Abstract
Rhodospirillum rubrum is a phototrophic purple nonsulfur bacterium known for its unique and well-studied nitrogen fixation and carbon monoxide oxidation systems and as a source of hydrogen and biodegradable plastic production. To better understand this organism and to facilitate assembly of its sequence, three whole-genome restriction endonuclease maps (XbaI, NheI, and HindIII) of R. rubrum strain ATCC 11170 were created by optical mapping. Optical mapping is a system for creating whole-genome ordered restriction endonuclease maps from randomly sheared genomic DNA molecules extracted from cells. During the sequence finishing process, all three optical maps confirmed a putative error in sequence assembly, while the HindIII map acted as a scaffold for high-resolution alignment with sequence contigs spanning the whole genome. In addition to highlighting optical mapping's role in the assembly and confirmation of genome sequence, this work underscores the unique niche in resolution occupied by the optical mapping system. With a resolution ranging from 6.5 kb (previously published) to 45 kb (reported here), optical mapping advances a “molecular cytogenetics” approach to solving problems in genomic analysis.
Rhodospirillum rubrum is one of two spiral Rhodospirillum species belonging to the alphaproteobacteria class of phototrophic purple nonsulfur bacteria (13, 14, 22). Found in aquatic environments such as lakes, streams, and standing water, R. rubrum is known for its carbon and nitrogen metabolism (6, 13, 14, 27), and its potential to produce hydrogen and biodegradable plastics (7, 10, 12, 23, 24, 28). Specifically, R. rubrum possesses the rare ability to oxidize carbon monoxide to carbon dioxide and has been the subject of studies seeking to understand the mechanisms and regulation of this process (6).
R. rubrum has proven to efficiently convert hydrogen to electrical current in fuel cells (23) and produce novel forms of biodegradable thermoplastics when grown on assorted β-hydroxycarboxylic acids and n-alkanoic acids (7). More recently, Handrick et al. (12) reported on R. rubrum's activator role in the degradation of polyhydroxybutyrate, a polymer of interest for its thermoplasticity and breakdown to water and carbon dioxide. Finally, from a biochemical standpoint, Zhang and others (29) studied the mechanisms behind R. rubrum's posttranslational regulation of nitrogenase activity.
To supplement understanding of R. rubrum's biology, the organism was sequenced by the Department of Energy Joint Genome Institute and finished by the Los Alamos Finishing Group. In order to aid in sequence assembly, three whole-genome restriction endonuclease maps of R. rubrum with resolutions ranging from 11 to 45 kb were created. Physical maps are an excellent means by which to independently validate sequence assembly, close sequence contig gaps, and resolve repeat-rich regions, which consistently confound sequence assembly methods (17, 20-22, 25). In comparison to other physical mapping techniques, the automation and resolution of optical mapping make the system ideal for addressing a wide range of problems in genomics, including the finishing and checking of microbial sequencing projects (4, 17, 30-32). In addition, the use of genomic DNA as the source of single molecules for mapping eliminates the need for libraries, PCR, or separations and makes optical mapping advantageous for whole-genome mapping and sequence assembly.
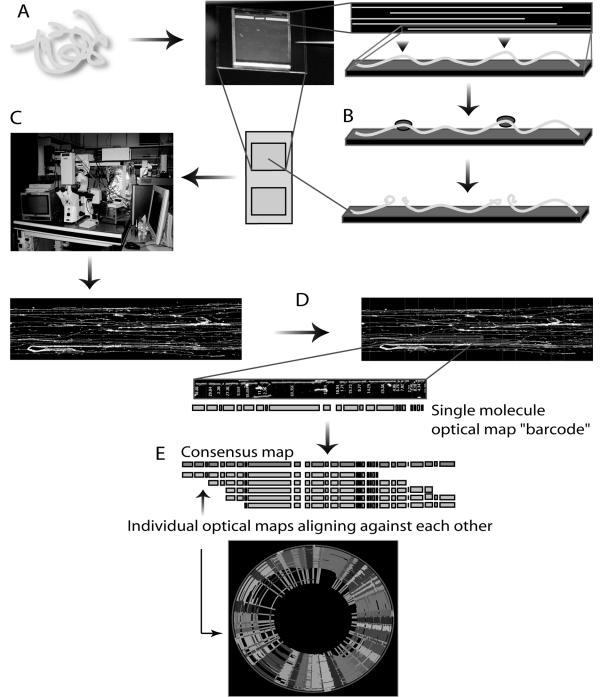
Optical mapping enables the construction of whole-genome restriction endonuclease maps from ensembles of single DNA molecules that have been elongated and immobilized on positively charged glass surfaces and subsequently cut with a restriction endonuclease (Fig. 1). The resulting single DNA molecule restriction endonuclease maps are stained with a fluorochrome and visualized by fluorescence microscopy. Because the order of restriction endonuclease fragments is retained on the optical mapping surface, there is no need for sorting fragments by size. The mass of each restriction endonuclease fragment is determined by integrated fluorescence intensity measurements. The single-molecule restriction endonuclease maps are assembled into contigs (2, 3, 16, 18), in a process similar to shotgun sequence assembly, that span entire microbial genomes. The depth of coverage minimizes mapping error and the overlapping cascades of optical maps create continuity of coverage across an entire genome.
FIG. 1.
Optical mapping process. A) Genomic DNA is extracted directly from cells and elongated and immobilized onto a positively charged glass surface through a combination of microfluidics and electrostatic interactions. B) A restriction enzyme cleaves the surface-bound molecules, which are then stained with a fluorescent dye. C) The digested single DNA molecules are imaged by a fully automated fluorescence microscopy scanning system that rapidly creates high-resolution images. D) Using the integrated fluorescence intensity measurements of the imaged restriction fragments, machine vision calculates the mass of each fragment. E) Ordered restriction maps, each representing a single DNA molecule, are assembled into contigs with consensus maps that span entire genomes.
