Abstract
Mammalian cells are able to repair chromosomal double-strand breaks (DSBs) both by homologous recombination and by mechanisms that require little or no homology. Although spontaneous homologous recombination is rare, DSBs will stimulate recombination by 2 to 3 orders of magnitude when homology is provided either from exogenous DNA in gene-targeting experiments or from a repeated chromosomal sequence. Using a gene-targeting assay in mouse embryonic stem cells, we now investigate the effect of heterology on recombinational repair of DSBs. Cells were cotransfected with an endonuclease expression plasmid to induce chromosomal DSBs and with substrates containing up to 1.2% heterology from which to repair the DSBs. We find that heterology decreases the efficiency of recombinational repair, with 1.2% sequence divergence resulting in an approximately sixfold reduction in recombination. Gene conversion tract lengths were examined in 80 recombinants. Relatively short gene conversion tracts were observed, with 80% of the recombinants having tracts of 58 bp or less. These results suggest that chromosome ends in mammalian cells are generally protected from extensive degradation prior to recombination. Gene conversion tracts that were long (up to 511 bp) were continuous, i.e., they contained an uninterrupted incorporation of the silent mutations. This continuity suggests that these long tracts arose from extensive degradation of the ends or from formation of heteroduplex DNA which is corrected with a strong bias in the direction of the unbroken strand.
DNA double-strand breaks (DSBs) compromise the genetic integrity of all organisms, since failure to repair a broken chromosome can result in loss of genetic information and inappropriate repair of DSBs can lead to defects such as chromosomal translocations. In striking contrast to the repair of DSBs in Saccharomyces cerevisiae, which occurs primarily by homologous recombination (11), repair of DSBs in mammalian cells has been assumed to occur primarily by nonhomologous recombination (12). The recent examination of DSB repair in mammalian cells, however, has contrasted with this view, demonstrating that repair of DSBs by homologous recombination can occur as a substantial proportion of repair events (15, 27, 28). Consistent with this, DSBs in mammalian cells can stimulate recombination to a high level (13), upwards of 1,000-fold or more, for allelic recombination (19a), intrachromosomal recombination (15, 28, 39), and gene targeting (6, 15, 27, 33).
Several models have been proposed for the repair of DSBs in mammalian cells and fungi, including the double-strand-break-repair model (38). In this model, the broken ends of chromosomal DNA are degraded to create a gap, which is repaired from an unbroken homologous template. The broken ends initiate recombination by invading the homologous template. As a result, heteroduplex DNA, consisting of paired strands from both the unbroken and broken recombination substrates, is present at the boundaries of the gap. Two Holliday junctions are formed at the boundaries of the heteroduplex and are subsequently resolved to give crossover or noncrossover products. Evidence from yeast meiotic and mitotic recombination studies suggests a modification of this model in which double-strand gaps are not formed; instead, conversion involves mismatch repair of heteroduplex DNA formed upon strand invasion and branch migration or by annealing subsequent to repair synthesis (30, 34). DSB repair by this mechanism is conservative.
Alternative models for DSB repair have been proposed, including a migrating D-loop model in which only one of the broken strands invades the homologous repair template and where DSB repair is coupled to recombination-dependent replication (9, 14, 18, 25). Studies of extrachromosomal plasmid recombination in mammalian cells have led to the proposal of a nonconservative model of homologous recombination, termed single-strand annealing (16). In the single-strand annealing model, both recombination substrates must be cleaved at or near the region of homology. This is a nonconservative mechanism, since sequence information is lost.
Homologous recombination can be suppressed by sequence divergence between recombining substrates. Substrates with amounts of sequence divergence limited to a few percent or less recombine at reduced levels compared to fully homologous sequences, whereas sequence divergence of 10 to 20% can nearly abolish recombination in some systems (29, 31, 42). Both the overall amount of heterology and the longest stretch of uninterrupted homology contribute to the suppression of recombination, with the latter factor likely having a more pivotal role (43). This suppression has the outcome of preventing recombination between DNA of diverged species, as well as of limiting recombination between highly diverged repetitive elements in genomes. It has been shown in several studies that some of the barrier to recombination between homologous, but not identical, DNAs can be partially overcome in mismatch repair mutants (2, 5, 7, 22, 24), implicating this system as a guardian of the genome against unwanted exchanges.
Since DSBs are such potent inducers of recombination, we have designed a system to examine the effect of heterology on DSB-induced recombination in mammalian cells. In this system, a cleavage site for the rare-cutting endonuclease I-SceI is integrated into the genome of cells (27). As expression of this endonuclease in vivo is not toxic (27a), an expression vector for the endonuclease is introduced along with homologous substrates from which to repair the chromosomal DSB. The repair substrates contain various amounts of heterology, from 0.1 to 1.2%. We have examined the effects of sequence divergence on both the frequency of recombination and the gene conversion tracts that result from DSB-induced recombination.
MATERIALS AND METHODS
Plasmid constructs and DNA manipulations.
The multiply marked pneo substrates pneo-5mu, pneo-7mu, and pneo-8mu were derived from pneo12 and intermediates produced during construction of pneo12 (39). To construct the pneo-mu plasmids, the 745-bp EagI/XbaI neo gene fragment from pneo12 or an intermediate plasmid was subcloned into the EagI/XbaI sites of BluescriptII KS+. For the homologous wild-type neo fragment in the control plasmid pneo-WT, the EagI/RsrII fragment from pMClneopA2 (41) replaced the corresponding EagI/RsrII fragment in pneo-5mu. The mouse embryonic stem (ES) cell line clone 12 contains a randomly integrated single copy of the S2neo gene (33). Integrated with the S2neo gene in the genome of this ES cell line are vector sequences which are homologous to the vector sequences in the pneo substrates. This homology extends for at least 700 bp downstream from the neo gene and is separated from the neo gene homology by approximately 100 bp of unrelated sequence, which is composed of 3′ neo gene sequences for S2neo and vector sequences for pneo.
Cell culture and transfections.
ES cells were cultured on gelatin-coated dishes, as previously described (26), in the presence of 103 U of leukemia inhibitory factor (ESGRO; Gibco Life Science) per ml. For each sample of cells for transfection, 2 × 107 cells in 1 ml of phosphate-buffered saline were electroporated with 25 μg of each uncut plasmid DNA in a 0.4-cm-electrode-gap cuvette (250 V, 960 μF). Electroporated cells were aliquoted into five 10-cm-diameter dishes. Colonies were selected in 200 μg of G418 (Geneticin; Gibco Life Science) per ml beginning 20 to 24 h after electroporation and were grown in selective medium for 10 to 14 days before colony counts or colony expansion.
