Abstract
The heterofermentative lactic acid bacteria Oenococcus oeni and Leuconostoc mesenteroides are able to grow by fermentation of pyruvate as the carbon source (2 pyruvate → 1 lactate + 1 acetate + 1 CO2). The growth yields amount to 4.0 and 5.3 g (dry weight)/mol of pyruvate, respectively, suggesting formation of 0.5 mol ATP/mol pyruvate. Pyruvate is oxidatively decarboxylated by pyruvate dehydrogenase to acetyl coenzyme A, which is then converted to acetate, yielding 1 mol of ATP. For NADH reoxidation, one further pyruvate molecule is reduced to lactate. The enzymes of the pathway were present after growth on pyruvate, and genome analysis showed the presence of the corresponding structural genes. The bacteria contain, in addition, pyruvate oxidase activity which is induced under microoxic conditions. Other homo- or heterofermentative lactic acid bacteria showed only low pyruvate fermentation activity.
Oenococcus oeni and Leuconostoc mesenteroides are heterofermentative lactic acid bacteria (LAB) that are closely related to each other and have previously been grouped within the same family due to many physiological and genetic similarities (6, 8, 9). O. oeni is used in wine fermentation for degradation of malic acid, whereas L. mesenteroides is used for fermentation of vegetables. Both bacteria convert hexoses by heterolactic fermentation to lactate, ethanol, and CO2. Ethanol formation from acetyl phosphate (or acetyl coenzyme A [acetyl-CoA]) is required for reoxidation of NAD(P)H, which is produced in the pentose phosphate pathway during hexose oxidation. In many heterolactic acid bacteria and in particular in O. oeni, NAD(P)H reoxidation by the ethanol pathway is slow because of low acetaldehyde dehydrogenase activity (16, 26, 27, 34). When, in addition, coenzyme A (HSCoA), and consequently acetyl-CoA, is limiting due to a shortage of the precursor pantothenate, the low capacity of NAD(P)H reoxidation by the ethanol pathway limits the metabolism and growth rate of the bacteria (26). For this reason, O. oeni, and to some extent also L. mesenteroides, uses alternative pathways for NAD(P)H reoxidation, such as the reduction of erythrose-4-phosphate to erythritol, to overcome the limitation (26, 34).
In addition to the alternative endogenous pathways of erythritol and glycerol formation for NAD(P)H reoxidation, the bacteria are able to use external electron acceptors for reoxidation of NAD(P)H. Fructose, O2, and pyruvate can be used as electron sinks by O. oeni and L. mesenteroides, yielding mannitol, H2O2, and lactate as the reduced end products (16, 23, 27). O. oeni, which has very low capacities for NAD(P)H reoxidation in the ethanol pathway, gains significantly in metabolic and growth rates by the use of external electron acceptors.
When pyruvate was supplied in excess as an electron acceptor, there were indications that pyruvate is not only used as an electron acceptor but also oxidized to acetate (27, 28). As shown here, pyruvate can be disproportionated to lactate and acetate at substantial rates and can be used as the sole substrate for fermentation and growth by the bacteria. The fermentation process and the enzymatic reactions of this new fermentation pathway of O. oeni and L. mesenteroides are characterized here.
MATERIALS AND METHODS
Bacteria and growth.
Oenococcus oeni B1 (26), Leuconostoc mesenteroides (DSMZ 20240), Lactobacillus lactis (DSMZ 20072), Lactobacillus plantarum (ATCC 8014), and Lactococcus lactis (DSMZ 20481) were used. Subcultures of the bacteria were grown in modified tomato juice medium (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], Braunschweig, Germany). Growth was performed at 30°C in anoxic Malo-Lactic Differential (MLD) medium (4) under N2 in sealed bottles (50 ml or 400 ml as given for the specific experiments) at pH 5.9 with the modification of Richter et al. (26, 27). Anoxic conditions were checked by the inclusion of resazurin (1 mg/liter) in control experiments. The media were inoculated with 2% (vol/vol) of the subcultures.
Cell homogenates and enzyme activities.
MLD medium (400 ml) containing the respective C source (80 mM pyruvate, 40 mM glucose, or 40 mM glucose plus 80 mM pyruvate) was inoculated with 8 ml of a subculture, made anoxic by degassing and gassing with N2 for three cycles and grown to the late-logarithmic growth phase. The bacteria were harvested (17,700 × g for 25 min at 4°C), washed, and resuspended in 10 ml potassium-phosphate buffer (50 mM at pH 7). The cells were broken in a cell mill (Vibrogen VI; Bühler) or by three passages through a French pressure cell (1,200 lb/in2). For treatment in the cell mill, the bacterial suspension (10 ml) was mixed with 20 g of zirconia/silica beads (0.1 mm; Roth) and incubated for three cycles of 3 min, with 30-s intervals between the cycles, at 4°C and 75 Hz. The cell homogenates were cleared by centrifugation at 11,400 × g for 15 min, and the supernatant was either frozen in aliquots at −20°C or used immediately. Protein was determined by the Bradford assay (3) using Rothiquant (Roth) in a microassay.
(i) PDH.
