Abstract
We report here the preparation of filamentous virus-like particles by the encapsulation of a linear or circular double-stranded DNA template with preassembled mushroom-shaped nanostructures having a positively charged domain. These nanostructures mimic the capsid proteins of natural filamentous viruses and are formed by self-assembly of coiled-coil peptides conjugated at opposite termini with cationic segments and poly(ethylene glycol) (PEG) chains. We found that a high molecular weight of PEG segments was critical for the formation of monodisperse and uniformly shaped filamentous complexes. It is proposed that electrostatic attachment of the nanostructures with sufficiently long PEG segments generates steric forces that increase the rigidity of the neutralized DNA template. This stiffening counterbalances the natural tendency of the DNA template to condense into toroids or buckle multiple times. The control achieved over both shape and dimensions of the particles offers a strategy to create one-dimensional supramolecular nanostructures of defined length containing nucleic acids.
Graphical Abstract
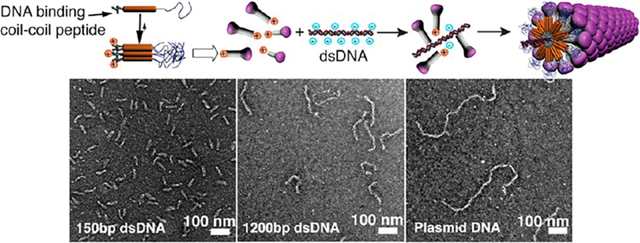
INTRODUCTION
The field of molecular self-assembly has continued to evolve in its capacity to design increasingly complex structures that are not accessible by traditional covalent organic synthesis.1 One important target that has been difficult is the one-dimensional self-assembly of building blocks with precise length. The growth of such assemblies occurs in an open mode and lacks the precision of filamentous viruses, closed systems like synthetic nanocages,2 or DNA-based nanostructures.3,4 A number of bottom-up approaches have been proposed to control the length of 1D self-assembled objects. For example, one can control supramolecular polymerization by capping,5 or the introduction of electrostatic repulsion between the monomers to frustrate the growth of the assemblies,6 which in both cases yield a broad statistical distribution of linear supramolecular polymers. A better control can be achieved using a vernier approach,7,8 yielding well-defined but low molecular weight linear supramolecular aggregates. So far, low polydispersity has been obtained only in organic solvents by the careful control of nucleation and growth kinetics of monodisperse cylindrical micelles formed by hydrophobic crystalline block copolymers.9 Therefore, preparing homogeneous 1D nanostructures by molecular self-assembly remains a great challenge, particularly in water.
One of nature’s great examples of perfectly monodisperse 1D assemblies is the tobacco mosaic virus (TMV), formed by directed self-assembly of proteins (capsomers) on a RNA template.10 Interestingly, TMV virions can be prepared in vitro just by mixing the viral RNA genome and the purified capsid proteins in aqueous solutions.11 Inspired by this process, we described previously a templating strategy involving a dumbbell-shaped template and a self-assembling peptide amphiphile.12 This approach yielded nonspherical aggregates with controlled dimensions measuring a few nanometers but was impractical to create precise 1D assemblies with the length of a filamentous virus particle. Synthetic analogs of viral capsid proteins have been produced by genetic engineering or chemical functionalization of the native protein to create viruses as templates for the development of new materials or bioactive nanostructures.13 Nucleic acids14 and negatively charged polyelectrolytes15 have been used as templates to reconstitute virus-like structures from native capsomers, but control of the dimensions was only obtained in the case of reconstituted closed icosahedral capsids. More recently, the coating of single DNA molecules with genetically engineered protein diblock copolymers was described,16 suggesting that the steric repulsion between the large colloidal stability domains (400 amino acids) on the DNA-binding proteins contributes to the stability of the complexes. To develop a system using easily accessible synthetic building blocks, we attempted first to create a monodisperse 1D assembly using a DNA template and simple DNA-binding peptides, but observed instead uncontrolled aggregation of molecules. We hypothesized that a preassembled nanostructure analogous to a viral protein, with low enough symmetry to contain a specific DNA-binding domain, might yield a virus-like monodisperse structure and avoid the uncontrolled aggregation we observed earlier. This Article describes a successful strategy to create the virus-like particles with defined length using as capsomers self-assembled noncentrosymmetric nanostructures analogous to supramolecular “mushrooms”.17,18 To create the mushroom nanostructure as the artificial capsomer, we designed molecules able to bind a nucleic acid 1D template. As depicted in Figure 1, the triblock molecules contain a spermine unit at one terminus (Sp), a known nucleic acid ligand,19 a coiled coil peptide (CC)20 known to aggregate into heptameric structures, and a long poly(ethylene glycol) chain that provides solubility in water (PEG2000 or PEG5000). We expected these molecules would form uniformly sized capsomer-like aggregates due to the synergy of a peptide predisposed to form finite heptameric aggregates20 and entropy-limited aggregation mediated by the long poly(ethylene glycol) segments.17,18 In the nanostructure designed here, a third contribution to finite aggregation would be the electrostatic repulsion among spermine segments.
Figure 1.