PCR analysis.
A region of the chromosomal neo gene in the ES clone 12 cells and in neo+ colonies was PCR amplified with primers Neo1 and Neo2 (27, 33). Amplified products were digested with I-SceI and appropriate restriction enzymes and electrophoresed on 0.8 to 1.3% agarose gels (25% agarose/75% Nusieve agarose).
RESULTS
Gene targeting with diverged repair substrates.
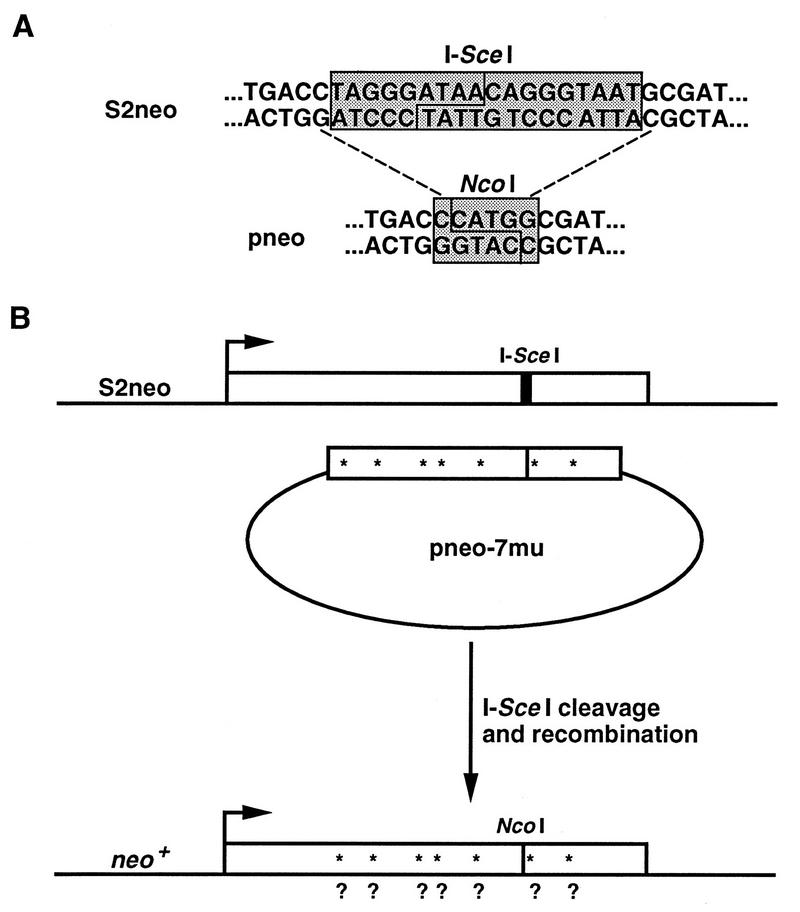
To analyze DSB-induced recombination between diverged sequences, we developed a gene-targeting assay in which the target chromosomal locus contains an I-SceI endonuclease cleavage site that can be repaired by diverged sequences in circular plasmids following an I-SceI-induced DSB at the locus. The chromosomal locus contains a neo gene (termed S2neo) which has been mutated by insertion of the 18-bp I-SceI cleavage site (Fig. 1A). The I-SceI site was inserted into an NcoI restriction site, with the mutation resulting in a 4-bp deletion of neo gene sequence, as well as the insertion of the 18-bp cleavage site. Restoration of a functional neo gene has been found to be dependent upon recombination with a correcting neo gene fragment (15, 33). The I-SceI endonuclease is transiently expressed in cells from a phosphoglycerate kinase 1 promoter in the expression vector pPGK3xnlsI-SceI (8).
FIG. 1.
DSB-induced gene targeting with diverged repair substrates. (A) Sequences of the chromosomal S2neo gene and of the correcting pneo plasmids at the position of the I-SceI cleavage site. The I-SceI endonuclease recognizes an 18-bp nonpalindromic site and cleaves to produce four-base 3′ overhangs, as indicated. The first three bases of the I-SceI site create a stop codon (TAG) in the neo coding sequence, making the S2neo gene a nonfunctional neo gene. The pneo plasmids have a wild-type neo gene sequence at this position, which is located at an NcoI restriction site. Note that the S2neo gene has a deletion of the four-base overhangs of the NcoI site which creates a nonrevertable deletion mutation. (B) DSB-induced gene targeting occurs when the chromosomal S2neo gene integrated in the genome of ES cells is cleaved in vivo at the I-SceI site (thick vertical bar) and is repaired from a transfected pneo plasmid. As an example, recombination is shown with plasmid pneo-7mu, which contains the correcting neo gene sequence at the NcoI site (thin vertical bar). The pneo-7mu repair substrate also contains seven silent mutations which create new restriction sites (asterisks) in the neo gene. Recombination yields a functional neo+ gene which can be analyzed for the conversion of the silent mutations (?).
The circular repair plasmids that were introduced contain internal neo gene fragments to restrict repair to a defined recombination pathway. With this design, selected recombination events are limited to conservative gene conversion events that do not involve crossing-over (Fig. 1B). Recombination with an associated crossover would result in neo gene disruption by plasmid sequences. We were particularly interested in gene conversion unassociated with crossing-over since this pathway has been predicted to be a major recombinational repair pathway for chromosomal DSBs in mitotically growing cells (20).