Here, three optical maps (XbaI, NheI, and HindIII) of the R. rubrum strain ATCC 11170 genome that aided in sequence assembly are presented. The high-resolution HindIII map confirmed the correct placement of the sequence contigs generated during the finishing process, and the lower-resolution NheI and XbaI maps confirmed the final sequence assembly. All three maps confirmed a sequence assembly error, the correction of which filled a gap in the sequence. With a resolution of 45 kb, the XbaI map is the lowest-resolution optical map reported to date. With a documented resolution of 6.5 kb (30) to 45 kb, optical mapping fills a critical niche in the resolution capabilities of genomic analysis systems. Here we show the utility of optical mapping's resolution range in microbial sequence assembly, and comment briefly on the advantages of optical mapping for solving additional problems in genomics.
MATERIALS AND METHODS
DNA preparation.
R. rubrum strain ATCC 11170 genomic DNA gel inserts (26) were prepared from a culture grown aerobically at 30°C in supplemented malate-ammonium medium supplemented with 10 μM NiCl2 (9) and stored in 0.5 M EDTA (pH 8.0). Prior to use, the DNA inserts were washed thoroughly overnight in TE (Tris-EDTA, pH 8.0) to remove excess EDTA. After melting the agarose inserts at 72°C for 7 min, the agarose was digested at 42°C for 2 h in β-agarase solution (100 μl TE, 1 μl [1 unit] NEB β-agarase [New England Biolabs, Beverly, MA]). The resulting concentrated DNA was diluted in TE to a concentration of a few pg/μl, to ensure minimal crowding of single DNA molecules on the optical mapping surfaces. Lambda DASH II bacteriophage DNA (Stratagene, La Jolla, CA) was added to the genomic DNA dilution to a concentration of 10 pg/μl as an internal standard for fragment sizing. The samples were mounted onto an optical mapping surface and inspected by fluorescence microscopy (details below) for molecular integrity and appropriate concentration.
Surface preparation.
Glass coverslips (22 by 22 mm, Fisher's Finest; Fisher Scientific, Pittsburgh, PA) were cleaned and derivatized as described previously (30). Surface properties were assayed by digesting lambda DASH II bacteriophage DNA with 40 units of XbaI, HindIII, and NheI diluted in 200 μl of digestion buffer with 0.2% Triton X-100 (Sigma, St. Louis, MO) at 37°C to determine optimal digestion times, which ranged from 30 to 120 min.
DNA mounting, overlay, digestion, and staining.
DNA molecules were mounted on derivatized glass surfaces by capillary action using a microfluidic device (8). Capillary flow elongates the DNA molecules; they are then immobilized by electrostatic interactions between the positively charged glass surface and negatively charged DNA molecules. Following DNA elongation and deposition, a thin layer of acrylamide (3.3% containing 0.02% Triton X-100 [Sigma, Pittsburgh, PA]) was applied to the surface. After polymerization, the surfaces were washed with 400 μl of TE for 2 min, followed by washing with 200 μl of digestion buffer for another 2 min. The digestion was then performed by adding 200 μl of digestion buffer with enzyme (20 μl of 10x buffer 2 [100 mM Tris, 500 mM NaCl, 100 mM MgCl2, 10 mM dithiothreitol, pH 7.9] [New England Biolabs, Beverly, MA], 176 μl high-purity water, 2 μl 2% Triton X-100 [Sigma, Pittsburgh, PA], and 4 μl of HindIII [New England Biolabs, Beverly, MA] or EcoRI [New England Biolabs, Beverly, MA] [10 unit/μl] or 2 μl XbaI [New England Biolabs, Beverly, MA] [20 units/μl]) to the surface and incubating in a humidified chamber at 37°C for 30 to 120 min.
Following digestion, the surfaces were washed twice by adding 400 μl of TE, waiting 2 to 5 min, and the solution was removed by aspiration. The surfaces were mounted onto a glass slide with 12 μl 0.2 μM YOYO-1 solution (containing 5 parts YOYO-1 [1,1′-[1,3-propanediylbis[(dimethyliminio)-3,1-propanediyl]]bis[4-[(3-methyl-2(3H)-benzoxazolylidene)-methyl]]-tetraiodide; Molecular Probes, Eugene, OR] in 95 parts of 14.3 M β-mercaptoethanol in 20% TE vol/vol). The edges of the glass surface were sealed to the glass slide with nail polish and incubated (4°C in the dark) for at least 20 min so the staining dye could diffuse before checking by fluorescence microscopy.
Image acquisition and processing.
The samples were imaged by fluorescence microscopy as previously described (17) using a 63x objective (Zeiss, Thornwood, NY) and a high-resolution digital camera (Princeton Instruments, Trenton, NJ). Single overlapping images, spanning the full length of the microfluidic channels, were collected, flattened, and superimposed by a fully automated image acquisition system, ChannelCollect (8). These flattened and overlapped superimages were then processed through the Pathfinder software (Rod Runnheim, unpublished), which identifies digested molecules to be made into single molecule maps. Features that are recognized as DNA molecules are denoted and created into an ordered restriction endonuclease map for that molecule. Comounted Lambda DASH II molecules were used to estimate the digestion rate and to provide internal fluorescence standards for accurately sizing the DNA fragments (1, 18, 20). Each digested genomic DNA molecule selected by Pathfinder becomes a single molecule optical map.
Optical map assembly.