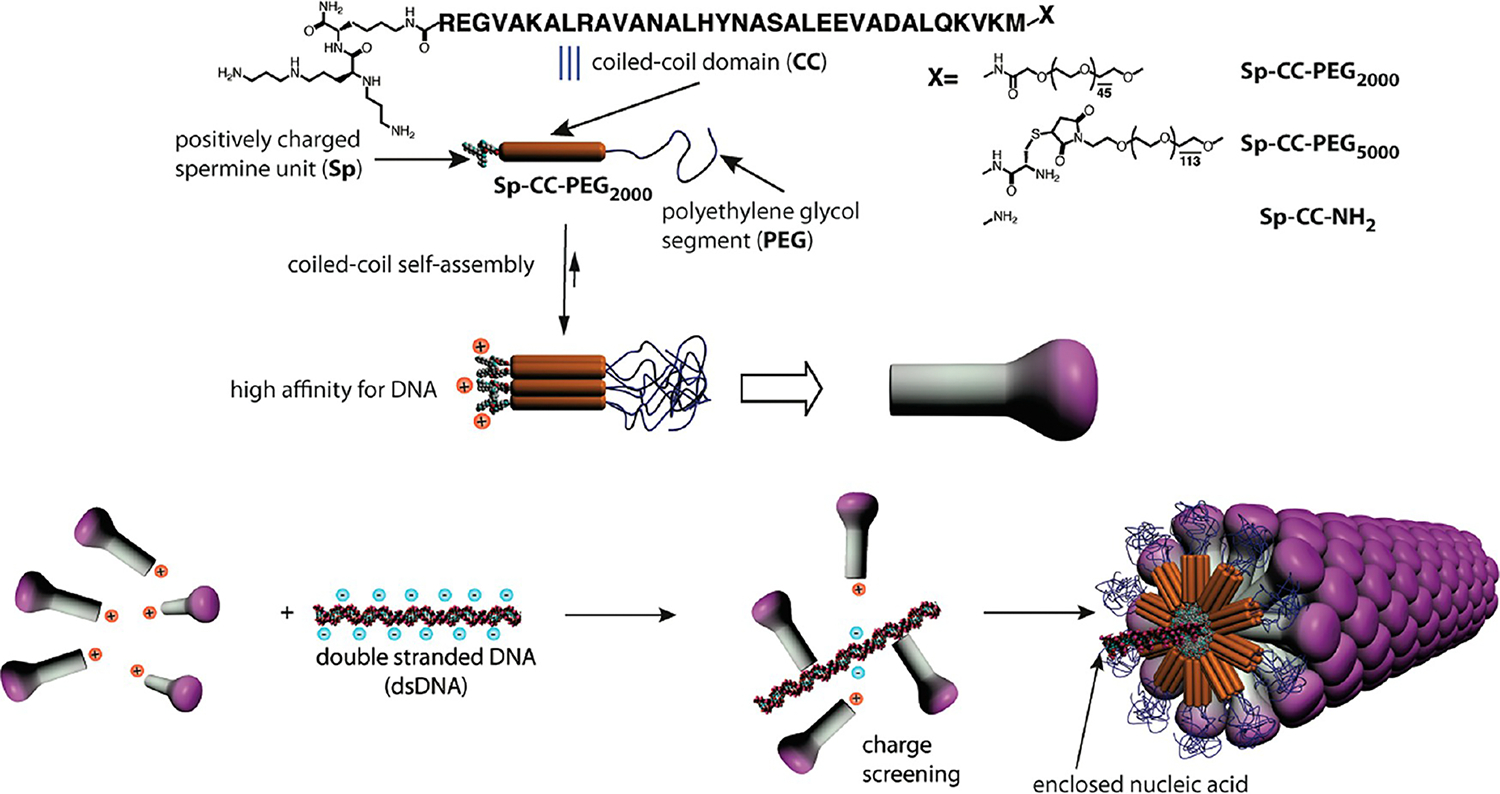
Schematic representation of the strategy to prepare synthetic filamentous viruses using a DNA template and self-assembled mushroom-shaped nanostructures created by the self-assembly of PEGylated coiled coil peptides modified by cationic spermine segments.
RESULTS AND DISCUSSION
Characterization of the Artificial Capsomers.
The self-assembly of the triblock molecules Sp-CC-PEG2000 and Sp-CC-PEG5000 into coiled-coil nanostructures was demonstrated using circular dichroism and analytical centrifugation. Circular dichroism (CD) shows >99% α-helical content for both Sp-CC-PEG2000 and Sp-CC-PEG5000 in phosphate buffer at pH 7.4 before and after addition of an excess of DNA (Figure 2A, Supporting Information S6).21 For Sp-CC-PEG2000, the proportion of ordered/disordered α-helix goes from 82:18 before to 100:0 after complexation with plasmid DNA, whereas for molecule Sp-CC-PEG5000, this ratio is 75:25 before and after addition of plasmid DNA. This slightly higher proportion of disordered α-helix conformation could be explained by the steric repulsion between the bulkier PEG5000 segments clustered at the N-terminus of the self-assembled coiled-coil peptides, as reported previously for PEG-coiled coil peptide conjugates.22
Figure 2.
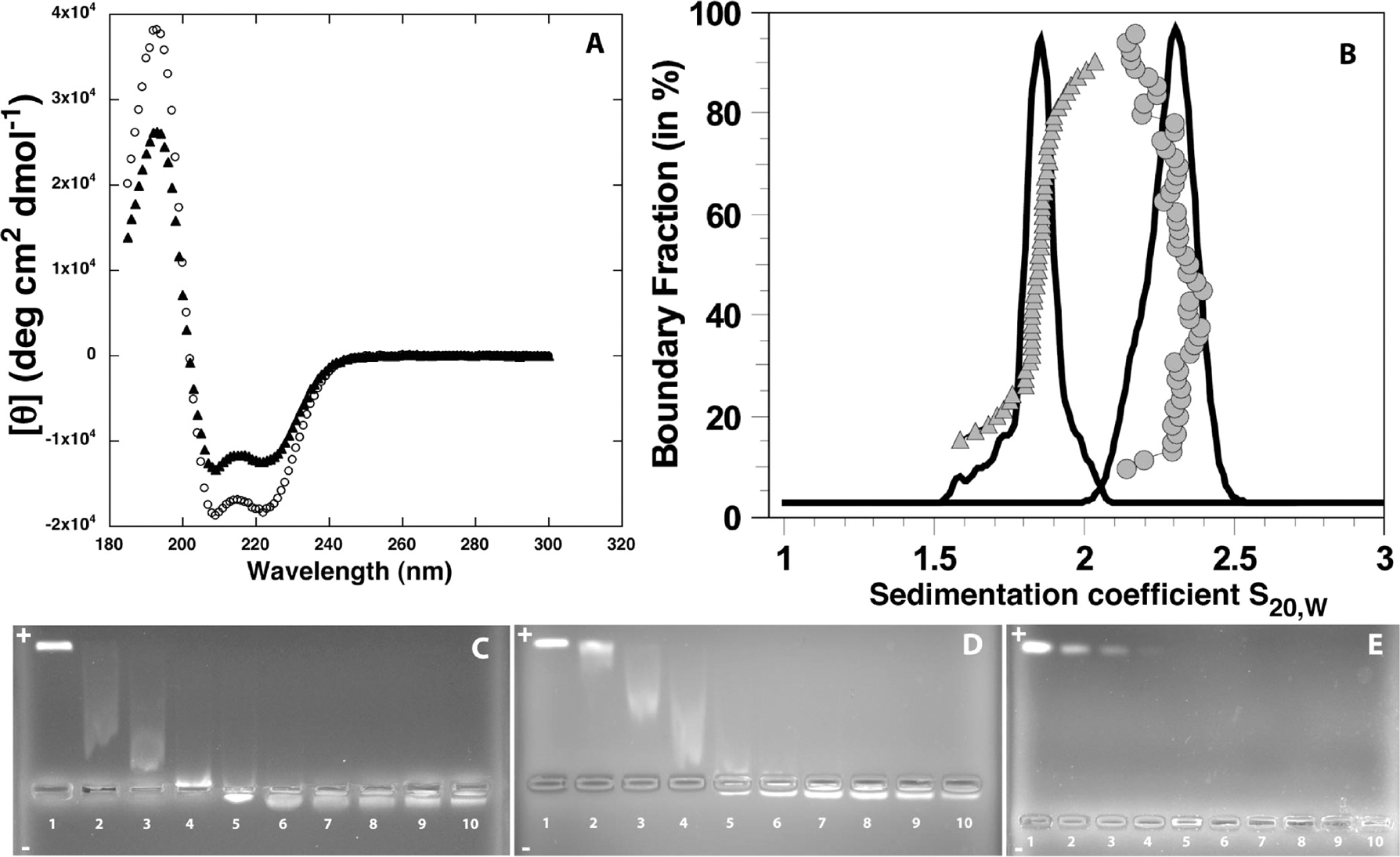
(A) Circular dichroism of peptides Sp-CC-PEG2000 (○) and Sp-CC-PEG5000 (▲) at 30 μM and pH = 7.4 in phosphate buffer (25 mM, 150 mM NaCl). (B) Van Holde–Weischet analysis and integral distribution for Sp-CC-PEG2000 (●) and Sp-CC-PEG5000 (▲). The g(s) distributions (solid line) obtained from a velocity experiment suggest the presence of a single, homogeneous solute. 2DSA/GA-MC analysis resulted in a sedimentation coefficient of 2.28 s for Sp-CC-PEG2000 and 1.92 s for Sp-CC-PEG5000. (C) Agarose gel electrophoresis of complexes of Sp-CC-PEG2000 and a 1200bp dsDNA fragment. Well 1, 200 ng of 1200 bp dsDNA fragment; wells 2–10, 200 ng of 1200bp dsDNA fragment+Sp-CC-PEG2000 at concentrations of 5.6, 11.3, 17, 22.6, 33.9, 45.2, 56.5, 79, and 102 μM, respectively, using a total volume of 20 μL. (D) Agarose gel electrophoresis of complexes of Sp-CC-PEG5000 and a 1200bp dsDNA fragment. Well 1, 200 ng of 1200bp dsDNA fragment; wells 2–10, 200 ng of 1200bp dsDNA fragment+Sp-CC-PEG5000 at concentrations of 3, 5.95, 8.9, 11.9, 17.8, 23.8, 29.75, 41.65, and 53 μM, respectively, total volume of 20 μL. (E) Agarose gel electrophoresis of complexes of Sp-CC-NH2 and a 300bp dsDNA fragment. Well 1, 200 ng of 300bp dsDNA fragment; wells 2–10, 200 ng of 300bp DNA fragment+Sp-CC-NH2 at concentrations of 2.5, 5, 7.5, 10, 15, 20, 25, 35, and 45 μM, respectively, using a total volume of 20 μL.