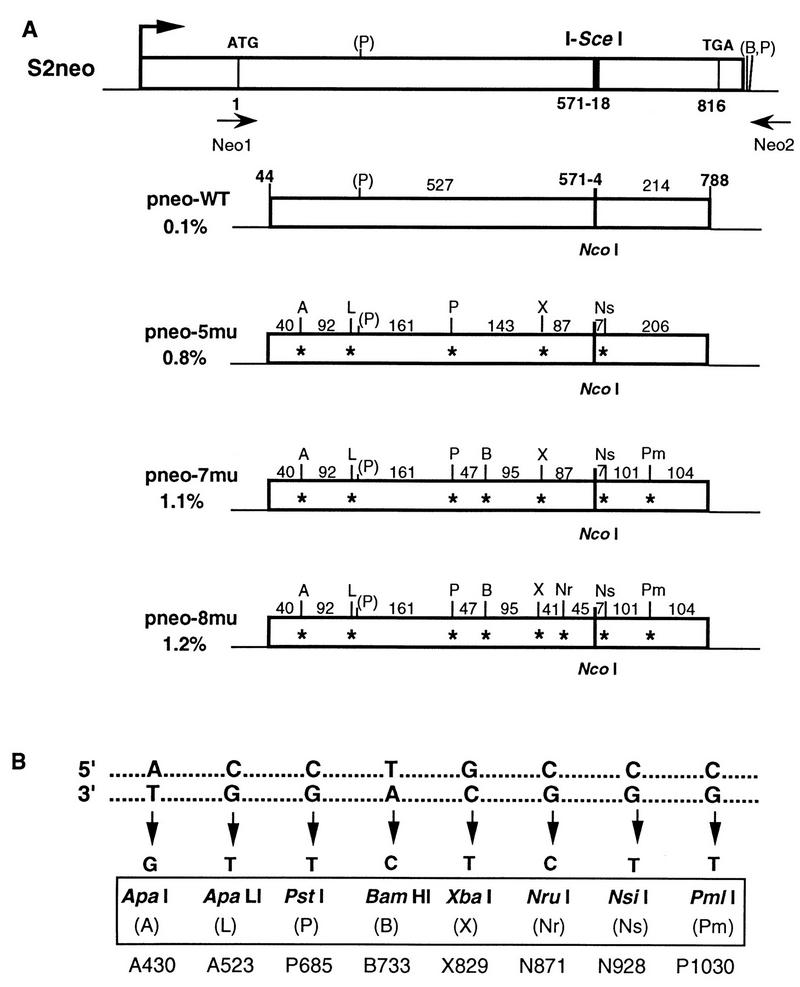
Each of the repair substrates, the pneo plasmids, contains a 745-bp internal neo gene fragment that is homologous with the chromosomal S2neo gene (Fig. 2). The diverged sequences in the repair substrates are base substitutions in the neo gene that are phenotypically silent mutations at third base codon positions, with each mutation creating a new restriction site (Fig. 2). In addition to the correcting sequence at the NcoI site, five to eight silent mutations were incorporated in the 745-bp neo gene fragment. As larger numbers of mutations are incorporated in the repair substrates, the stretches of perfect homology in the neo fragment decrease and the overall percent divergence of the fragment increases. Thus, in pneo-5mu, which contains five silent mutations, the longest stretch of perfect homology is 206 bp and there is an overall divergence of 0.8%. neo gene fragments in pneo-7mu and pneo-8mu contain seven and eight silent mutations, respectively, with the overall divergence of pneo-7mu being 1.1% and that of pneo-8mu being 1.2%. The longest stretch of perfect homology is 161 bp for both pneo-7mu and pneo-8mu, although in pneo-8mu another stretch of perfect homology adjacent to the cleavage site is shortened from 87 to 45 bp by the additional point mutation. Plasmid pneo-WT serves as a control in these experiments, differing from the S2neo gene only by having a wild-type sequence at the NcoI site.
FIG. 2.
Silent mutations in the diverged repair substrates. (A) Positions of the silent mutations and corresponding new restriction sites in the donor pneo substrates. The length of homology between the neo fragment in the pneo plasmid and the chromosomal S2neo gene is 745 bp. The translation start, I-SceI cleavage site, and termination codon in the S2neo gene are also indicated. The percentages of divergence of the internal neo gene fragments in pneo-WT, pneo-5mu, pneo-7mu, and pneo-8mu relative to the same region in S2neo are shown. For simplicity, the heterology at the NcoI/I-SceI sites is considered to be one change within the 745-bp segment, although recombination at this position results in loss of the 18-bp cleavage site and gain of 4 bp at the NcoI site. The distances in base pairs between the 1-bp silent mutations (asterisks) are indicated. Neo1 and Neo2 are the primers that were used for PCR amplification of the chromosomal neo gene. Abbreviations for the new restriction sites that are generated by the silent mutations are shown in panel B. Note that there is a naturally occurring PstI site (P) within the neo gene fragment as well as BamHI and PstI sites (B, P) downstream of the neo gene. The promoter (arrow) for the S2neo gene is derived from polyomavirus (strain F441) and the herpes virus thymidine kinase gene (41). (B) Silent mutations in the diverged pneo substrates which create new restriction sites. The top duplex indicates the sequence of the wild-type neo gene at the positions where silent mutations were introduced. The sequence of the mutations is given for the bottom strand. Restriction sites created as a result of the introduced mutations are indicated, with abbreviations used for the sites shown in parentheses. The designations given by Taghian and Nickoloff (39) for the mutations are shown at the bottom.
In addition to the neo gene homology, the pneo plasmids also have homology with the chromosome downstream of the S2neo gene. This homology is at least 700 bp of bacterial vector sequences. Because there is approximately 100 bp of unrelated sequence separating the two homologous segments (composed of 3′ neo gene sequences for S2neo and vector sequences for pneo), it is not clear if the downstream homology affects DSB repair.
Effect of heterology on the frequency of DSB-induced chromosomal recombination.
To determine the effect of heterology on the frequency of DSB-induced gene targeting, we used a mouse ES cell line which has a single copy of S2neo integrated into the genome (33). This ES cell line, termed clone 12, was cotransfected with each of the pneo substrates and the I-SceI expression vector. Cells were selected in G418 starting 1 day after transfection. G418r (neo+) recombinant colonies were counted 10 to 14 days later.