The assay of Reed and Willms (25), which depends on the conversion of pyruvate to acetyl-CoA, NADH, and CO2 in the presence of NAD (acetyl-CoA formation via pyruvate dehydrogenase [PDH]), was used. Acetyl-CoA is converted in a coupled assay by phosphotransacetylase to acetyl phosphate, and NAD is regenerated by inclusion of lactate dehydrogenase. Formation of acetyl phosphate is determined colorimetrically by reaction with hydroxylamine and Fe(III), which results in formation of a reddish brown dye that was measured discontinuously at 540 nm (14). The formation of acetyl phosphate (or of the dye complex) corresponds to the activity of pyruvate oxidation and is expressed in μmol of acetyl-CoA or acetyl phosphate formed/min/g of protein. In the same setup, degradation of pyruvate was tested by high-performance liquid chromatography (HPLC) (see below), and it was verified that pyruvate degradation depended on the presence of NAD.
(ii) Pox.
Pyruvate oxidase (acetyl phosphate-forming, or Pox) activity (pyruvate + O2 + Pi → acetyl phosphate + H2O2 + CO2) was tested by the formation of acetyl phosphate in the presence of O2 and pyruvate, but without added phosphotransacetylase or lactate dehydrogenase. The requirement for O2 and the independence from HSCoA allow differentiation from pyruvate dehydrogenase. Acetyl phosphate formation was tested with hydroxylamine and Fe(III) as described above for pyruvate dehydrogenase.
(iii) Other enzyme assays.
Lactate dehydrogenase was assayed photometrically as the pyruvate-dependent oxidation of NADH (34). Phosphotransacetylase activity was followed photometrically at 233 nm by the formation of acetyl-CoA from acetyl phosphate plus HSCoA (Eacetyl-CoA = 4.4 mM−1 cm−1) using 260 nm as a reference wavelength (Specord S10; Zeiss, Jena, Germany) according to Klotzsch (13). Acetate kinase was tested by the decrease of NADH in a coupled assay with pyruvate kinase and lactate dehydrogenase (2).
All activities were assayed at least in duplicate (variation, <30%), and average values are given. The activities correspond to the conversion of 1 μmol of pyruvate (pyruvate dehydrogenase, pyruvate oxidase, and lactate dehydrogenase), the formation of 1 μmol of acetyl-CoA (phosphotransacetylase), or the conversion of 1 μmol of acetate or acetyl phosphate (acetate kinase) per g of protein/min at 30°C.
Fermentation substrates and products.
For determination of fermentation balances in growing bacteria, MLD medium (50 ml) was inoculated with 2% (vol/vol) of a subculture of the bacteria. Growth was performed under anoxic conditions, which were controlled by inclusion of resazurin. At time zero (inoculation) and at various times after inoculation, samples were withdrawn and centrifuged at 11,000 × g for 5 min. The cell-free supernatants were analyzed for the substrates and products by HPLC separation on an Aminex HPX87H column (26). The assays were performed in duplicate (or repeated if the values differed by more than 15%), and the mean values are given. In addition, d- and l-lactate were determined photometrically by the d- and l- lactate dehydrogenase-dependent reduction of NAD at the expense of lactate (34). For fermentation balances, cultures that had converted >7 mM pyruvate were used. All fermentation reactions were performed twice or more, and typical results are shown. In some experiments, a medium without a C source was inoculated and incubated with the bacteria for 12 to 24 h under anoxic conditions to ensure degradation of fermentable substrates in the medium. Growth was then started by addition of the substrate after cessation of bacterial growth in the medium (generally at an optical density at 578 nm [OD578] of 0.2 to 0.3).
For measurement of fermentation balances in resting cells, the bacteria were first grown in MLD medium to the late-exponential growth phase with the respective substrate. The bacteria were then washed in morpholinepropanesulfonic acid (MOPS) salts medium (pH 7.4) and resuspended in the same medium under anoxic conditions (26, 27). The bacteria (OD578, ∼6) were incubated in the medium under anoxic conditions with the respective carbon source. At time zero and after various times, samples were withdrawn, and the substrates and products were determined from the supernatant by HPLC analysis.
Database search and identification of genes encoding the relevant enzymes.
Proteome data of the partly and fully sequenced genomes were obtained from the U.S. Department of Energy Joint Genome Institute (JGI) and the European Bioinformatics Institute (EBI). Identification of the genes includes functional domain searches and BLAST analysis. Functional domains characteristic for each enzyme were determined using the reference sequences from the following organisms: L. mesenteroides (PDH, Pox, and pyruvate formate lyase [PFL]), Escherichia coli (phosphotransacetylase, acetate kinase, and d-lactate dehydrogenase), and Enterococcus faecium (pyruvate ferredoxin oxidoreductase [PFOR]). The proteomes were screened for the functional domains identified. The analysis was carried out by means of InterProScan (18). For verification, the hit sequences were analyzed using BLAST. The presence of an open reading frame (ORF) indicates that the corresponding domain is present and that the BLAST hit showed the functional annotation.
RESULTS AND DISCUSSION
Growth of Oenococcus oeni and Leuconostoc mesenteroides with pyruvate.