We then investigated the coiled-coil formation further by analytical ultracentrifugation, which gives access to the absolute molecular weight of the aggregates in solution and subsequently their aggregation number. Sedimentation coefficient (S20,w) distributions were calculated using the van Holde–Weischet analysis.23 Sedimentation analysis from both equilibrium and velocity experiments (Supporting Information S7)21 conducted on Sp-CC-PEG2000 revealed the presence of a single species. Specifically, for Sp-CC-PEG2000 at a concentration of 30 μM, the van Holde–Weischet analysis suggested the presence of a homogeneous solution. From further characterization of this solute using the method known as two-dimensional spectrum analysis and genetic algorithm–Monte Carlo (2DSA/GA-MC), we determined a molecular weight of 33.74 kDa (95% confidence intervals: 33.70, 33.78 kDa) and a weight-average sedimentation coefficient of 2.28S (Figure 2B), indicating a slight concentration-dependent nonideality (Supporting Information S7).21 These molecular weights were confirmed by equilibrium experiments that also suggest the presence of a homogeneous 34.67 kDa oligomer (Supporting Information S7)21 (RMSD = 4.64 × 10−03). These molecular weight values are consistent with a hexameric assembly of Sp-CC-PEG2000 when we expected the GCN4-pAA peptide sequence to favor the formation of a heptameric coiled-coil.20 However, as both sedimentation velocity and sedimentation equilibrium experiments only measure the buoyant molecular weight of the sample, even a slight error (5%) in the estimate of the partial specific volume (estimated at 0.81 mL/mg) could result in fitting the data to a molecular weight consistent with a heptameric aggregate (Supporting Information S7).21 Therefore, our results are consistent with the formation of either a hexameric or a heptameric coiled-coil aggregate, a behavior consistent with the self-assembly of the GCN4-pAA sequence. The van Holde–Weischet analysis for samples of Sp-CC-PEG5000 at 15, 30, and 60 μM suggested the presence of homogeneous solutions with a weight-average sedimentation coefficient of 2.05, 1.81, and 1.98 at 15, 30, and 60 μM (see Figure 2B for representative data). Monte Carlo analysis of the velocity experiments (Supporting Information S7)21 on Sp-CC-PEG5000 at these concentrations further confirmed that this coiled-coil peptide forms oligomers, of which the major components have an average degree of oligomerization between 6.5 and 7.3 for an average molecular weight of 9229 g mol−1 for Sp-CC-PEG5000. Within the experimental error of this experiment, these values are consistent with the formation of heptameric coiled-coil aggregates.