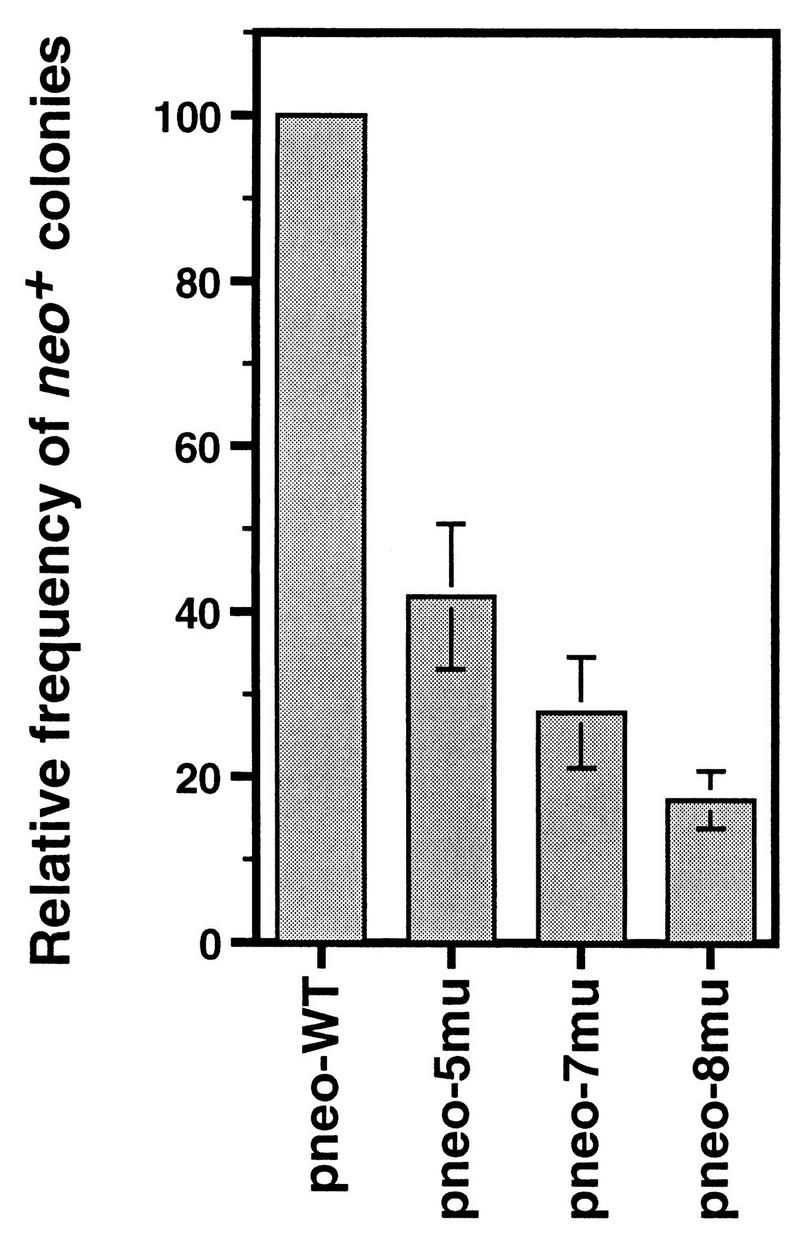
In the cotransfection of pneo-WT and the I-SceI expression vector, a frequency of up to 4.7 × 10−4 neo+ colonies was obtained from the transfected cell population (Table 1). In each of five independent experiments with the diverged repair substrates and the I-SceI expression vector, the frequency of neo+ colonies relative to the pneo-WT control was found to decrease as the amount of heterology of the pneo substrate increased (Table 1; Fig. 3). With 0.8% divergence in pneo-5mu and 1.1% divergence in pneo-7mu, mean 2.5-fold and 4-fold decreases in recombination were observed, respectively. The largest effect, seen with 1.2% divergence in pneo-8mu, was a sixfold decrease.
TABLE 1.
Gene targeting in ES cells with diverged repair substrates
| Transfected plasmid(s) | Frequency of neo+ coloniesa
|
||||
|---|---|---|---|---|---|
| Expt 1 (% WT)b | Expt 2 (% WT) | Expt 3 (% WT) | Expt 4 (% WT) | Expt 5 (% WT) | |
| pneo-WT + pPGK3xnlsI-SceI | 1.40 × 10−4 (100) | 1.44 × 10−4 (100) | 1.15 × 10−4 (100) | 4.69 × 10−4 (100) | 4.63 × 10−4 (100) |
| pneo-5mu + pPGK3xnlsI-SceI | 0.39 × 10−4 (28) | 0.71 × 10−4 (49) | 0.61 × 10−4 (53) | 1.77 × 10−4 (38) | 1.90 × 10−4 (41) |
| pneo-7mu + pPGK3xnlsI-SceI | 0.28 × 10−4 (20) | 0.58 × 10−4 (40) | 0.33 × 10−4 (29) | 1.14 × 10−4 (24) | 1.22 × 10−4 (26) |
| pneo-8mu + pPGK3xnlsI-SceI | 0.18 × 10−4 (13) | 0.27 × 10−4 (19) | 0.27 × 10−4 (23) | 0.72 × 10−4 (15) | 0.73 × 10−4 (16) |
| pneo-WT + pPGKlacZ | <1 × 10−7 | NDc | ND | <1 × 10−7 | ND |
| pPGK3xnlsI-SceI | <1 × 10−7 | ND | ND | <1 × 10−7 | ND |
| pMC1neod | 5.4 × 10−4 | ND | 4.8 × 10−4 | 7.8 × 10−4 | 1.4 × 10−3 |
The frequency of neo+ colonies was determined by dividing the number of neo+ colonies obtained by the number of electroporated cells (2 × 107), correcting for 50% viability of cells generally observed following electroporation under these conditions.
% WT, percent frequency of neo+ colonies derived from transfection of the diverged pneo repair substrate with pPGK3xnlsI-SceI divided by the frequency of neo+ colonies derived from transfection of pneo-WT with pPGK3xnlsI-SceI.
ND, not determined.
Total number of colonies from pMC1neo transfection were estimated by plating dilutions of electroporated cells (1:6 to 1:60).
FIG. 3.
Relative gene-targeting efficiencies of diverged repair substrates. The frequency of neo+ colonies was plotted for the cotransfections of each of the pneo substrates with pPGK3xnlsI-SceI relative to the pneo-WT with pPGK3xnlsI-SceI control. In each case, the mean of five independent experiments is shown with the standard deviation indicated by an error bar.
In these experiments, no neo+ colonies were obtained from cotransfections of the pneo plasmids with a control pPGKlacZ vector (Table 1 and data not shown). Similarly, we did not detect neo+ clones from cells which were transfected with the pneo substrates alone (data not shown). These results show that gene targeting of the pneo plasmid is inefficient in the absence of a chromosomal DSB (<10−7 of the electroporated cells) but is stimulated more than 3 orders of magnitude by a DSB in the target locus. No neo+ colonies resulted from transfection of cells with the I-SceI expression vector alone, indicating that restoration of a neo+ gene is dependent upon a correcting neo gene fragment. Taken together these data clearly demonstrate that all of the recombinants detected from cotransfection of the pneo plasmids and the I-SceI expression vector were derived from DSB-induced recombination. In addition to the ES clone 12 cell line, we have performed similar experiments in another ES cell line which has a single S2neo gene integrated at a different position in the genome. Similar results have been obtained with this clone (data not shown), demonstrating that our results are independent of chromosomal context of the DSB site.
Gene conversion tracts from the pneo-8mu repair substrate.
For insight into the mechanism of gene conversion, we examined the incorporation of the silent mutations into the chromosome of the recombinants. Genomic DNA was isolated from neo+ colonies, and PCR amplification was performed with neo gene primers that would hybridize to the chromosomal neo gene but not to the pneo plasmid DNA (Fig. 2A) (27, 33). This strategy would prevent amplification of plasmid DNA that had integrated randomly in the genome. Endonuclease digests were performed on the amplified products to verify that the chromosomal S2neo gene had undergone DSB-promoted recombination and to determine the extent of gene conversion beyond the position of the I-SceI cleavage site.