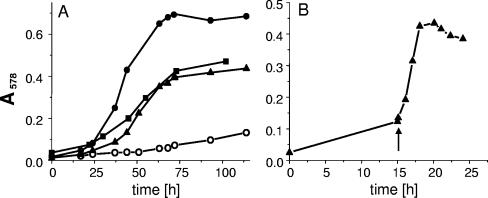
Oenococcus oeni and Leuconostoc mesenteroides are able to use pyruvate as an electron acceptor during growth on hexoses, and the pyruvate is then reduced to lactate (23, 27, 28). When pyruvate was supplied in a two- to fourfold excess over hexoses, as much as one-quarter of the pyruvate was converted to acetate (27, 28). Therefore, we tested whether pyruvate can serve as a substrate for growth. O. oeni showed significant growth in MLD medium with pyruvate compared to growth without an added carbon source (Fig. 1A). Due to the nutritional requirements of O. oeni and L. mesenteroides, complex media are required for growth. The MLD medium that was used for growth is supposed to supply only essential nutrients but no fermentable substrate. Growth in the MLD medium depended on pyruvate (Fig. 1B). The medium was first depleted by incubation with bacteria, but without addition of pyruvate, which resulted in very poor growth (OD578, ≤0.15). After addition of pyruvate, the growth rate was stimulated dramatically, and an additional OD578 of 0.3 to 0.4 was obtained for L. mesenteroides (Fig. 1B) and O. oeni (not shown). The stimulation of the growth rate with pyruvate was comparable to that with glucose (Table 1), but the cell densities achieved with glucose were higher. Supply of pyruvate plus glucose, where pyruvate is used as an electron acceptor for glucose oxidation, resulted in high growth rates, as described previously (27, 28). The response of L. mesenteroides to pyruvate as a carbon source or as an electron acceptor was very similar to that of O. oeni (Table 1). The molar growth yields of these bacteria with pyruvate amounted to 4 and 5.3 g (dry weight)/mol pyruvate, respectively, about half the growth yields with glucose (Table 1). Therefore, the ATP yield with pyruvate can be estimated as 0.5 mol of ATP/mol of pyruvate.
FIG. 1.
Growth on pyruvate in MLD medium (A) and in “predigested” MLD medium (B). The substrates for growth were pyruvate (40 mM in panel A and 20 mM in panel B) (▴), 40 mM glucose (▪), 10 mM glucose plus 20 mM pyruvate (•), or no additional carbon source (○). Experiment A is shown for Oenococcus oeni, experiment B for Leuconostoc mesenteroides. In panel B, pyruvate was added (arrow) after growth of the bacteria in MLD medium for 15 h without an additional carbon source.
TABLE 1.
Parameters for growth of Oenococcus oeni and Leuconostoc mesenteroides on pyruvate and comparison to growth on glucosea
| Carbon source | μ (h−1)
|
Y (g [dry wt]/mol)
|
||
|---|---|---|---|---|
| O. oeni | L. mesenteroides | O. oeni | L. mesenteroides | |
| Pyruvate | 0.053 | 0.42 | 4.0 | 5.3 |
| Glucose | 0.024 | 0.28 | 8.3 | 10.5 |
| Glucose + pyruvate | 0.083 | 0.47 | 17.9 | 22.0 |
Growth was performed under anoxic conditions in MLD medium with the carbon source indicated. Growth rate (μ) and growth yield (Y) were determined from the experiments for which results are shown in Fig. 1. Each value is the mean for two independent experiments.
Lactate and acetate as the end products of pyruvate fermentation.
Fermentation balances were determined with bacteria growing in MLD medium on pyruvate after preceding consumption of fermentable substrates in the medium. This precaution should help to avoid the formation of additional products from components of the MLD medium. Under these conditions, O. oeni as well as L. mesenteroides fermented pyruvate to approximately stoichiometric amounts of lactate and acetate (and presumably CO2) (Table 2), suggesting disproportionation of 1 mol of pyruvate to 0.5 mol of lactate and of acetate. Neither ethanol, erythritol, glycerol, nor other fermentation products were detected in significant amounts. The recovery of 83 to 97% of the carbon in lactate, acetate, and CO2 confirms that no other products were formed in substantial amounts and that pyruvate was the only substrate for growth.
TABLE 2.
Fermentation balances of O. oeni and L. mesenteroides for growth on pyruvate or glucose under anoxic conditions or in resting cellsa
| Condition | Fermentation balance (mol of product/mol of pyruvate consumed)
|
% Cc | |||
|---|---|---|---|---|---|
| Lactate | Acetate | Ethanol | CO2b | ||
| Growing bacteria | |||||
| O. oeni B1 | 0.44 | 0.47 | <0.04 | 0.47 | 91 |
| L. mesenteroides | 0.50 | 0.33 | <0.04 | 0.33 | 83 |
| Resting bacteria | |||||
| O. oeni B1 | 0.38 | 0.47 | <0.04 | 0.47 | 85 |
| L. mesenteroides | 0.45 | 0.52 | <0.04 | 0.52 | 97 |
Fermentation balances of growing bacteria were determined in MLD medium after consumption of fermentable substrates in the medium by the bacteria. Pyruvate (20 mM) was added as the substrate for growth after digestion of the medium without added substrate. The fermentation balance of the resting bacteria was determined by incubating washed bacteria (OD578, 6) in MOPS buffer with 10 mM pyruvate under anoxic conditions.