The custom-written software Gentig (1-3, 16-18) overlapped the single-molecule restriction endonuclease maps by aligning restriction endonuclease sites based on fragment sizes. Gentig assembles the individual molecule restriction endonuclease maps into a contig that spans the entire genome. Bayesian inference estimates the probability that two single-molecule restriction endonuclease maps, while subject to various data errors stemming from sizing, missing restriction endonuclease sites (missing cuts), and spurious restriction endonuclease sites (false cuts), may have been derived from the proposed placement. A known statistical distribution of the error sources is required for the Bayesian approach, as is fine-tuning of parameters such as standard deviation, digestion rate, false cut, and false match probability. These parameters can be reestimated from the data using a limited number of iterations of Bayesian probability density maximization.
Once these parameters have been accurately estimated from the data, an efficient dynamic programming algorithm computes the best offset and alignment between a pair of maps. The accuracy of an optical map as its own entity is estimated by Gentig's ability to assess a set of hypothetical maps against the optical map data set and, using error models, report a false-positive probability (2). For circular genomes, this is reported as the false circularization probability. The value represents the probability that the circular contig created by Gentig is a false positive.
DNA sequencing.
Two randomly sheared libraries of the R. rubrum strain ATCC 11170 genome were produced with 3-kb inserts (plasmids) and 40-kb inserts (fosmids). These libraries were sequenced to a total depth of approximately 11x and all reads were quality assessed and trimmed for vector sequence before being used for assembly.
For the 3-kb DNA shearing and plasmid subcloning, approximately 3 to 5 μg of isolated DNA was randomly sheared to 3- to 4-kb fragments (25 cycles at speed code 12) in a 100-μl volume using a HydroShear (GeneMachines, San Carlos, CA). The sheared DNA was immediately blunt end-repaired at room temperature for 40 min using 6 U of T4 DNA Polymerase (Roche, Basel, Switzerland), 30 U of DNA polymerase I Klenow Fragment (New England Biolabs, Beverly, MA), 10 μl of 10 mM deoxynucleoside triphosphate mix (Amersham Biosciences, Piscataway, NJ), and 13 μl of 10x Klenow buffer in a 130 μl total volume. After incubation the reaction was heat inactivated for 15 min at 70°C, cooled to 4°C for 10 min and then frozen at −20°C for storage. The end-repaired DNA was run on a 1% TAE (Tris-acetate-EDTA)-agarose gel for ∼30 to 40 min at 120 V. Using ethidium bromide stain and UV illumination, 3- to 4-kb fragments were extracted from the agarose gel and purified using QIAquick gel extraction kit (QIAGEN, Valencia, CA). Approximately 200 to 400 ng of purified fragment was blunt-end ligated for 40 min into the SmaI site of 100 ng of pUC18 cloning vector (Roche, Basel, Switzerland) using the Fast-Link DNA ligation kit (Epicentre, Madison, WI).
Following standard protocols, 1 μl of ligation product was electroporated into DH10B Electromax cells (Invitrogen, Carlsbad, CA) using the Gene Pulser II electroporator (Bio-Rad, Hercules, CA). Transformed cells were transferred into 1,000 μl of SOC-medium and incubated at 37°C in a rotating wheel for 1 h. Cells (usually 20 to 50 μl) were spread on LB agar plates, 22 by 22 cm, containing 100 μg/ml of ampicillin, 120 μg/ml of isopropylthiogalactopyranoside (IPTG), and 50 μg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Colonies were grown for 16 h at 37°C. Individual white recombinant colonies were selected and picked into 384-well microtiter plates containing LB/glycerol (7.5%) medium containing 50 μg/ml of ampicillin using the Q-Bot multitasking robot (Genetix, Dorset, United Kingdom).
To test the quality of the library, 24 colonies were directly PCR amplified with pUCM13 −28 and −40 primers using standard protocols. Libraries were considered high quality if they had >90% 3-kb inserts. For more details see http://www.jgi.doe.gov/sequencing/protocols/General3kbLibraryCreationSOP.doc and http://www.jgi.doe.gov/sequencing/protocols/FosmidLibraryCreationSOP.DOC.
For the plasmid amplification and sequencing steps, 2-μl aliquots of saturated Escherichia coli DH10B cultures containing pUC18 vector with random 3- to 4-kb DNA inserts grown in LB/glycerol (7.5%) medium containing 50 μg/ml of ampicillin were added to 8 μl of a 10 mM Tris-HCl (pH 8.2), 0.1 mM EDTA denaturation buffer. The mixtures were heat lysed at 95°C for 5 min and then placed at 4°C for 5 min. To these denatured products 10 μl of a rolling circle amplification (RCA) reaction mixture (TempliPhi DNA sequencing template amplification kit, Amersham Biosciences, Piscataway, NJ) were added. The amplification reactions were carried out at 30°C for 12 to 18 h. The amplified products were heat inactivated at 65°C for 10 min then placed at 4°C until used as the template for sequencing.
Aliquots of the 20 μl of amplified plasmid RCA products were sequenced with standard M13 −28 or −40 primers. The reactions contained 1 μl of RCA product, 4 pmol of primer, 5 μl of distilled H2O, and 4 μl of DYEnamic ET terminator sequencing kit (Amersham Biosciences, Piscataway, NJ). Cycle sequencing conditions were 30 rounds of 95°C for 25 seconds, 50°C for 10 seconds, 60°C for 2 min, and then held at 4°C. The reactions were then purified by a magnetic bead protocol [for more details see http://www.jgi.doe.gov/sequencing/protocols/DYEnamicET-TerminatorCycleSequencing(10ulrxn)SOP.doc] and run on a MegaBACE 4000 (Amersham Biosciences, Piscataway, NJ). Alternatively, 1 μl of the RCA product was sequenced with 2 pmol of standard M13 −28 or −40 primers, 1 μl 5x buffer, 0.8 μl H2O, and 1 μl BigDye sequencing kit (Applied Biosystems, Foster City, CA) at 1 min denaturation and 25 cycles of 95°C for 30 seconds, 50°C for 20 seconds, 60°C for 4 min, and finally held at 4°C. The reactions were then purified by a magnetic bead protocol and run on an ABI PRISM 3730 (Applied Biosystems, Foster City, CA) capillary DNA sequencer. Detailed protocols for fosmid library creation, fosmid DNA isolation and cleanup procedure can be found at http://www.jgi.doe.gov/Internal/protocols/prots_production.html.