DNA Binding Studies.
The ability of modified peptides Sp-CC-PEG2000, Sp-CC-PEG5000, and Sp-CC-NH2 to bind nucleic acids was demonstrated by agarose gel shift assays, with different ratios of peptide to dsDNA template (Figure 2C–E). In this assay, the reduction of the negative net charge of the dsDNA template upon binding to the positively charged peptides and the formation of higher molecular weight complexes reduce mobility toward the anode (top) in the agarose gel. After staining with ethidium bromide, we indeed observed for Sp-CC-PEG2000 and Sp-CC-PEG5000 the progressive retardation of the band associated with the dsDNA/peptide complexes with respect to the free dsDNA template. In both cases, a further increase in peptide concentration led to a positive electrophoretic charge for the complexes as they migrate toward the cathode. This charge inversion indicates a complete neutralization of the negative charges of the dsDNA template. In the case of Sp-CC-NH2, no free DNA could be detected in the sample at a peptide concentration of 10 μM corresponding to 200 peptide molecules for one molecule of 1200bp dsDNA or 0.66 peptides per base pair. This suggests that Sp-CC-NH2 is a better condensing agent for dsDNA than Sp-CC-PEG2000 and Sp-CC-PEG5000 requiring concentrations of 23 and 18 μM, respectively, to completely neutralize one molecule of 1200bp dsDNA (approximately 1400–1800 peptide molecules, which corresponds to 1.2–1.5 peptides per base pair). A higher ratio of peptide/dsDNA molecule could indicate either a lower affinity for dsDNA or a denser coating of the bound dsDNA molecules by the ligands. In addition, we showed that the binding of Sp-CC-PEG2000 is not affected by physiological salt concentrations (150 mM NaCl) by gel electrophoresis.21 In the following TEM experiments, we ensured a complete neutralization of the dsDNA templates by using a large excess of all three ligands at a concentration of 95 μM.
Structural Characterization of the Complexes: Effect of the PEG Segment Length.
Both conventional and cryogenic microscopy as well as small-angle X-ray scattering (SAXS)21 were utilized to image and characterize the complexes formed between the preassembled nanostructures and linear or circular dsDNA templates having different lengths to probe the effect of the size and topology of the templates on the morphology of the complexes. Figure 3 shows complexes formed between a plasmid pbr322 DNA template and each of the three molecules under conditions for which no free DNA could be detected by gel electrophoresis. As shown in Figure 3A, an amorphous mass is observed when the plasmid is mixed with positively charged molecules only containing the spermine and peptide segments (Sp-CC-NH2). In the case of Sp-CC-PEG2000, discrete complexes are observed, presumably containing only one DNA template (Figure 3B,E). The presence of a single DNA molecule is assumed based on previous reports, indicating that PEGylation of DNA-polycation complexes can prevent aggregation, and promote the formation of complexes containing a single DNA molecule.24–29 While discrete, the structures formed between the pbr322 plasmid and Sp-CC-PEG2000 are heterogeneous and consist of rods, branched, and toroidal complexes in a proportion of 36%, 23%, and 41%, respectively. Therefore, the majority of the complexes formed in this case have either undefined or toroidal morphologies (Figure 3B,D). This result is consistent with the heterogeneous morphologies reported for PEGylated DNA-polycation complexes24,29 and the phenomenon of DNA condensation30 by multivalent cations, which is the transition of DNA molecules from an extended coil to smaller ordered particles, usually highly ordered toroids or rods containing several DNA chains.30 Interestingly, we found that complexes formed with Sp-CC-PEG5000 showed a dramatic improvement in homogeneity in terms of both dimension and morphology. Interestingly, for the complex pbr322/Sp-CC-PEG5000, we observed only filamentous complexes with an average length of about 727 nm (Figure 3C,D) and an extremely low polydispersity index of 1.005. The length polydispersity index (PDI) was calculated from the ratio of the number Ln and weighted Lw length average by analogy to the standard molecular weight polydispersity for polymers.31 The calculated length of the pbr322 plasmid in a supercoiled conformation is 721 nm,21 within 1% of the observed length. This is a clear indication that the circular dsDNA template is included in the complexes in a tight supercoiled conformation, and that for circular dsDNA, the theoretical length of the extended template Lth should correspond to the length of the supercoiled plasmid.21 We then prepared complexes of supercoiled plasmids of different sizes using Sp-CC-PEG5000 as the ligand (Figures 4 and Supporting Information S42). The analysis of puC19/Sp-CC-PEG5000 and pKLAC1-malE/Sp-CC-PEG5000 again revealed homogeneous filamentous complexes. Furthermore, in these two additional cases, the length of the complexes essentially corresponds to the expected length of the supercoiled plasmids. For complexes pKLAC1-malE/Sp-CC-PEG5000, the average length of the complexes was 1619 nm, very close to the calculated length of this plasmid in a supercoiled conformation (1677 nm). These long complexes were difficult to analyze, because the flexible filaments tend to collapse on the carbon film of the TEM grid, making it difficult to distinguish between branched structures and linear complexes having overlapping segments. Therefore, for TEM analysis of this sample, only complexes having an extended conformation were counted. For puC19/Sp-CC-PEG5000, pbr322/Sp-CC-PEG2000, and pKLAC1-malE/Sp-CC-PEG5000, toroid formation was also effectively suppressed, and the filamentous morphology was the dominant morphology, which is a dramatic improvement over literature precedents for PEGylated cation/DNA complexes.24,29
Figure 3.