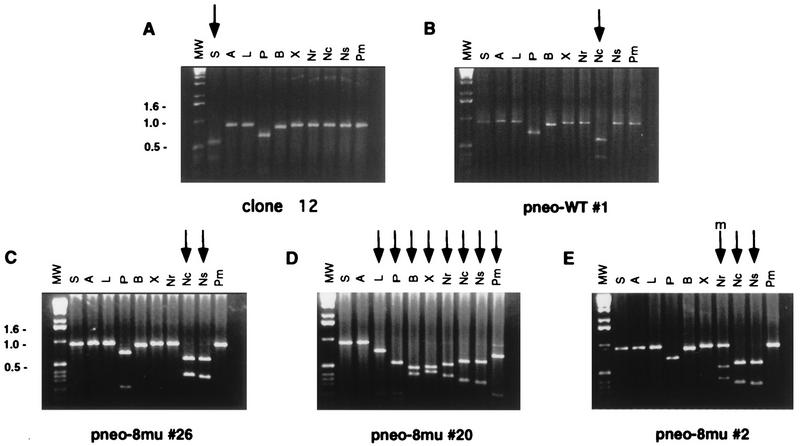
To restore neo gene function, each neo+ colony was expected to have lost the I-SceI site and incorporated an NcoI site at that position. We analyzed 40 neo+ colonies derived from cotransfection of the pneo-8mu plasmid with the I-SceI expression vector. As expected, in each of the neo+ colonies the I-SceI site was lost from the chromosomal S2neo locus and replaced with the NcoI site (Fig. 4 and 5A), verifying that the neo+ colonies resulted from gene targeting. The incorporation of the 4 bp at the NcoI site occurred with the loss of the four-base I-SceI overhangs, as well as the loss of 5 bp upstream and 9 bp downstream of the I-SceI site (Fig. 1A).
FIG. 4.
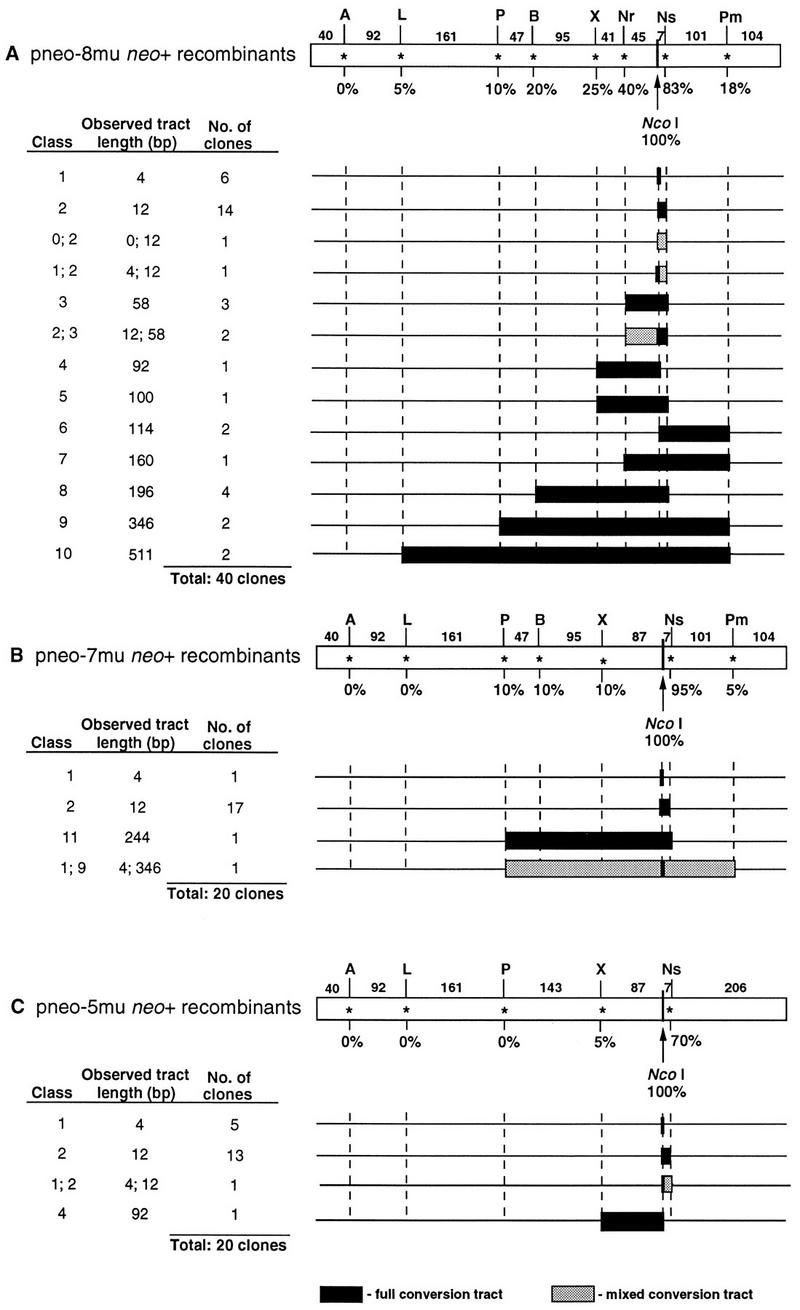
PCR analysis of parental ES cells and neo+ clones derived from cotransfection of pneo substrates with the I-SceI expression vector. The neo gene coding region was amplified by PCR as a 0.9-kb fragment from genomic DNA with primers Neo1 and Neo2 and electrophoresed on agarose gels following digestion by the indicated endonucleases. Endonucleases are abbreviated as follows: I-SceI, S; ApaI, A; ApaLI, L; PstI, P; BamHI, B; XbaI, X; NruI, Nr; NcoI, Nc; NsiI, Ns; and PmlI, Pm. The 1-kb ladder molecular weight marker used is also indicated (MW). Note that due to naturally occurring PstI and BamHI sites (Fig. 2), the amplified fragments in each of the clones are shifted when cut with these enzymes. (A) Amplified fragment from the S2neo gene in the parental ES clone 12 cells. The fragment is cleaved by I-SceI, as expected, but it is not cleaved by NcoI or the restriction enzymes whose sites were created in the pneo plasmids. (B) Amplified neo fragment from a neo+ colony from cotransfection of pneo-WT and the I-SceI expression vector. The fragment is cleaved by NcoI but not by I-SceI or the other restriction enzymes. (C to E) Amplified neo gene fragments from selected neo+ colonies from cotransfection of the pneo-8mu substrate and the I-SceI expression vector, showing a short gene conversion tract (C), a long tract (D), and a mixed tract (E). The amplified fragment from each of the neo+ clones is not cleaved by I-SceI but is cleaved by NcoI and various restriction enzymes, as indicated by the arrows. In clone pneo-8mu #20 (D), the small amount of DNA not cleaved by PmlI is not reproducible. The “m” over the arrow in panel E indicates partial cleavage by the restriction enzyme.