In the sequence finishing process, all drafted reads were assembled together with SPS Phrap (SPSOFT, Albuquerque, NM). Repetitive regions of the genome were resolved with repFinisher (Cliff S. Han, unpublished). Autofinish (11) was used in the first cycle of finishing to select sequencing reactions. Remaining gaps and low quality regions closed with primer walking on subclones or by shattering PCR fragments covering the gaps.
Optical maps versus DNA sequence-based maps.
Alignments between the optical maps and DNA sequenced-based maps from the seven finishing-stage sequence contigs were created with the MapViewer software (OpGen, Inc., Madison, WI), a Perl/Tk application that provides an intuitive graphical interface for optical map analysis. In addition to creating and displaying alignments of optical maps, MapViewer allows the user to manipulate the relative positions and orientations as well as the scale of the optical maps to better understand these alignments. The map alignments are generated with a dynamic programming algorithm that finds the optimal alignment of two restriction endonuclease maps according to a scoring model that incorporates fragment sizing errors, false and missing cuts, and missing small fragments. For a given alignment, the score is proportional to the log of the length of the alignment, penalized by the differences between the two maps, such that longer, better matching alignments will have a higher score.
Using Gentig, the XbaI, NheI, and HindIII maps were aligned separately with the DNA sequence-based HindIII, XbaI, and NheI maps generated from the finished sequence. These initial alignments enabled determination of missing fragments, false cuts, or missing cuts. The relative sizing error for each fragment in the optical maps was calculated from the formula [100% × (optical map fragment size - corresponding DNA sequenced-based map fragment size)/corresponding DNA sequence-based map fragment size] and was plotted against the DNA sequence-based map fragment sizes to show the relationship between fragment size and relative error.
RESULTS
Acquisition of optical map data and construction of optical maps.
The three restriction endonuclease maps presented here were created via whole-genome shotgun optical mapping (1-3, 16, 18). This mapping strategy has parallels to whole genome shotgun sequencing; large numbers of optical maps, analogous to “sequence reads,” are assembled to cover any given locus.
The resolution of the optical maps affects the contig rate and average molecule size in the contig (Table 1). For the XbaI map, a total of 405 digested molecules were imaged and processed. Of this total, 204 were included in the whole-genome map contig, giving a contig rate of 50%. This low contig rate can be explained in part by the low resolution of the optical map. A map with an average fragment size of 44.73 kb requires very large genomic DNA molecules for contig assembly, due to the number of fragments in a single-molecule map required for confidence in merging that map into a map contig. The average size of molecules in the XbaI contig was about 900 kb, whereas the average size of collected DNA molecules was 637.69 kb. In addition, the digestion rate was calculated at 76.01%, which is lower than the target digest rate of 80% or higher. While still acceptable, a digest rate of 76.01% will reduce the density of apparent restriction endonuclease sites, or markers, thus increasing the difficulty in creating a contig. However, even with the low rate of contig formation, the total mass of molecules in the contig was 184.23 Mb, which corresponds to about 42-fold coverage.
TABLE 1.
R. rubrum optical mapping data
| Parameter | Enzyme
|
||
|---|---|---|---|
| XbaI | NheI | HindIII | |
| Data collection | |||
| No. of molecules | 405 | 409 | 932 |
| Avg molecule size (kb) | 637.69 | 635.72 | 411.76 |
| Total mass (Mb) | 258.26 | 260.01 | 383.76 |
| Molecules in the contigs | |||
| No. of molecules | 204 | 345 | 623 |
| Avg molecule size (kb) | 903.07 | 731.30 | 405.49 |
| Total mass (Mb) | 184.23 | 252.30 | 252.68 |
| Contig rate (%) | 50 | 84 | 67 |
| Genome size (kb) | 4,323.08 | 4,223.13 | 4,456.35 |
| Average fragment size (kb) | 44.73 | 31.53 | 10.95 |
| Average fragment size standard deviation (kb) | 4.31 | 2.83 | 1.16 |
| Circularization false probability | 0.00738 | 0.00329 | 0.00440 |
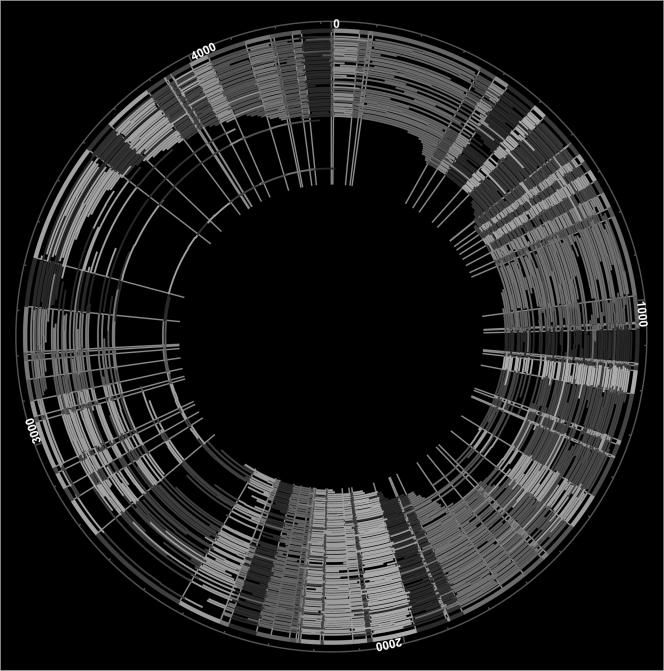
Figure 2 shows the finished XbaI map, which resembles a classical macrorestriction endonuclease map in terms of resolution. Notably, the whole-genome contig was circularized without gaps, and a typical restriction endonuclease fragment was calculated from the average of about 30 fragments. The sizing error per single fragment, or precision, was calculated from the set of restriction endonuclease fragments used to determine each consensus fragment. The average standard deviation of fragment size about the mean was 4.31 kb. The size of the R. rubrum XbaI circular contig was 4,323.08 kb, and was calculated by summing the restriction endonuclease fragments in the XbaI consensus map. This map is the lowest-resolution optical map created to date.