Effect of the PEG segment length on complex morphologies with circular plasmid DNA. (A) Complex between Sp-CC-NH2 and plasmid pbr322, (B) complex between Sp-CC-PEG2000 and plasmid pbr322, and (C) complex between Sp-CC-PEG5000 and plasmid pbr322. (D) Relative distribution of the three different complex morphologies observed, branched (B), toroidal (T), and rodlike (R), for (A), (B), and (C). (E) Histogram length distribution of rodlike complexes between Sp-CC-PEG2000 and plasmid pbr322; this distribution is centered around Lth/2, which is indicative of the folding of the DNA template. (F) Histogram length distribution of rodlike complexes between Sp-CC-PEG5000; this distribution is centered around Lth, corresponding to the theoretical length of the extended supercoiled plasmid DNA template. For all samples, the plasmid pbr322 (200 ng) was added to the peptide at a concentration of 95 μM in 20 μL of 25 mM Tris buffer (pH 7.4, 25 mM). Scale bar = 200 nm for all images.
Figure 4.
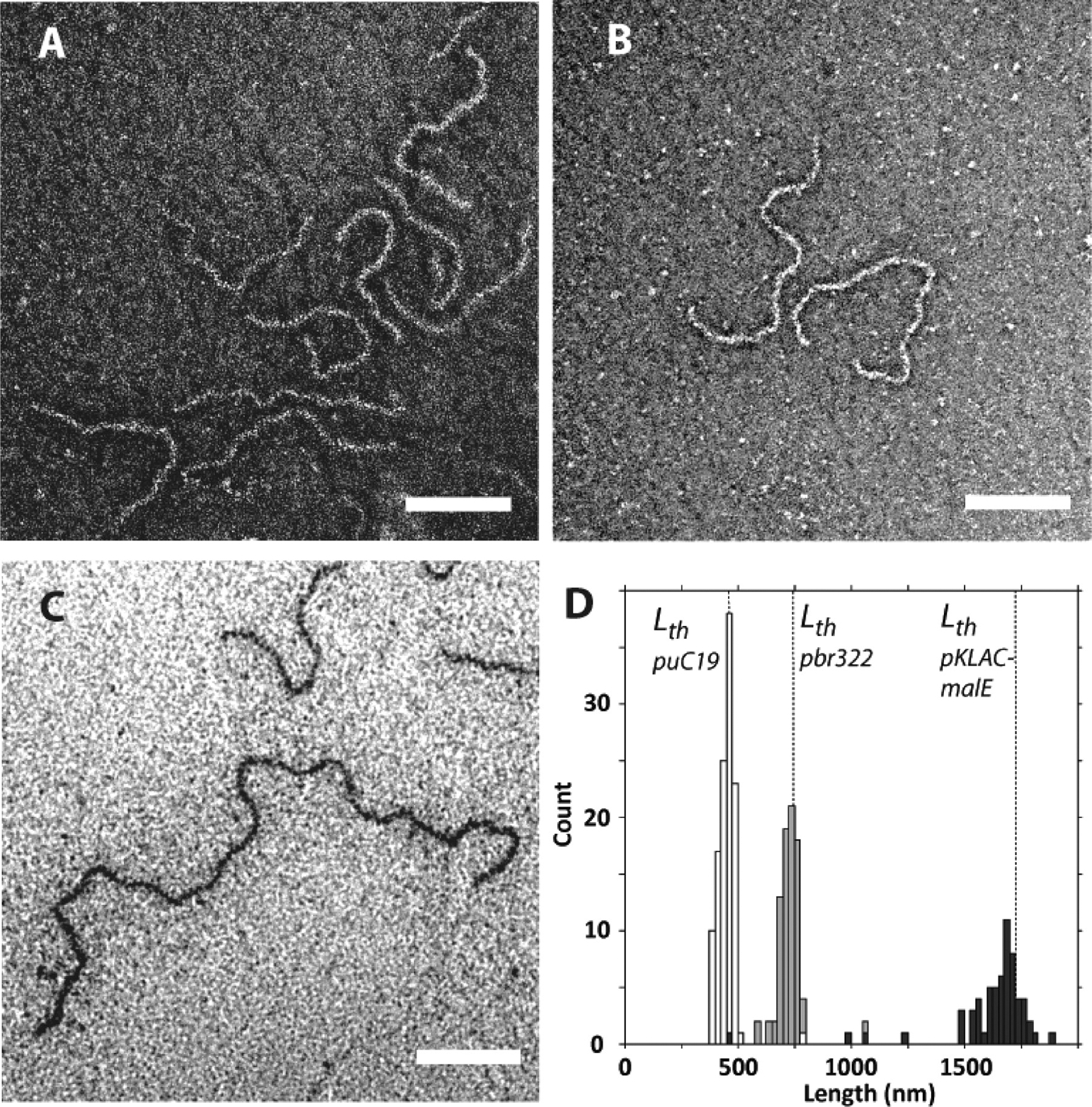
Effect of the size of the plasmid DNA template on the peptide/DNA complex length. (A) Complex between Sp-CC-PEG5000 and puC19 plasmid (2686 bp), (B) complex between Sp-CC-PEG5000 and pbr322 plasmid (4361 bp), (C) complex between Sp-CC-PEG5000 and pKLAC1-malE plasmid (10153 bp), and (D) combined length distributions for complexes of Sp-CC-PEG5000 with puc19 plasmid (white bars; 2686 bp), pbr322 plasmid (gray bars; 4361 bp), and pKLAC1-malE plasmid (black bars; 10153 bp). For all samples, the plasmid (200 ng) was added to the peptide at a concentration of 95 μM in 20 μL of 25 mM Tris buffer (pH 7.4, 25 mM). Scale bar = 200 nm for all images.
We observed a low polydispersity index (1.011) for the subpopulation of rodlike pbr322/Sp-CC-PEG2000 complexes, even though the sample as a whole was heterogeneous. While the expected theoretical length Lth of the supercoiled plasmid pbr322 was about 721 nm, the observed distribution was centered around 361 nm, which is the predicted length for a supercoiled DNA template folded in half (Figure 3E). To better understand the effect of the length of the template as well as the influence of the ligand on this folding behavior, we conducted a detailed study using linear dsDNA templates of different lengths. This extension to linear dsDNA could also make our approach applicable to a broader range of dsDNA templates, which might be suitable to get smaller artificial viruses or to encapsulate different kinds of nucleic acids such as siRNA.
Extension to Linear dsDNA and Discussion on the Folding of the Complexes.