Calculated from the amounts of acetate formed.
Carbon yield of the fermentation products (lactate, acetate, CO2) compared to the amount of pyruvate consumed.
Similar fermentation experiments were performed with resting bacteria in cell suspensions. Bacteria pregrown on pyruvate degraded pyruvate to lactate and acetate (and presumably CO2) as the major products. The products were found in ratios very similar to those of the growing bacteria (Table 2), confirming pyruvate disproportionation to lactate, acetate, and CO2 by the bacteria. An enzymatic assay showed that the lactate excreted by O. oeni was exclusively (>98%) d-lactate.
Use of pyruvate by other homo- and heterolactic acid bacteria.
In a similar way, Lactobacillus plantarum, Lactobacillus lactis, and Lactococcus lactis were incubated under anaerobic conditions in MLD medium with pyruvate as the sole C source. The bacteria showed only poor growth stimulation by pyruvate (OD578, ≤0.2). L. plantarum and Lactobacillus lactis metabolized pyruvate at low rates. The products were lactate and acetate; other products, such as formate, were not detected in significant amounts. When pyruvate was supplied in the presence of glucose, Lactobacillus lactis and L. plantarum cometabolized comparable amounts of glucose and pyruvate. Lactate was the major end product, suggesting that pyruvate was used predominantly as an electron acceptor. Lactococcus lactis, on the other hand, showed no growth stimulation by pyruvate as the sole substrate in MLD medium, and the pyruvate was not metabolized when pyruvate was supplied as the only substrate or in addition to glucose. In cell suspensions, neither L. plantarum, Lactobacillus lactis, nor Lactococcus lactis was able to convert pyruvate as the sole substrate at a rate comparable to that of O. oeni or L. mesenteroides. In summary, the Lactobacillus strains can use pyruvate as an electron sink and for pyruvate fermentation, but the rates, in particular that of pyruvate fermentation, are very low. Lactococcus, on the other hand, was not able to use pyruvate for fermentation or as an electron acceptor.
Genes for potential enzymes of pyruvate fermentation.
The genome sequences of O. oeni (strain PSU-1) and L. mesenteroides (strain ATCC 8293) were screened for genes encoding potential enzymes of a pyruvate fermentation pathway (Table 3). In O. oeni and L. mesenteroides, genes homologous to genes encoding pyruvate dehydrogenase, phosphotransacetylase, acetate kinase, and d-lactate dehydrogenase were found. The sequence predicted a classical NAD-dependent enzyme with an E1α E1β E2 E3 subunit composition (pdhABCD genes) typical for gram-positive bacteria (11). The genes encoding phosphotransacetylase (pta homologous), acetate kinase (ackA homologous), and d-lactate dehydrogenase (ldhA homologous) shared similarity with the corresponding genes of E. coli and other bacteria. For some of the enzymes, more than one gene cluster was found, suggesting the presence of isoenzymes (see Table 3).
TABLE 3.
Genes encoding potential enzymes of pyruvate fermentation by O. oeni and L. mesenteroidesa
| Enzyme | Gene(s) in:
|
Orthologous gene in E. coli | |
|---|---|---|---|
| O. oeni | L. mesenteroides | ||
| PDH | |||
| E1α | S3 G1158 | S56 G1496 | NPb |
| E1β | S3 G1159 | S56 G1497 | NP |
| E2 | S3 G1160 | S56 G1498 | aceF |
| E3 | S3 G1161 | S56 G1499 | lpdA |
| S6 G1574 | |||
| Phosphotransacetylase | S2 G766 | S25 G712 | pta |
| Acetate kinase | S6 G1856 | S36 G1026 | ackA |
| S42 G1163 | |||
| d-Lactate dehydrogenase | S5 G1762 | S24 G696 | ldhA |
| S1 G56 | S49 G1324 | ||
| Pyruvate oxidase (acetyl phosphate forming) | S37 G1370 | S1 G63 | poxB |
| S17 G518 | |||
| S25 G968 | |||
| S25 G969 | |||
| S25 G970 | |||
The genes of O. oeni and of L. mesenteroides were identified by functional domain searches and BLAST analysis using orthologous genes of E. coli or the other bacteria encoding the enzymes. The table gives the gene designations in O. oeni PSU-1 and L. mesenteroides ATCC 8293 according to the genomic database (http://www.jgi.doe.gov/) including the paralogs.
NP, no homologous genes present.
In both bacteria, in addition, genes encoding pyruvate oxidase of the Leuconostoc Pox type were found. Lactic acid bacteria are known to contain pyruvate oxidase which produces acetyl phosphate from pyruvate (pyruvate + O2 + Pi → acetyl phosphate + H2O2 + CO2) (29, 30). In O. oeni genes for four or five Pox type pyruvate oxidases were present, whereas in L. mesenteroides only one ORF could be identified. Genes encoding other enzymes for pyruvate decarboxylation, such as pyruvate ferredoxin oxidoreductase from anaerobic bacteria, pyruvate:quinone oxidoreductase (Pqo from Corynebacterium glutamicum or PoxB from E. coli), PFL, or pyruvate decarboxylase, were not detected in the genome of O. oeni or L. mesenteroides.