FIG. 5.
Gene conversion tracts from neo+ recombinants. Observed gene conversion tracts from neo+ clones derived from cotransfection of ES clone 12 cells with the I-SceI expression vector and repair substrates pneo-8mu (A), pneo-7mu (B), and pneo-5mu (C). The recombinants were derived from two experiments. The full-length homology region in the respective neo fragment is shown (open rectangles). The positions of the silent mutations for each pneo plasmid are shown above the homology regions with the distance between the mutations indicated in base pairs. When gene conversion tracts are calculated, an additional base pair is added for the conversion of the silent mutation. Below each silent mutation is the percent conversion of the mutation in the gene conversion tracts. The correcting NcoI site, shown below the tract, occurs in 100% of neo+ clones, since it restores a functional neo+ gene. Gene conversion tracts extending to the last incorporated mutation and mixed conversion tracts (along with the two classes constituting them) are shown for each of the recombinants. The 0 class indicates no conversion with retention of the I-SceI site.
If silent mutations from the donor pneo plasmids had been coconverted with the NcoI site, the amplified products were expected to be digested by restriction enzymes recognizing the corresponding new restriction sites. The incorporation of the silent mutations in the 40 recombinants examined was found to inversely correlate with the distance from the NcoI site (Fig. 5A). The mutation nearest to the NcoI site, located 8 bp downstream at the NsiI site, was found to be incorporated at a high frequency, i.e., in 83% of the recombinants. The next closest mutation, at the NruI site (46 bp upstream), was found to be incorporated in 40% of the recombinants, while the mutation at the XbaI site (88 bp upstream) was found in 25% of the recombinants. Each of the remaining mutations was incorporated into 20% or fewer of the recombinants, with the mutation at the ApaLI site (394 bp upstream) being incorporated into just two of the clones. The farthest mutation, at the ApaI site (487 bp upstream), was not found in any of the 40 recombinants examined. When checked, Southern blot analysis of neo+ recombinant clones gave the same results to those obtained by PCR analysis for the incorporation of the silent mutations (data not shown).
All of the gene conversion tracts were found to be continuous from the cleavage site, such that each of the mutations between the outermost converted restriction site and the NcoI site was converted. The longer gene conversion tracts (≥58 bp) were mostly bidirectional, extending both 5′ and 3′ from the cleavage site. The ability of one side to recombine did not appear to interfere with the ability of the other side to recombine. For example, the clones which have the longest gene conversion tracts (i.e., the class 9 and 10 recombinants) are bidirectional. A few of the gene conversion tracts were unidirectional, for example, the gene conversion tract in the class 4 recombinant extended 88 bp 5′ to the NcoI site while the NsiI mutation, which is only 8 bp 3′ to the NcoI site, was not converted. Similarly, in class 6 recombinants, the gene conversion tract extended 110 bp 3′ to the NcoI site, while the NruI mutation 46 bp 5′ to the NcoI site was not converted.
In most of these recombinants, there was no ambiguity as to the incorporation of the silent mutations. The amplified product appeared either completely cleaved or completely uncleaved when analyzed by agarose gel electrophoresis (Fig. 4). However, for four recombinants, the amplified products were found to be approximately half cleaved by one or more restriction enzymes (Fig. 4E and data not shown). The partial cleavage was reproducible from independent PCR amplifications of genomic DNA and may reflect either segregation of unrepaired mismatches in heteroduplex DNA after recombination or independent repair events in daughter cells of a cell that had been transfected with the I-SceI expression vector. For one mixed recombinant clone (i.e., a mixed class 1 and 2 recombinant) the amplified product was only partially cleaved by NsiI. For two other mixed recombinant clones (mixed class 2 and 3 recombinants) the amplified product was only partially cleaved by NruI, although for both of these clones the product was completely cleaved by NsiI. The amplified product of another mixed recombinant clone (a mixed class 0 and 2 recombinant) was partially cleaved by NsiI; however, it was also partially cleaved by I-SceI and NcoI, suggesting that the clone may contain a duplication of the neo gene in the chromosome with one of the genes being converted.
Gene conversion tracts from other pneo repair substrates.
neo+ colonies were examined that were derived from cotransfection of the I-SceI expression vector with the other pneo plasmids. Colonies derived from cotransfection of the I-SceI expression vector and pneo-WT were examined as a control. Except for the incorporation of the NcoI site and loss of the I-SceI cleavage site, no other restriction site change to the chromosomal locus was observed (Fig. 4B and data not shown).
For each of the neo+ colonies derived from cotransfection of the I-SceI expression vector and either pneo-7mu (20 recombinants) or pneo-5mu (20 recombinants), the I-SceI site in the chromosomal neo locus was converted to an NcoI site (Fig. 5B and C). As observed with the recombinants derived from transfection of the pneo-8mu plasmid, the frequency of incorporation of the silent mutations on either side of the NcoI site for these clones was dependent on the distance of the mutation from the I-SceI cleavage site. The mutation at the NsiI site 8 bp from the NcoI site was incorporated into 83% of the recombinants (19 of 20 from pneo-7mu and 14 of 20 from pneo-5mu), whereas the most distant mutations were incorporated in 10% or less of the recombinants. All gene conversion tracts derived from these plasmids were also continuous.
As observed with transfections with pneo-8mu, a few recombinants, one from pneo-5mu and one from pneo-7mu, had amplified products that were only partially cleaved by some restriction enzymes. The amplified product from the mixed clone generated by pneo-5mu transfection was only partially cleaved by NsiI (Fig. 5C), and the amplified product from the mixed clone generated by pneo-7mu was partially cleaved by five enzymes, PstI, BamHI, XbaI, NsiI, and PmlI (Fig. 5B). Cleavage by NcoI in both cases was complete. As with the mixed clones described above, these mixed clones likely arose either from heteroduplex formation with inefficient mismatch repair or from a cell that had been transfected with the I-SceI expression vector and whose daughter cells had independent repair events.