Complexes between short linear 150bp dsDNA fragments and Sp-CC-PEG2000 (150bp/Sp-CC-PEG2000) and Sp-CC-PEG5000 (150bp/Sp-CC-PEG5000) were found to have a rodlike morphology and showed predominantly the length of the DNA template (Figure 5A,B). The dimensions of these complexes were determined to be 53 nm in length for 150bp/Sp-CC-PEG2000.21 This result was confirmed by SAXS experiments,21 which clearly showed the appearance of rodlike complexes after mixing a 150 bp dsDNA template with an excess of either peptide Sp-CC-PEG2000 or Sp-CC-PEG5000. No folding or toroid formation was observed in this case, because 150bp dsDNA fragments are known not to undergo DNA condensation.32
Figure 5.
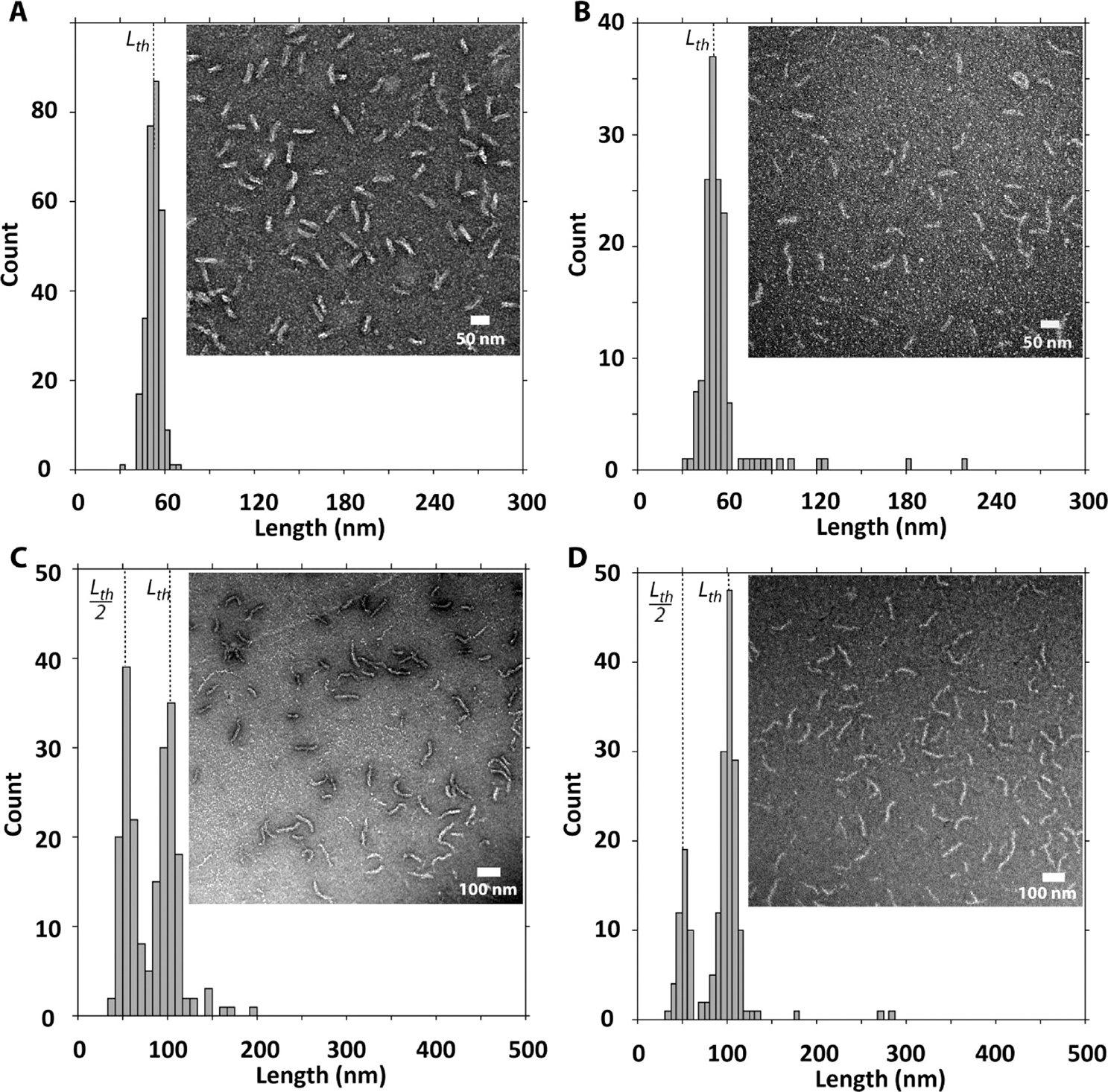
Effect of the length of the PEG segment on the peptide-linear double-stranded DNA complex morphology. (A) Complex between Sp-CC-PEG2000 and 150 bp dsDNA fragment, (B) complex between Sp-CC-PEG5000 and 150 bp dsDNA fragment, (C) complex between Sp-CC-PEG2000 and 300 bp dsDNA fragment, and (D) complex between Sp-CC-PEG5000 and 300 bp dsDNA fragment. For all samples, the dsDNA (200 ng) was added to the peptide at a concentration of 95 μM in 20 μL of 25 mM Tris buffer (pH 7.4, 25 mM).