Enzyme activities of the pyruvate fermentation pathway.
The genes found in O. oeni and L. mesenteroides (Table 3) suggest a pathway for pyruvate fermentation as shown in Fig. 2, including pyruvate decarboxylation by pyruvate dehydrogenase, phosphoryl transfer from acetyl phosphate to ADP by acetate kinase, and NADH reoxidation by lactate dehydrogenase. The presence of the corresponding enzyme activities was tested in cell extracts of the bacteria after anoxic growth (Table 4). O. oeni contained high activities of lactate dehydrogenase, acetate kinase, and phosphotransacetylase after growth on pyruvate or glucose; only lactate dehydrogenase activity was diminished after growth on pyruvate. Acetate kinase showed the lowest activities. L. mesenteroides generally has higher growth rates than O. oeni and contains higher activities, and the activities were slightly decreased after growth on pyruvate.
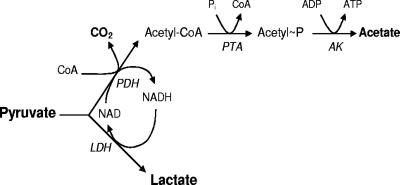
FIG. 2.
Scheme showing the intermediates in the pyruvate fermentation pathway of O. oeni and L. mesenteroides. The scheme shows the intermediates, the enzymes, and the supposed genes in O. oeni. LDH, d-lactate dehydrogenase; PTA, phosphotransacetylase; AK, acetate kinase.
TABLE 4.
Activities of enzymes related to pyruvate fermentation of O. oeni and L. mesenteroidesa
| Enzyme and growth condition | Activity (μmol of substrate/ g of protein/min) in:
|
|
|---|---|---|
| O. oeni | L. mesenteroides | |
| Lactate dehydrogenase | ||
| Pyruvate | 2,300 | 19,940 |
| Glucose | 10,400 | 35,080 |
| Acetate kinase | ||
| Pyruvate | 240 | 415 |
| Glucose | 194 | 910 |
| Phosphotransacetylase | ||
| Pyruvate | 5,810 | 13,150 |
| Glucose | 6,100 | 18,000 |
| Pyruvate dehydrogenase (acetyl phosphate formation) | ||
| Pyruvate | ≤6 | 102 |
| Glucose | ≤6 | 12 |
| Pyruvate dehydrogenase (pyruvate degradation) | ||
| Pyruvate | 208 | 425 |
| Glucose | <1 | 286 |
| Pyruvate oxidase | ||
| Pyruvate | <1 | <1 |
| Pyruvate + O2 | <1 | 98 |
Enzymes were measured in cell homogenates obtained from the bacteria after anoxic growth in MLD medium with pyruvate or glucose as the substrate.
Pyruvate dehydrogenase was tested in cell extracts in a coupled assay by the formation of acetyl phosphate, since measurement of NADH formation is susceptible to interference in cell homogenates due to the presence of NADH-oxidizing enzymes. L. mesenteroides contained much higher (8.5-fold) PDH activity in pyruvate-grown than in glucose-grown bacteria. In the same assay, O. oeni showed no activity of acetyl phosphate (or acetyl-CoA, respectively) formation. However, when pyruvate dehydrogenase was assayed under the same conditions by the decrease of pyruvate, the enzyme was found with substantial activity in pyruvate-grown (but not in glucose-grown) O. oeni. L. mesenteroides showed the same type of activity, which was again higher in the pyruvate-grown bacteria.
Pyruvate oxidase activity was determined as the O2-dependent conversion of pyruvate to acetyl phosphate. O. oeni contained no activity of this type, even after microoxic growth of the bacteria on pyruvate. L. mesenteroides, on the other hand, contained enzyme activity after microoxic, but not after anaerobic, growth on pyruvate, suggesting that this enzyme, which requires O2 for function, is formed or active only after (micro)aerobic growth.
Pyruvate-cleaving enzymes in LAB: different functions for pyruvate oxidase, pyruvate formate lyase, and pyruvate dehydrogenase.
LAB and the related bifidobacteria were screened for genes encoding enzymes for pyruvate cleavage. The genes were identified by a BLAST search and confirmed by the presence of the functional domains. Genes or gene clusters encoding PDH (pdhABCD genes), Pox (poxB gene),PFL (pflB or pflD gene), and PFOR (por gene, encoding the NifJ type of PFOR) were identified (Table 5). None of the bacteria contained genes for pyruvate decarboxylase or a pyruvate oxidase of the Pqo type. Most of the bacteria contained more than one gene or gene cluster encoding the PDH, Pox, PFL, and PFOR enzymes (Table 5). The most frequent enzyme was Pox, followed by PDH and PFL. The roles of the PDH, Pox, and PFL enzymes in the metabolism of the LAB are known at least in part, as discussed below, whereas for PFOR of Lactococcus lactis no function can be assigned.