DISCUSSION
We have examined the effect of heterology on recombinational repair of chromosomal DSBs in mammalian cells. We find that sequence divergence in the range of 0.8 to 1.2% in the 745-bp homologous fragment in our gene-targeting plasmid decreased the frequency of DSB-induced recombinational repair, with a divergence of 1.2% resulting in an approximately sixfold decrease in gene targeting. A substantial majority (80%) of the 80 recombinants examined had observed gene conversion tracts of 58 bp or less. All of the gene conversion tracts were continuous, incorporating all of the mutations between the cleavage site and the outermost converted site. Gene conversion tracts 58 bp or longer were mostly bidirectional.
A summary of the gene conversions in the recombinant clones from the three heterologous substrate plasmids is shown in Fig. 6. We found that the majority of recombinants from each plasmid transfection had gene conversion tracts of at least 12 bp, incorporating both the NcoI and NsiI sites. In the 17% of recombinants (14 of 80) that did not incorporate the mutation at the NsiI site, the endpoint of the conversion tract was located within the 7 bp between these two sites. In these recombinants there was either no or very little degradation of the chromosome ends prior to or during recombination. The next mutation from the cleavage site, located 46 bp upstream at the NruI site in the pneo-8mu substrate, was converted in 40% recombinants examined. Clones which have converted no more than the three mutations at the NcoI, NsiI, and NruI sites (i.e., the class 1, 2, and 3 clones) (Fig. 5A) comprise 68% of the recombinants repaired from pneo-8mu, giving observed gene conversion tracts of 58 bp or less. The lack of conversion at the next mutations (at the XbaI and PmlI sites) thus demarcates a maximum gene conversion tract of 200 bp (i.e., for the class 3 clones). The longest observed gene conversion tract was 511 bp and was seen in just 2 of the 80 clones. This gene conversion tract contained seven of the eight silent mutations, with the farthest mutation, located 93 bp upstream (at an ApaI site), not being converted. Although the distance likely affects the incorporation of this mutation, its location only 40 bp from the homology border also may have had an impact. Reduced conversion for markers near homology borders has been seen for spontaneous and DSB-induced recombination in yeast (1, 37).
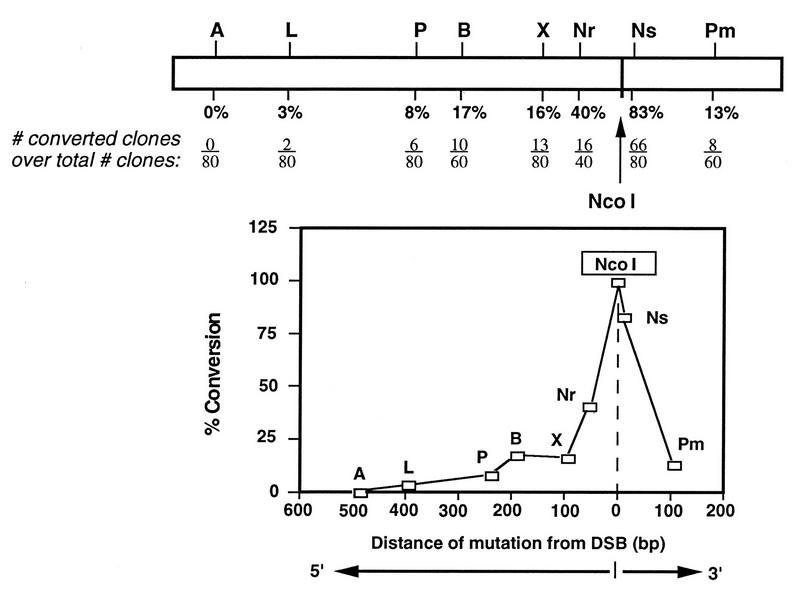
FIG. 6.
Summary of gene conversion frequencies for each silent mutation. Below each silent mutation is the percent conversion from the pneo-5mu, pneo-7mu, and pneo-8mu substrates. The number of clones converted at each mutation is divided by the total number of clones analyzed for each mutation. For the NcoI site, there is a conversion tract of at least 4 bp. Using these data, the conversion frequency for each mutation was plotted as a function of distance from the DSB.
Plotting the percent conversion at each of the silent mutations against the distance of the mutation from the cleavage site gives the distribution shown in Fig. 6. There is a steep decline in the amount of conversion close to the cleavage site. At greater distances from the cleavage site (around 100 bp), the degree of incorporation of the mutations has a much shallower decline. There does not appear to be a marked directional preference or end effect, as mutations located at approximately similar distances on opposite sides of the cleavage site (i.e., at the XbaI and PmlI sites) are incorporated at similar frequencies. In a recent study, Taghian and Nickoloff report that conversion by chromosomal DSBs between direct repeats gave almost identical bimodal and symmetric distribution (39).
Two possible mechanisms can be invoked to account for the gene conversion tracts from the DSB site. One is mismatch correction following heteroduplex formation between one strand of the neo gene on the cleaved chromosome and the complementary strand of the neo fragment on the donor plasmid. In S. cerevisiae it is likely that the 3′ strand of the cleaved chromosome would form heteroduplex DNA with an uncleaved partner, since DSBs in yeast are processed to form 3′ single-stranded tails (4, 35, 36). The occurrence of a few clones with partially converted mutations is consistent with the idea that heteroduplex DNA is formed in at least some instances. (It cannot be excluded, however, that these clones arose from two independent repair events that occurred after DNA replication in cells expressing I-SceI.) An alternative mechanism involves conversion in a double-stranded gap, in which both strands of the DNA duplex on each side of the break are removed by nucleases. In this case, information available for repair must derive entirely from the neo fragment on the donor pneo plasmid. Formation of a double-strand gap at the I-SceI break site would be consistent with the continuous nature of the gene conversion tracts we observed. With gap formation, longer tracts would reflect extensive degradation and shorter tracts would reflect limited amounts of degradation.
The observation that heterology affects recombination frequencies, however, argues for a significant contribution of heteroduplex formation to gene conversion events. Since we find that at least 17% of the recombinants have maintained the parental sequence to within 7 bp of the I-SceI recognition site, at least a portion of the chromosome ends are protected from any more than a few base pairs of degradation. If all the chromosome ends are protected from degradation, formation of heteroduplex DNA between the two substrates would also give rise to continuous tracts if it were coupled with a strong bias to correct mismatches in the direction of the donor substrate. Such a bias would be signaled by the chromosomal break (19, 22, 23). The bias would have to overcome the usual directionality in the repair of mismatched bases. For example, the G/T mismatch at the XbaI site would be expected to be rarely converted to A/T (≤10% of repairs [3]), the mutation in the donor plasmid, yet it is converted in 16% of the recombinants. Since most or all DSB-induced conversion in yeast reflects mismatch repair of heteroduplex DNA (20, 23) and many of the features of yeast conversion tracts are similar to those we observe here (see below and reference 37), mismatch repair of heteroduplex DNA may predominate over double-strand gap formation in mammalian cells as well.