Interestingly, in the case of 300bp/Sp-CC-PEG2000, the length distribution obtained by TEM analysis was again consistent with a folding mechanism for condensation, as observed for pbr322/Sp-CC-PEG2000. Indeed, the observed distribution was bimodal with maxima centered around the expected length (Lth) as well as the length predicted for a DNA template that is folded once (Lth/2) (Figure 5C). Thus, the length distribution of rods is not random but corresponds to a distribution of “quantized” lengths having a length L = Lth/(n + 1) of the plasmid DNA contour length, where n is the folding number. Interestingly, for the 300bp/Sp-CC-PEG5000 complexes, the population formed around the folded template is smaller than the proportion of the same population for 300bp/Sp-CC-PEG2000 complexes (Figure 5C,D). This implies that in this case the forces opposing the folding are stronger when nanostructures having a longer PEG segment are used to complex the DNA. Because the only difference between Sp-CC-PEG2000 and Sp-CC-PEG5000 is the N-terminal fragment, we propose that the steric repulsion between PEG domains in the preassembled mushroom aggregates results in an increase of the persistence length of the complexes lp and prevents their transition into shorter rods. This hypothesis was confirmed by the length distribution of complexes between the more sterically demanding ligand Sp-CC-PEG5000 and 600 or 1200bp dsDNA. We found that the maximum folding number for 600bp/Sp-CC-PEG5000 complexes is n = 1, as compared to n = 3 for 600bp/Sp-CC-PEG2000 complexes. Similarly, the 1200bp/Sp-CC-PEG5000 complexes showed a length distribution centered around the template folded once (n = 1, 204 nm). We therefore conclude that the DNA binding nanostructures formed by Sp-CC-PEG2000 and Sp-CC-PEG5000 interfere with the folding process of the bound DNA template. These observations are consistent with the quantized folding model for DNA condensation proposed by Kataoka et al.24 In this model, the discrete folded states in DNA–polycation complexes originate from the Euler bending modes of initial rodlike DNA–polycations complexes under a crushing force . For an internal DNA segment, the critical buckling force was determined by Manning to be in the following form:33
where corresponds to the bending stiffness of the segment of length . The value for is not known in our system, but it is expected to depend on the persistence length of the complexes, and therefore should be dependent on the ligand’s structure, according to the following expression:33
where is Boltzmann’s constant. As in the Euler buckling model, increasing the folding number requires loads higher than to be applied to the initial rod structure,24 an increase of , and the elastic bending stiffness of the rod has a direct effect on the extent of the folding.
Alternatively, another sequential folding mechanism has been reported to explain the different stages of plasmid DNA condensation by polylysine conjugates.34 In this case, increasing the folding number would result in an increase in the charge density of the folded DNA template, which in turn would have to be compensated by an increase in the ligand density, resulting in turn in increasingly stiffer rods. For sterically demanding ligands, the optimum degree of folding might therefore be determined by the balance between the steric repulsion of the ligands and the electrostatic interactions with the folded DNA template.
It is also important to note that the selectivity of the folding process observed in the case of complexes with linear dsDNA is consistent with the observations made for complexes with supercoiled plasmid DNA. Indeed, due to the spatial proximity between opposing plasmid segments, an extended supercoiled plasmid DNA template can be considered as an analog to a linear dsDNA template folded once,21 and having both of its ends ligated. In addition, it has been shown that the screening of the electrostatic repulsion between the juxtaposed supercoiled DNA segments by divalent salts causes the transition from a loosely to a tightly interwound superhelix to occur.35 As an apparent complete charge neutralization of the DNA backbone was observed by gel electrophoresis upon binding of the supercoiled plasmid templates to our artificial capsomers (Figure 2C–E), it is reasonable to expect the same transition to occur in our system. In other words, supercoiling preorganizes the circular DNA template and facilitates its transition into a tightly interwound superhelix conformation equivalent to a prefolded linear dsDNA template with a folding number n = 1. Any subsequent folding of the supercoiled plasmid would yield a template with a folding number equivalent to 3 or more for linear dsDNA. In line with our observations, this implies that complexes between a supercoiled plasmid DNA and Sp-CC-PEG2000 would fold only once (Figure 3B,E), whereas complexes with Sp-CC-PEG5000 should not fold because folding numbers higher than one are not expected according to the model established for linear dsDNA.
Increasing the bulk of the N-terminal fragments in Sp-CC-PEG5000 nanostructures not only favored the formation of essentially monodisperse filamentous nanostructures, but also suppressed toroid formation. We found that when the length of the template was increased above 300 bp, the morphology and dimensions of the complexes between DNA fragments and Sp-CC-PEG2000 became more heterogeneous as more toroids were observed for 600bp/Sp-CC-PEG2000 and 1200bp/Sp-CC-PEG2000 complexes (Figure 6A,C, Supporting Information Table S1). In contrast, complexes formed between linear dsDNA fragments longer than 300bp and Sp-CC-PEG5000 showed consistently homogeneous rods having predominantly a length corresponding to the DNA template folded once without toroid formation (see Figure 6B,D, Supporting Information Table S1). The morphology of the complexes is also not affected by the presence of physiological salt concentration,21 making our approach suitable for applications requiring physiological buffers. Therefore, our approach can be extended to the preparation of monodisperse filamentous complexes from linear dsDNA templates. As previously discussed, when supercoiled plasmids were complexed with Sp-CC-PEG5000, homogeneous rodlike complexes were observed as well, having in this case the length of the extended supercoiled template.
Figure 6.

Effect of the length of the PEG segment on the peptide-linear double-stranded DNA complex morphology. (A) Complex between Sp-CC-PEG2000 and 600 bp dsDNA fragment, (B) complex between Sp-CC-PEG5000 and 600 bp dsDNA fragment, (C) complex between Sp-CC-PEG2000 and 1200 bp dsDNA fragment, and (D) complex between Sp-CC-PEG5000 and 1200 bp dsDNA fragment. For all samples, the dsDNA (200 ng) was added to the peptide at a concentration of 95 μM in 20 μL of 25 mM Tris buffer (pH 7.4, 25 mM).
CONCLUSION
The strategy described here allows precise control of the dimensions of filamentous virus-like particles templated by DNA. We showed that tuning steric forces among nanoscale ligands binding to DNA templates makes it possible to obtain homogeneous supramolecular rod-like objects either with homogeneously folded dsDNA or extended supercoiled plasmids reaching lengths of up to 1.6 μm. One-dimensional supramolecular objects with precise length could have tunable characteristics such as permeability, polyvalency, and extended circulation times in biological systems, among others. At the same time, they can carry precise cargoes of molecules or serve as templates and building blocks for more complex structures. Therefore, the principles learned here to create filamentous supramolecular structures with precise length could be of general interest in the design of future therapies and materials.