FIG. 2.
Whole-genome circular XbaI map of R. rubrum. The outermost circle (thick line) represents the consensus map created by Gentig from the single-molecule maps shown as arcs. The single-molecule maps were made from single DNA molecules digested with XbaI. The color ordering of the fragments is for contrast and is therefore random.
With an average fragment size of 31.53 kb, the NheI map has a resolution in between those of the XbaI and HindIII maps. A total of 409 molecules were collected and processed to form the NheI map. Of the total, 345, or 84% of the molecules, went into the circular contig. The average size of the collected molecules was 635.72 kb, only 2 kb smaller than the average size of molecules collected for the XbaI map. However, the average size of molecules in the contig was 731.30 kb, almost 200 kb smaller than the average size of the molecules in the XbaI contig. As the average fragment size of the NheI map is smaller, a molecule of a given size will have more restriction endonuclease fragments, and smaller molecules can be included in the contig. Thus, in comparison to the XbaI map, the increased NheI contig rate is due in part to the smaller average fragment size. In addition, the digestion rate was calculated to be 87%, which means that the patterns were more informative and accurate in this map. The mass of the molecules in the contig was 252.30 Mb; this represents approximately 60-fold coverage. The contig circularized without gaps, and about 42 fragments were used to calculate a given fragment mass in the consensus map. Based on the final NheI consensus map, the average standard deviation of the fragment size about the mean was 2.83 kb. Summing the masses of all of the restriction endonuclease fragments in the consensus map gave a total genome size of 4,223.13 kb.
Finally, 932 molecules were collected for the production of the HindIII map. With an average fragment size of 10.95 kb, this map is the highest resolution of the set of maps presented here. Of the 932 molecules, 623, or 67% of them went in to the final contig. The smaller average fragment size loosened the requirements for molecule size in the contig; the average size of molecules in the contig was 405.49 kb. The total mass represented by the molecules in the contig was 252.68 Mb, which corresponds to about 57-fold coverage, based on the HindIII contig size of 4,456.35 kb. Again, the size of the HindIII contig was calculated by summing the masses of the restriction endonuclease fragments in the consensus map. The digestion rate of the molecules in the contig was calculated to be 78.9%. An average of about 31 fragments was used to calculate the mass of each fragment in the consensus map. For each fragment in the consensus map, the standard deviation about the mean was 1.16 kb.
In all of the maps, the high coverage ensured accurate calling of restriction endonuclease sites, fragment sizing, and sizing of the entire circularized genome map. Below, the accuracy of the optical maps compared to the sequence is assessed. The false circularization probability for the XbaI, NheI, and HindIII maps was 0.00738, 0.00329, and 0.00440, respectively. Since the XbaI map had the lowest coverage, contig rate, and digestion rate, it is not surprising that the false circularization probability for this map is the highest. However, for all the maps, the false probability values were well below 0.05, which is considered the upper limit for confident map circularization (30). The restriction endonuclease patterns generated by the XbaI, NheI, and HindIII maps appeared random; no particular restriction endonuclease patterns or structural features were observed.
Use of optical maps in sequence assembly.
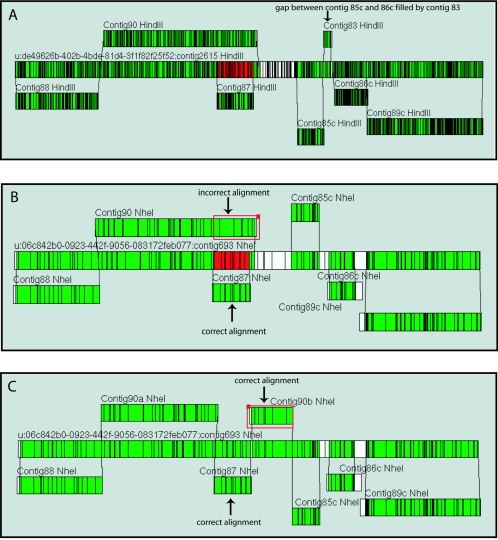
All of the optical maps were made in order to guide and verify the R. rubrum genome sequence assembly process. Near the end of the finishing effort, nine sequence contigs were generated ranging in size from 2311 base pairs to 1,465,886 base pairs. Alignment of the optical maps against the DNA sequence-based maps of the sequence contigs gave three independent indications of sequence contig assembly and order. Two of the sequence contigs did not align against the optical maps. They were contig 84, the plasmid sequence contig, and contig 82c, which, with a size of 2.311 kb, was too small to align with the optical maps. Six of the seven remaining contigs aligned to both the XbaI and NheI maps. Only the HindIII map, with its higher resolution, was able to align all seven sequence contigs, including contig 83 with its small size of 80.404 kb (Fig. 3A).
FIG. 3.
Use of optical maps in confirming assembly and order of sequence contigs. In silico maps of the sequence contigs were made and aligned against the whole-genome optical maps located in the center of each diagram. A) The high-resolution HindIII map enabled alignment of the small (∼80 kb) sequence contig (contig 83) between contigs 85c and 86c. Aside from the redundant alignment of contigs 87 and 90, no gaps remained in alignment between the optical map and sequence. B) The NheI map gave the clearest representation of the redundant alignment, shown in red in the optical map, of contigs 87 and 90. The alignment of contig 87 had a better match to the optical map than do the rightmost eight fragments in contig 90 (shown in the red square). C) Removing the rightmost eight fragments from contig 90 to make contigs 90a and 90b and reversing the orientation of 90b permitted alignment to the gap between contig 87 and contig 85c. Alignments were made with MapViewer Software (OpGen, Inc, Madison WI.).