TABLE 5.
Pyruvate-cleaving enzymes in lactic acid and related bacteria as deduced from genome sequencesa
| Bacteriumb | Orthologc of:
|
Homo- or heterofermentative | |||
|---|---|---|---|---|---|
| PDH | Pox | PFL | PFOR | ||
| O. oeni | PDH | Pox1 to 5 | NP | NP | Heterofermentative |
| L. mesenteroides | PDH | Pox | NP | NP | Heterofermentative |
| Lactobacillus brevis | PDH | NP | NP | NP | Heterofermentative |
| Lactobacillus plantarum | PDH | Pox1 to 5 | PflB1 and 2 | NP | Facultatively heterofermentative |
| Lactobacillus casei | PDH | Pox1 to 3 | NP | NP | Facultatively heterofermentative |
| Lactobacillus gasseri | NP | Pox | NP | NP | Homofermentative |
| Lactobacillus delbrueckii subsp. bulgaricus | NP | NP | NP | NP | Homofermentative |
| Lactobacillus johnsonii | NP | Pox | NP | NP | Homofermentative |
| Lactococcus lactis | PDH | Pox | Pfl | PFOR | Homofermentative |
| Lactococcus cremoris | PDH | Pox | Pfl | NP | Homofermentative |
| Pediococcus pentosaceus | PDH | Pox1 and 2 | NP | NP | Homofermentative |
| Bifidobacterium longum | NP | NP | Pfl | NP | |
The presence of PDH, pyruvate oxidase PoxB from Leuconostoc (forming acetyl phosphate + H2O2), PFL, and PFOR (por gene) orthologs is indicated. The positives were found by BLAST search and contain the functional domains characteristic for each enzyme.
Fully sequenced genomes are underlined; the other genomes are draft versions.
NP, no homologous genes present.
(i) Pyruvate oxidase.
Pox of the LAB catalyzes formation of acetyl phosphate (pyruvate + Pi + O2 → acetyl phosphate + CO2 + H2O2) (29) and was found in most of the homo- and heterofermentative bacteria. The enzyme is advantageous in (micro)aerobic growth, resulting in increased acetate production and higher ATP or cell yields, in particular under glucose limitation (15, 19, 20, 24, 30). Bacteria of the Lactobacillales group with respiratory capacity (Streptococcus strains) do not need the Pox enzyme for this purpose and lack the enzyme (not shown). By combined action of Pox and of a (nonrespiratory) NADH oxidase (12, 16), glucose is oxidized to acetate and CO2 (idealized reaction: glucose + 4 O2 → 2 acetate + 2 CO2 + 4 H2O2). This reaction yields 4 and 3 mol of ATP/mol of glucose for homo- and heterolactic bacteria, respectively, compared to 2 and 1 mol of ATP/mol of glucose in (anaerobic) homo- and heterolactic glucose fermentation.
(ii) Pyruvate formate lyase.
PFL is the key enzyme for mixed acid fermentation, with acetate, ethanol, and formate as the products in addition to lactate. PFL is found only in LAB capable of homofermentative growth (Table 5), which use this enzyme to shift to mixed acid formation under glucose limitation to increase the ATP yields (3 compared to 2 mol of ATP/mol of glucose in homolactic fermentation) (7, 17, 33). The restriction of PFL to the homofermentative LAB suggests that this is the major or only role for the enzyme in LAB.
(iii) Pyruvate dehydrogenase.
The PDH of gram-positive bacteria consists of four subunits (PdhABCD) and is strongly inhibited by NADH (30-32). Since the NADH/NAD+ ratio is high during glucose fermentation, PDH is supposed to be active mainly under aerobic conditions. Therefore, in (anoxic) glucose fermentation, pyruvate is mainly metabolized by lactate dehydrogenase and PFL (12). In the presence of O2, on the other hand, pyruvate is metabolized in significant amounts by PDH. The surplus NADH is reoxidized by NADH oxidase. In this way glucose is metabolized to acetate plus CO2 (1 glucose + 4 O2 → 2 acetate + 2 CO2 + 4 H2O2), yielding 4 or 3 mol of ATP/mol of glucose, respectively, from the glycolytic or pentose phosphate pathway, compared to 2 or 1 mol of ATP in homo- or heterolactic fermentation. The broad distribution of the PDH enzyme in the LAB and its aerobic expression suggest an important role for PDH (together with NADH oxidase) in pyruvate oxidation under oxic conditions.
The PDH-plus-NADH oxidase-dependent pathway functions alternatively to pyruvate oxidase with the same ATP yields. It is not known to what extent either pathway is used in the presence of O2 or whether the two pathways are used for different purposes. A similar situation exists in Escherichia coli, which contains PDH and pyruvate oxidase PoxB (pyruvate:quinone reductase). These two enzymes function with different energetic efficiencies, and their alternative function is not completely clear (1, 10). In O. oeni and other LAB which are limited in cellular HSCoA contents due to pantothenate auxotrophy (26), use of the HSCoA-independent pyruvate oxidase pathway could be advantageous.
Growth of heterolactic acid bacteria by pyruvate fermentation: a new function for pyruvate dehydrogenase.