Supplementary Material
ASSOCIATED CONTENT
Supporting Information
Materials and methods. Transmission electron microscopy controls. Negative and positive staining of the complexes between dsDNA templates and peptides Sp-CC-NH2, Sp-CC-PEG2000, and Sp-CC-PEG5000. Effect of the peptides design on the DNA toroids formation. Cryogenic transmission electron microscopy. Circular dichroism. Analytical ultracentrifugation. Small-angle X-ray scattering studies. Calculation of the theoretical length of supercoiled plasmids. Additional TEM images showing the effect of the size of the plasmid DNA template on the peptide/DNA complex length. Stability of the complexes to physiological salt concentration. Table for the statistical analysis of the complexes. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Energy-Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Award No. (DE-FG02–00ER45810) (Biomolecular Materials Program), and by the National Science Foundation under Award No. (DMR-1006713). DOE funded work on the characterization of self-assembled structures and NSF funded the synthetic work for the systems investigated. Y.R. received partial support from a Lavoisier Fellowship from the French Ministry of Foreign Affairs, and T.M. was supported by a National Science Foundation Graduate Research Fellowship. The authors thank the Biological Imaging Facility (BIF) at Northwestern for the use of TEM equipment.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Lehn J-M Science 2002, 295, 2400. [DOI] [PubMed] [Google Scholar]
- (2).Yoshizawa M; Klosterman JK; Fujita M Angew. Chem., Int. Ed. 2009, 48, 3418. [DOI] [PubMed] [Google Scholar]
- (3).Lo PK; Altvater F; Sleiman HF J. Am. Chem. Soc. 2010, 132, 10212. [DOI] [PubMed] [Google Scholar]
- (4).Seeman NC Nature 2003, 421, 427. [DOI] [PubMed] [Google Scholar]
- (5).Lortie F; Boileau S; Bouteiller L; Chassenieux C; Lauprêtre F Macromolecules 2005, 38, 5283. [Google Scholar]
- (6).Besenius P; Portale G; Bomans PHH; Janssen HM; Palmans ARA; Meijer EW Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 17888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hunter CA; Tomas SJ Am. Chem. Soc. 2006, 128, 8975. [DOI] [PubMed] [Google Scholar]
- (8).O’Sullivan MC; Sprafke JK; Kondratuk DV; Rinfray C; Claridge TDW; Saywell A; Blunt MO; O’Shea JN; Beton PH; Malfois M; Anderson HL Nature 2011, 469, 72. [DOI] [PubMed] [Google Scholar]
- (9).Wang X; Guerin G; Wang H; Wang Y; Manners I; Winnik MA Science 2007, 317, 644. [DOI] [PubMed] [Google Scholar]
- (10).Klug A Philos. Trans. R. Soc., B 1999, 354, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Fraenkel-Conrat H; Williams RC Proc. Natl. Acad. Sci. U.S.A. 1955, 41, 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bull SR; Palmer LC; Fry NJ; Greenfield MA; Messmore BW; Meade TJ; Stupp SI J. Am. Chem. Soc. 2008, 130, 2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lee L; Niu Z; Wang Q Nano Res. 2009, 2, 349. [Google Scholar]
- (14).de la Escosura A; Janssen PGA; Schenning APHJ; Nolte RJM; Cornelissen JJ L. M. Angew. Chem., Int. Ed. 2010, 49, 5335. [DOI] [PubMed] [Google Scholar]
- (15).Minten IJ; Ma Y; Hempenius MA; Vancso GJ; Nolte RJM; Cornelissen JJ L. M. Org. Biomol. Chem. 2009, 7, 4685. [DOI] [PubMed] [Google Scholar]
- (16).Hernandez-Garcia A; Werten MWT; Stuart MC; de Wolf FA; de Vries R Small 2012, 8, 3490. [DOI] [PubMed] [Google Scholar]
- (17).Stupp SI; LeBonheur V; Walker K; Li LS; Huggins KE; Keser M; Amstutz A Science 1997, 276, 384. [DOI] [PubMed] [Google Scholar]
- (18).Sayar M; Stupp SI Macromolecules 2001, 34, 7135. [Google Scholar]
- (19).Patel MM; Anchordoquy TJ Biophys. J. 2005, 88, 2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liu J; Zheng Q; Deng Y; Cheng C-S; Kallenbach NR; Lu M Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Materials, methods, and additional figures are available as Supporting Information.
- (22).Vandermeulen GWM; Tziatzios C; Klok H-A Macromolecules 2003, 36, 4107. [Google Scholar]
- (23).Demeler B; van Holde KE Anal. Biochem. 2004, 335, 279. [DOI] [PubMed] [Google Scholar]
- (24).Osada K; Oshima H; Kobayashi D; Doi M; Enoki M; Yamasaki Y; Kataoka KJ Am. Chem. Soc. 2010, 132, 12343. [DOI] [PubMed] [Google Scholar]
- (25).DeRouchey J; Walker GF; Wagner E; Radler JO J. Phys. Chem. B 2006, 110, 4548. [DOI] [PubMed] [Google Scholar]
- (26).Katayose S; Kataoka K Bioconjugate Chem. 1997, 8, 702. [DOI] [PubMed] [Google Scholar]
- (27).Vinogradov S; Batrakova E; Li S; Kabanov A Bioconjugate Chem. 1999, 10, 851. [DOI] [PubMed] [Google Scholar]
- (28).Vachutinsky Y; Kataoka K Isr. J. Chem. 2010, 50, 175. [Google Scholar]
- (29).Itaka K; Yamauchi K; Harada A; Nakamura K; Kawaguchi H; Kataoka K Biomaterials 2003, 24, 4495. [DOI] [PubMed] [Google Scholar]
- (30).Bloomfield VA Biopolymers 1997, 44, 269. [DOI] [PubMed] [Google Scholar]
- (31)., , .
- (32).Widom J; Baldwin RL J. Mol. Biol. 1980, 144, 431. [DOI] [PubMed] [Google Scholar]
- (33).Manning GS Biophys. J. 2006, 91, 3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Golan R; Pietrasanta LI; Hsieh W; Hansma HG Biochemistry 1999, 38, 14069. [DOI] [PubMed] [Google Scholar]
- (35).Bednar J; Furrer P; Stasiak A; Dubochet J; Egelman EH; Bates AD J. Mol. Biol. 1994, 235, 825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