All three comparisons of optical map to sequence supported a problematic assembly of sequence contig 90. Alignment of the NheI map to the sequence contigs best illustrates this (Fig. 3B). Contig 87 and the rightmost eight fragments of contig 90 align to the region in red in the NheI map. Inspection shows a cleaner alignment of the region with contig 87. Removing the rightmost eight fragments from contig 90 and inverting the orientation produced a solid alignment with the gap in the NheI map that was between contig 90 and contig 85c (Fig. 3C). Our realization of this problematic assembly confirmed the Los Alamos finishing group's suspicions of an erroneous assembly in this region. Elsewhere in the genome, there was good agreement between sequence contigs and optical maps.
The finished sequence (GenBank accession number AAAG00000000) contained a 4.4-Mb circular chromosome (contig 94, exact size is 4,352,726 base pairs) and a 54 kb plasmid (contig 93, exact size is 54,412 base pairs). Minor differences have been found between the optical maps and finished sequence and are described below.
Assessment of optical mapping errors.
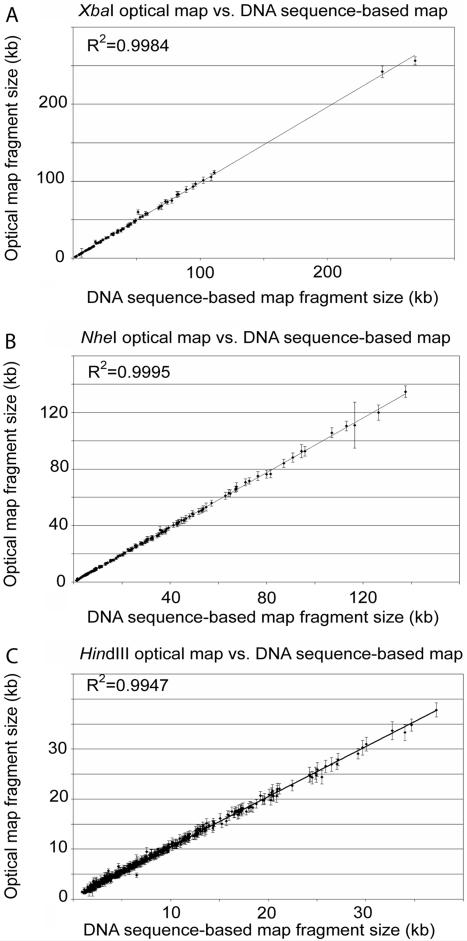
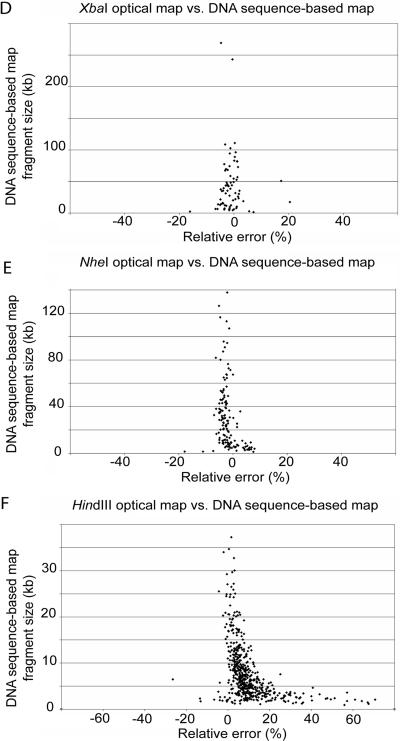
Comparisons between sequence and optical mapping data were made in order to evaluate the errors and accuracy in the XbaI, NheI, and HindIII maps (Fig. 4). The relative sizing error was calculated by the alignment of optical maps with the DNA sequence-based maps made from the finished sequence (Fig. 4A to F). The error bars in Figs. 4A, 4B, and 4C reflect the standard deviation about the means of the restriction endonuclease fragment sizes used in calculating the consensus map fragments. In general, a high degree of correspondence was evident between the optical map and DNA sequence-based map fragment sizes. The regression values for the trendlines are 0.9985, 0.9995, and 0.9947 for the XbaI, NheI, and HindIII maps, respectively.
FIG. 4.
Comparisons of the XbaI, NheI, and HindIII optical maps to sequence data. (A, B, and C) Plots of optical map fragment sizes versus the DNA sequence-based map fragment sizes from the finished sequence for XbaI (A), NheI (B), and HindIII (C). The error bars represent the standard deviation of optical map fragment size about the mean. (D, E, F) Plots of the relative fragment sizing error [100 × (optical map fragment size - corresponding DNA sequence-based map fragment size)/corresponding DNA sequence-based map fragment size] versus DNA sequence-based map fragment size for XbaI (D), NheI (E), and HindIII (F).
Figures 4D, 4E, and 4F are scatter plots showing the relationship between absolute relative fragment sizing error (optical map versus DNA sequence-based map) and restriction endonuclease fragment size. For the XbaI map, the average relative sizing error (see figure caption) was 6.20% for fragments smaller than 5 kb and 2.87% for fragments larger than 5 kb. For fragments smaller than 5 kb in the NheI map, the average relative sizing error was 5.87%, and it was 3.11% for fragments larger than 5 kb. Finally, for the HindIII map, the average relative sizing error was 16.70% for fragments smaller than 5 kb and 8.05% for fragments larger than 5 kb. The positive skew in the relative error plots for small fragments in Fig. 4 are discussed in greater detail in the following section.
