T R, Cole M D. Rapid cytoplasmic turnover of c-mycmRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabnick K S, Housman D E. Determinants that contribute to cytoplasmic stability of human c-fosand β-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klausner R D, Rouault T A, Harford J B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 21.Krowczynska A, Yenofsky R, Brawerman G. Regulation of messenger RNA stability in mouse erythroleukemia cells. J Mol Biol. 1985;181:231–239. doi: 10.1016/0022-2836(85)90087-7. [DOI] [PubMed] [Google Scholar]
- 22.Li X A, Beebe D C. Messenger RNA stabilization in chicken lens development: a reexamination. Dev Biol. 1991;146:239–241. doi: 10.1016/0012-1606(91)90463-d. [DOI] [PubMed] [Google Scholar]
- 23.Liebhaber S A, Wang Z, Cash F E, Monks B, Russell J E. Developmental silencing of the embryonic ζ-globin gene: synergistic action of the promoter and 3′-flanking region combined with stage-specific silencing by the transcribed segment. Mol Cell Biol. 1996;16:2637–2646. doi: 10.1128/mcb.16.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodish H F, Small B. Different lifetimes of reticulocyte messenger RNA. Cell. 1976;7:59–65. doi: 10.1016/0092-8674(76)90255-5. [DOI] [PubMed] [Google Scholar]
- 25.Maata A, Elkholm E, Penttinen R P. Effect of the 3′-untranslated region on the expression levels and mRNA stability of alpha1(1) collagen gene. Biochim Biophys Acta. 1995;1260:294–300. doi: 10.1016/0167-4781(94)00207-j. [DOI] [PubMed] [Google Scholar]
- 26.Morales J, Russell J E, Liebhaber S A. Destabilization of human α-globin mRNA by translation anti-termination is controlled during erythroid differentiation and paralleled by phased shortening of the poly(A) tail. J Biol Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 27.Mullner E W, Kuhn L C. A stem-loop in the 3′ untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988;53:815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- 28.Peltz S W, Brewer G, Bernstein P, Hart P, Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1:99–126. [PubMed] [Google Scholar]
- 29.Pondel M D, Proudfoot N J, Whitelaw C, Whitelaw E. The developmental regulation of the human ζ-globin gene in transgenic mice employing β-galactosidase as a reporter gene. Nucleic Acids Res. 1992;20:5655–5660. doi: 10.1093/nar/20.21.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross J, Pizarro A. Human beta and delta globin messenger RNAs turn over at different rates. J Mol Biol. 1983;167:607–617. doi: 10.1016/s0022-2836(83)80101-6. [DOI] [PubMed] [Google Scholar]
- 32.Ross J, Sullivan T D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- 33.Russell J E, Liebhaber S A. Molecular genetics of thalassemia. Adv Genome Biol. 1993;2:283–353. [Google Scholar]
- 34.Russell J E, Liebhaber S A. The stability of human β-globin mRNA is dependent on structural determinants positioned within its 3′ untranslated region. Blood. 1996;87:5314–5323. [PubMed] [Google Scholar]
- 35.Russell J E, Morales J, Liebhaber S A. The role of globin mRNA stability in the control of globin gene expression. Prog Nucleic Acid Res Mol Biol. 1997;57:249–287. doi: 10.1016/s0079-6603(08)60283-4. [DOI] [PubMed] [Google Scholar]
- 36.Sabath D E, Koehler K M, Yang W Q, Patton K, Stamatoyannopoulos G. Identification of a major positive regulatory element located 5′ to the human ζ-globin gene. Blood. 1995;85:2587–2597. [PubMed] [Google Scholar]
- 37.Sabath D E, Spangler E A, Rubin E M, Stamatoyannopoulos G. Analysis of the human ζ-globin gene promoter in transgenic mice. Blood. 1993;82:2899–2905. [PubMed] [Google Scholar]
- 38.Sharpe J A, Chan-Thomas P S, Lida J, Ayyub H, Wood W G, Higgs D R. Analysis of the human alpha globin upstream regulatory element (HS-40) in transgenic mice. EMBO J. 1992;11:4565–4572. doi: 10.1002/j.1460-2075.1992.tb05558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shyu A B, Greenberg M E, Belasco J G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 40.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–234. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 41.Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves D. A dominant control region for human β-globin locus conferring integration site-independent gene expression. Nature. 1989;33:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 42.Tang W, Luo H-Y, Albitar M, Patterson M, Eng B, Waye J S, Liebhaber S A, Higgs D R, Chui D H K. Human embryonic ζ-globin chain expression in deletional α-thalassemias. Blood. 1992;80:517–522. [PubMed] [Google Scholar]
- 43.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol Cell Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Liebhaber S A. Complementary change in cis determinants and trans factors in the evolution of an mRNP stability complex. EMBO J. 1996;15:5040–5051. [PMC free article] [PubMed] [Google Scholar]
- 44a.Wang, Z., and S. A. Liebhaber. Unpublished data.
- 45.Weiss I M, Liebhaber S A. Erythroid cell-specific mRNA stability elements in the α2-globin 3′ nontranslated region. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss I M, Liebhaber S A. Erythroid cell-specific determinants of α-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whittemore L-A, Maniatis T. Postinduction turnover of beta-interferon gene expression. Mol Cell Biol. 1990;10:1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wisdom R, Lee W M F. The protein coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 49.Yagi M, Gelinas R, Elder J T, Peretz M, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Chromatin structure and developmental expression of the human α-globin cluster. Mol Cell Biol. 1986;6:1108–1116. doi: 10.1128/mcb.6.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]