O. oeni and L. mesenteroides are able to grow with pyruvate under anoxic conditions. The pyruvate is converted to lactate, acetate, and CO2 by the following pathway (see also Fig. 2): 2 pyruvate → 1 d-lactate + 1 acetate + 1 CO2. One molecule of pyruvate is decarboxylated to acetyl-CoA, which allows generation of ATP by acetate kinase. The ATP yield (0.5 mol/mol of pyruvate) is in agreement with the growth yields. The pathway was confirmed by the presence of the corresponding genes for the enzymes in the genomes of the bacteria (http://www.jgi.doe.gov/). The key enzyme of the pathway is PDH. The broad distribution of the PDH enzyme within LAB, most of which are not able to ferment pyruvate (Table 5), and its aerobic expression suggest that the principal function of the enzyme is pyruvate oxidation combined with the oxidation of NADH. The function of PDH in pyruvate fermentation presumably represents a “misuse” or adaptation for a pathway that is normally not intended for PDH. Pyruvate oxidase, which is also present in the bacteria, requires O2 for function and is therefore not suitable for pyruvate fermentation.
From the growth yield and rate, it can be calculated (see reference 26) that O. oeni and L. mesenteroides require activities of 250 and 1,330 U/g of protein for each of the enzymes of the pyruvate fermentation pathway. In O. oeni, only PDH (pyruvate-degrading activity) has slightly lower activity (208 U/g protein). L. mesenteroides also contains the enzyme activities, and only acetate kinase and pyruvate dehydrogenase activities are lower than expected. Pyruvate fermentation is not widespread among lactic acid bacteria. Thus, the heterofermentative L. plantarum and the homofermentative Lactobacillus lactis ferment pyruvate at low rates, but the reaction does not support substantial growth.
Only a few other fermentation reactions are known to use PDH instead of PFOR or PFL for cleavage of pyruvate. Thus, Bacillus subtilis ferments glucose in the absence of electron acceptors via pyruvate to lactate, acetate, and some other products. The acetate production depends on PDH, which normally functions in aerobic metabolism (5, 21, 22). Eubacterium pyruvativorans is one of the few bacteria growing by fermentation of external pyruvate (35). The fermentation products are short-chain fatty acids, and the fermentation presumably requires PFOR. Thus, E. pyruvativorans seems to be the only other bacterium described in addition to O. oeni and L. mesenteroides that is capable of pyruvate fermentation, but the fermentation reactions are different.
Acknowledgments
The work was supported by a grant from the Stiftung Rheinland-Pfalz für Innovation and by the Fonds der Chemischen Industrie to G.U.
The expert support of D. Vlad for some experiments is gratefully acknowledged.
REFERENCES
- 1.Abdel-Hamid, A. M., M. M. Attwood, and J. R. Guest. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483-1498. [DOI] [PubMed] [Google Scholar]
- 2.Bergmeyer, H. U. 1983. Acetate kinase from Escherichia coli, p. 126-129. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 3rd ed., vol. II. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cavin, J. F., H. Prevost, J. Lin, P. Schmitt, and C. Davies. 1989. Medium for screening Leuconostoc oenos strains defective in malolactic fermentation. Appl. Environ. Microbiol. 55:751-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz Ramos, H., T. Hoffmann, M. Marino, H. Nedjari, E. Presecan-Siedel, O. Dreesen, P. Glaser, and D. Jahn. 2000. Fermentative metabolism of Bacillus subtilis: Physiology and regulation of gene expression. J. Bacteriol. 182:3072-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dicks, L. M., F. Dellaglio, and M. Collins. 1995. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 45:395-397. [DOI] [PubMed] [Google Scholar]
- 7.Fordyce, A. M., V. L. Crow, and T. D. Thomas. 1984. Regulation of product formation during glucose or lactose limitation in nongrowing cells of Streptococcus lactis. Appl. Environ. Microbiol. 48:332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garvie, E. I. 1967. Leuconostoc oenos sp. nov. J. Gen. Microbiol. 48:431-438. [DOI] [PubMed] [Google Scholar]
- 9.Garvie, E. I. 1986. Genus Leuconostoc, p. 1071-1975. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 10.Guest, J. R., A. M. Abdel-Hamid, G. A. Auger, L. Cunningham, R. A. Henderson, R. S. Machado, and M. M. Attwood. 2004. Physiological effects of replacing the PDH complex of E. coli by genetically engineered variants or by pyruvate oxidase, p. 387-405. In F. Jordan and M. S. Patel (ed.), Thiamine: catalytic mechanisms and role in normal and disease states. Marcel Dekker, Inc., New York, N.Y.