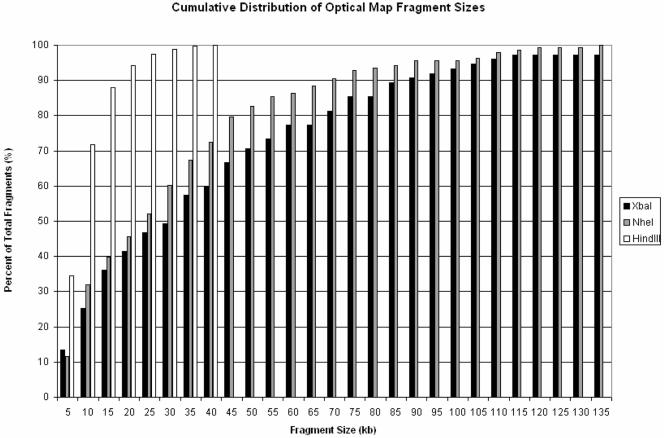
Figure 5 shows the cumulative distribution of fragment sizes for the three optical maps. Only the XbaI map has fragments greater than 135 kb, and thus this value was chosen as an endpoint in the figure to facilitate visual comparison of the three maps' fragment size distributions. Each bar represents the cumulative percentage of consensus map fragments in 5-kb intervals. For each of the maps, the distribution is roughly exponential, as expected. One key difference between the HindIII and the lower-resolution XbaI and NheI maps is the proportion of fragments smaller than 5 kb and 10 kb. In the HindIII map, about 35% of all fragments in the consensus map are smaller than 5 kb; 72% of fragments are smaller than 10 kb, and 100% of fragments are under 40 kb (the largest fragment is 37.82 kb). These numbers are in stark contrast to those for the NheI and XbaI maps. In the NheI map, only 12% of fragments are smaller than 5 kb, and 32% of fragments are smaller than 10 kb. Similarly, in the XbaI map, only 13% of fragments are smaller than 5 kb and 25% of fragments are smaller than 10 kb. For the XbaI map, there were only two additional fragments greater than 135 kb: a 242.20-kb fragment and a 256.30-kb fragment (not shown). The increased average relative sizing error for small fragments (Fig. 4F) seen in the HindIII map may be due to the high proportion of fragments 2 kb or smaller, many in tandem with each other, in this high-resolution map.
FIG. 5.
Cumulative distribution of optical map fragment sizes for the three optical maps. For each of the three whole-genome R. rubrum optical maps, the percentage of fragments within the size range from 0 to 135 kb is plotted. Each bar represents the cumulative percentage of consensus map fragments in 5-kb intervals. From the figure, the drastic differences in map fragment sizes, particularly for fragments less than 10 kb, are evident.
Comparing optical maps to the sequence.
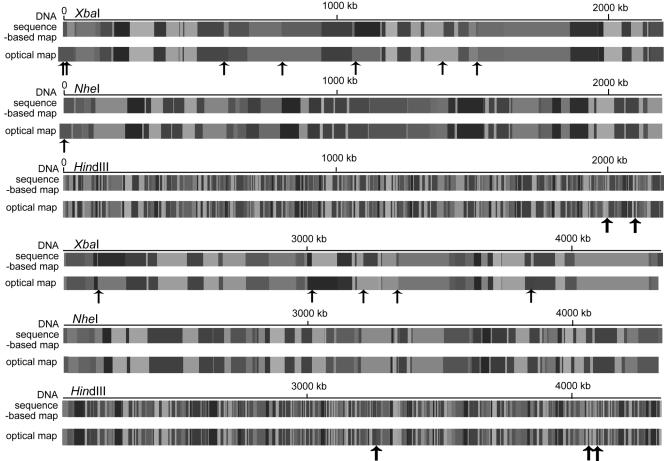
An assessment of the previously described errors in the context of optical map to sequence alignment is necessary for distinguishing random errors from those that may consistently point to discrepancies between optical maps and sequence. Figure 6 shows the linearized XbaI, NheI, and HindIII alignments of the consensus optical map to the corresponding DNA sequence-based map, in order to show the exact locations of discrepancies between the sequence and the optical maps.
FIG. 6.
Linear view of consensus optical maps with DNA sequence-based maps. Solid black arrows represent the locations of missing cuts in the consensus optical maps. The XbaI and NheI consensus optical maps extend to the left of the origin of the DNA sequence-based map because both optical maps had a missing cut at that position.
The alignment of the XbaI map with the DNA sequence-based map showed that there were no false cuts (apparent in the optical map but not in the DNA sequence-based map) and 12 missing cuts (apparent in the DNA sequence-based map but not the optical map) out of a total of 100 XbaI cuts in the DNA sequence-based map. Optical maps normally do not report restriction endonuclease fragments smaller than 500 bp, and, due to the resolution of optical mapping, reporting of fragments smaller than 1 kb is incomplete (21). The XbaI map had no missing fragments over 500 bp. Out of 100 fragments, the DNA sequence-based map showed two fragments smaller than 500 bp, and two fragments between 500 bp and 1 kb.
In comparison to the DNA sequence-based map, the NheI map showed no false cuts and one missing cut out of a total of 145 cuts in the DNA sequence-based map. There were four missing fragments, over 500 bp, in the NheI map. The DNA sequence-based map had no fragments smaller than 500 bp, and three fragments smaller than 1 kb, out of a total of 145 fragments.
Finally, the HindIII map showed no false cuts and five missing cuts in comparison to the 684 cuts in the DNA sequence-based map. Of the 684 fragments, 664 were greater than 500 bp. Of these fragments, 125 were missing in the HindIII optical map. Fifty-eight of the missing fragment loci, corresponded to DNA sequence-based fragments >500 bp and ≤1 kb, 59 to fragments >1 kb and ≤2 kb, and the remaining eight to fragments >2 kb and <3 kb.