We cannot rule out the possibility that conversion occurs by both mechanisms, i.e., heteroduplex correction and double-strand gap formation, with shorter tracts arising from heteroduplex correction of preserved chromosome ends and longer tracts from chromosome breaks which have eluded end protection mechanisms. The bimodal nature of the conversion tracts would be consistent with two distinct mechanisms, although it does not exclude the possibility that only one mechanism (i.e., heteroduplex correction, see above) is occurring. Examination of gene conversion in mismatch repair mutants will be necessary to address this question.
The 2.5- to 6-fold decrease in the frequency of recombination with the diverged substrates demonstrates the effect of sequence divergence on the efficiency of DSB-induced recombination. In previously described gene-targeting experiments, 0.6% sequence divergence was found to reduce spontaneous recombination 20-fold (40), a much greater reduction than seen here. In studies examining spontaneous intrachromosomal recombination, 1% sequence divergence was found to have a much less dramatic effect, at least in some cases (43). In these experiments, the minimal length of uninterrupted homology was determined to be a more important factor governing the frequency of recombination than the absolute amount of sequence divergence (43). The minimal length of uninterrupted homology required for efficient recombination was estimated to be between 232 and 134 bp, since a decrease from 232 bp of uninterrupted homology to 134 bp resulted in a 20-fold reduction in recombination. In our experiments, the length of uninterrupted homology was also within this range, e.g., 206 bp (pneo-5mu) and 161 bp (pneo-7mu and pneo-8mu). By contrast, we see only a two- to threefold reduction in recombination between pneo-5mu and pneo-8mu, indicating that DSBs may partially overcome the barrier to homology length requirements in recombination between diverged sequences.
In each of our experiments, we found a decrease in the frequency of recombination as the divergence of the repair substrates increased (Fig. 3). This trend suggests that additional mutations both close to and far away from the cleavage site can incrementally impact on recombination frequencies. The one additional mutation in plasmid pneo-8mu is 46 bp from the DSB and interrupts the 87-bp tract of perfect homology upstream of the I-SceI cleavage site, implying that the length of perfect homology adjacent to a DSB may be an important factor governing the efficiency of DSB-promoted recombination.
Interestingly, the DSB-induced gene conversion tracts we obtained in ES cells are similar to those found in DSB-induced recombination in S. cerevisiae. In yeast, gene conversion tracts are short (200 to 300 bp) and the majority of tracts are continuous, although one primary difference is that gene conversion tracts in yeast are primarily unidirectional (37). The similarity to DSB-induced recombination in yeast may be thought surprising, since nonhomologous repair contributes significantly to DSB repair in mammalian cells (10, 15, 17, 21, 27) yet is rare in yeast. However, the recent identification of mammalian homologs to genes involved in recombination in yeast (32), along with the work presented here, suggests that recombination pathways between the organisms may be highly conserved.
ACKNOWLEDGMENTS
We thank Roger Johnson and Andy Pierce for comments on the manuscript.
This work was supported by grants from the National Science Foundation (MCB-9419507) and the American Cancer Society (NP-82674) to M.J. and a grant from the National Cancer Institute to J.A.N. (CA54079). C.R. is a Leukemia Society of America Postdoctoral Fellow.
REFERENCES
- 1.Ahn B-Y, Dornfeld K J, Fagrelius T J, Livingston D M. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol Cell Biol. 1988;8:2442–2448. doi: 10.1128/mcb.8.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailis A M, Arthur L, Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics. 1990;126:535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown T C, Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988;54:705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- 4.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 5.Chambers S R, Hunter N, Louis E J, Borts R H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choulika A, Perrin A, Dujon B, Nicolas J-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 8.Donoho, G., M. Jasin, and P. Berg. Unpublished results.
- 9.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godwin A R, Bollag R J, Christie D-M, Liskay R M. Spontaneous and restriction enzyme-induced chromosomal recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:12554–12558. doi: 10.1073/pnas.91.26.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haber J E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. BioEssays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 12.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 13.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 14.Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 15.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Chromosomal double-strand break repair in Ku80 deficient cells. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin F-L, Sperle K, Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990;10:103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukacsovich T, Yang D, Waldman A S. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI. Nucleic Acids Res. 1994;22:5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 19a.Moynahan M E, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 21.Phillips J W, Morgan W F. Illegitimate recombination induced by DNA double-strand breaks in a mammalian chromosome. Mol Cell Biol. 1994;14:5794–5803. doi: 10.1128/mcb.14.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priebe S D, Westmoreland J, Nilsson-Tillgren T, Resnick M A. Induction of recombination between homologous diverged DNAs by double-strand gaps and breaks and role of mismatch repair. Mol Cell Biol. 1994;14:48024814. doi: 10.1128/mcb.14.7.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray B L, White C I, Haber J E. Heteroduplex formation and mismatch repair of the “stuck” mutation during mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5372–5380. doi: 10.1128/mcb.11.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 25.Resnick M. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976;59:97–109. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 26.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Washington, D.C: IRL Press; 1987. pp. 71–112. [Google Scholar]
- 27.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Rouet P, Smih F, Jasin M. Expression of site-specific endonuclease stimulates recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sargent R G, Brenneman M A, Wilson J H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider W P, Nichols B P, Yanofsky C. Procedure for production of hybrid genes and proteins and its use in assessing significance of amino acid differences in homologous tryptophan synthetase α polypeptides. Proc Natl Acad Sci USA. 1981;78:2169–2173. doi: 10.1073/pnas.78.4.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 31.Shen P, Huang H V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinohara A, Ogawa T. Homologous recombination and the role of double-strand breaks. Trends Biochem. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 33.Smih F, Rouet P, Romanienko P J, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl F. Meiotic recombination in yeast: coronation of the double-strand-break repair model. Cell. 1996;87:965–968. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- 35.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 37.Sweetser D B, Hough H, Whelden J F, Arbuckle M, Nickoloff J A. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol Cell Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 39.Taghian, D. G., and J. A. Nickoloff. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 17:6386–6393. [DOI] [PMC free article] [PubMed]
- 40.te Riele H, Maandag E R, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 42.Waldman A S, Liskay R M. Differential effects of base-pair mismatch on intrachromsomal versus extrachromsomal recombination in mouse cells. Proc Natl Acad Sci USA. 1987;84:5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldman A S, Liskay R M. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]