- 11.Henderson, C. E., and R. N. Perham. 1980. Purification of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus and resolution of its four component polypeptides. Biochem. J. 189:161-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, N. B., C. R. Melchiorsen, K. V. Jokumsen, and J. Villadsen. 2001. Metabolic behaviour of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Appl. Environ. Microbiol. 67:2677-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klotzsch, H. R. 1969. Phosphotransacetylase from Clostridium kluyveri. Methods Enzymol. 13:381-386. [Google Scholar]
- 14.Lipmann, F., and C. Tuttle. 1945. A specific micromethod for determination of acyl phosphates. J. Biol. Chem. 159:21-28. [Google Scholar]
- 15.Lorquet, F., P. Goffin, L. Muscariello, J. Baudry, V. Ladero, M. Sacco, M. Kleerebezem, and P. Hols. 2004. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 186:3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maicas, S., S. Ferrer., and I. Pardo. 2002. NAD(P)H regeneration is the key for heterolactic fermentation of hexoses in Oenococcus oeni. Microbiology 148:325-332. [DOI] [PubMed] [Google Scholar]
- 17.Melchiorsen, C. R., K. Jokumsen, J. Villadsen, H. Israelsen, and J. Arnau. 2001. The level of pyruvate-formate lyase controls the shift from homolactic to mixed-acid product formation in Lactococcus lactis. Appl. Microbiol. Biotechnol. 58:338-344. [DOI] [PubMed] [Google Scholar]
- 18.Mulder, N. J., R. Apweiler, T. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. Orchard, M. Pagni, D. Peyruc, C. Ponting, J. Selengut, F. Servant, C. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, M. G., and S. Condon. 1984. Correlation of oxygen utilization and hydrogen peroxide accumulation with oxygen induced enzymes in Lactobacillus plantarum cultures. Arch. Microbiol. 138:44-48. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, M. G., L. O'Connor, D. Walsh, and S. Condon. 1985. Oxygen dependent lactate utilization by Lactobacillus plantarum. Arch. Microbiol. 141:75-79. [DOI] [PubMed] [Google Scholar]
- 21.Nakano, M. M., Y. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth in Bacillus subtilis: identification of fermentation end products and genes required for growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a strict aerobe (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 23.Nuraida, L., I. Grigolava, J. D. Owens, and G. Campbell-Platt. 1992. Oxygen and pyruvate as external electron acceptors for Leuconostoc spp. J. Appl. Bacteriol. 72:517-522. [Google Scholar]
- 24.Ramos, A., A. R. Neves, R. Ventura, C. Mayock, P. López, and H. Santos. 2004. Effect of pyruvate kinase overproduction on glucose metabolism of Lactococcus lactis. Microbiology 150:1103-1111. [DOI] [PubMed] [Google Scholar]
- 25.Reed, L. J., and C. R. Willms. 1966. Purification of the pyruvate dehydrogenase complex (Escherichia coli). Methods Enzymol. 9:247-265. [Google Scholar]
- 26.Richter, H., D. Vlad, and G. Unden. 2001. Significance of pantothenate for glucose fermentation by Oenococcus oeni and for suppression of the erythritol and acetate production. Arch. Microbiol. 175:26-31. [DOI] [PubMed] [Google Scholar]
- 27.Richter, H., I. Hamann, and G. Unden. 2003. Use of the mannitol pathway in fructose fermentation of Oenococcus oeni due to limiting redox regeneration capacity of the ethanol pathway. Arch. Microbiol. 179:227-233. [DOI] [PubMed] [Google Scholar]
- 28.Richter, H., A. A. De Graaf, I. Hamann, and G. Unden. 2003. Significance of phosphoglucose isomerase for the shift between heterolactic and mannitol fermentation of fructose by Oenococcus oeni. Arch. Microbiol. 180:465-470. [DOI] [PubMed] [Google Scholar]
- 29.Sedewitz, B., K. H. Schleifer, and F. Götz. 1984. Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum. J. Bacteriol. 160:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sedewitz, B., K. H. Schleifer, and F. Götz. 1984. Physiological role of pyruvate oxidase in the aerobic metabolism of Lactobacillus plantarum. J. Bacteriol. 160:462-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snoep, J. L., M. J. Teixeira de Mattos, M. Starrenburg, and J. Hugenholtz. 1992. Isolation, characterization, and physiological role of the pyruvate dehydrogenase complex and α-acetolactate synthase of Lactococcus lactis subsp. lactis bv. diacetylactis. J. Bacteriol. 174:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snoep, J. L., M. R. de Graef, A. Westphal, A. de Kok, T. J. de Mattos, and O. Neijssel. 1993. Differences in sensitivity to NADH of purified dehydrogenase complex of Enterococcus faecalis, Lactococcus lactis, Azotobacter vinelandii and Escherichia coli: implications for their activity in vivo. FEMS Microbiol. Lett. 114:279-283. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, T. D., K. W. Turner, and V. L. Crow. 1980. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J. Bacteriol. 144:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veiga-da-Cunha, M., H. Santos, and E. von Schaftingen. 1993. Pathway and regulation of erythritol formation in Leuconostoc oenos. J. Bacteriol. 175:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace, R. J., N. McKain, N. R. McEwan, E. Miyagawa, L. C. Chaudhary, T. P. King, N. D. Walker, J. H. Apajalahti, and C. J. Newbold. 2003. Eubacterium pyruvativorans sp. nov., a novel non-saccharolytic anaerobe from the rumen that ferments pyruvate and amino acids, forms caproate and utilizes acetate and propionate. Int. J. Syst. Evol. Microbiol. 53:965-970. [DOI] [PubMed] [Google Scholar]