Comparing the locations of the missing cuts and missing fragments revealed no consistent errors among the three optical maps. Thus, errors appear to be random and not associated with any major discrepancy between the sequence and the optical maps.
DISCUSSION
The goal of whole-genome optical restriction endonuclease mapping of R. rubrum strain ATCC 11170 was to aid in sequence assembly and finishing. The enzymes XbaI, NheI, and HindIII were selected because of their different cutting frequencies. The advantages of both low- and high-resolution optical maps in sequence assembly are demonstrated here. The high-resolution HindIII map was able to align and order the seven sequence contigs (not including the ∼2-kb contig) generated at the end of the finishing effort without gaps. While the error in contig 90 was evident in the HindIII optical map to sequence alignment, in this case, the lower-resolution XbaI and NheI maps best displayed this error and how it could be corrected. Yet in general, an array of different-resolution optical maps is advantageous for addressing discrepancies in genome sequences. All three maps were used to confirm the final 4.353-Mb sequence contig generated by the Los Alamos finishing group.
The finished R. rubrum strain ATCC 11170 sequence size of 4.353 Mb is closest to the estimate of 4.323 Mb provided by the XbaI map. The overall sizing error for the XbaI map is 0.7%, which is smaller than the error associated with other whole-genome physical maps generated by pulsed-field gel electrophoresis (32). The sizing errors for the NheI map and HindIII map were 3% and 2%, respectively. Yet, the alignment of the NheI and HindIII optical maps against the DNA sequence-based maps showed no apparent overall size discrepancies, and thus this error most likely stems from the summation and increased error associated with small fragments. As the number of fragments summed to calculate genome size increases, so does the error associated with this calculation. As such, the low-resolution XbaI map should, and does, give the most accurate estimate of genome size.
The high number of missing fragments in the HindIII map and increased sizing error of small fragments illustrate the challenges optical mapping faces for scoring of small fragments. Of the 125 missing fragments in the HindIII optical map, 116 corresponded to DNA sequence-based map fragments less than or equal to 2 kb. This corresponds to a small fragment loss rate of 75%, as the DNA sequence-based map contained 154 fragments less than or equal to 2 kb. By contrast, the fragment loss rate for fragments >2 and ≤3 kb was 12% (8 out of 69 fragments were missing), and zero for fragments greater than 3 kb.
A key element of the optical mapping system is the elongation and immobilization of single DNA molecules onto glass surfaces. Immobilization via electrostatic interactions between the negatively charged DNA and positively charged glass surface must be subtle enough to enable biochemical reactions, such as a restriction endonuclease digest, yet strong enough to retain the resulting fragments. The loss of fragments 2 kb and smaller reflects the difficulty in retaining small fragments in their exact position on the surface after a restriction endonuclease digest but also in identifying and correctly sizing the fragments during image acquisition and subsequent processing. The error models in the optical map assembly software (Gentig) take into account the likelihood of losing small fragments and enable alignment against the sequence, as seen here in the HindIII map, despite the significant small fragment loss.
An increased positive sizing error is seen in both the HindIII map and NheI map for small fragments. One possible explanation is the likelihood of overestimating the size of small fragments when they are scored. In other words, when a small fragment is marked, it is unlikely that the fragment would be undersized, and thus errors in this size range do not balance each other as well as they do for the larger. New efforts in DNA mounting and small-fragment sizing with the Pathfinder software are currently under way in order to improve retention and scoring of small fragments.
With an average fragment size of 44.73 kb, the XbaI map represents the lowest-resolution optical map created. There are significant advantages of a low-resolution map. First, a low-resolution map requires very large single molecule for assembly into a whole-genome contig. As average fragment sizes increases, so does the molecule size required for achieving a unique pattern of restriction endonuclease fragments for accurate map assembly. Here, the average size of molecules in the XbaI map approached 1 Mb. This scale approaches the lower-resolution limit of more global cytogenetic methods that reveal chromosomal insertions, deletions, rearrangements, etc. (5, 15, 19). With a documented resolution between 6.5 kb (32) and 45 kb (reported here), optical mapping's niche falls between low-resolution, global methods, such as comparative genomic hybridization, and very high-resolution genotyping systems. This “molecular cytogenetics” approach has enormous potential for aiding in large genome (such as mammalian) sequencing projects as well as for identifying genomic variation in the form of insertions, deletions, and repetitive elements, a difficult and often evasive task.
With the ability to qualify conclusions drawn from low-resolution cytogenetic techniques and contextualize the information gleaned from high-resolution genotyping tools, the optical mapping system can be particularly powerful when used in conjunction with other methods. We are currently pursuing these directions with the optical mapping system, as well as working on improvements for larger molecules and improved small fragment retention for the goal of widening the range of optical mapping's resolution.
Here, three optical maps that have aided in sequence assembly and validation of R. rubrum have been shown. In addition, we have widened the resolution range of the optical mapping system and contextualized this contribution to genomic analysis. Continual improvements and new applications of the optical mapping system are under way. For example, in a recent comparative genomics study, optical mapping revealed novel genomic insertions and rearrangements in Shigella flexneri in addition to genomic differences between sequenced strains of Escherichia coli and Yersinia pestis that were aligned as maps (31). Optical mapping's role in sequencing projects has expanded to larger, more complex genomes, such as the ∼34-Mb genome of the diatom Thalassiosira pseudonana (4). Optical mapping projects will continue to encompass increasingly challenging questions, with the goal of providing new insights on genome structure and organization that will potentiate the capabilities of higher and lower-resolution genomic analysis systems.
Acknowledgments
This work was supported by Department of Energy grant DE-FC02-01ER63175 and NIH grants 2 R01 HG000225-10, NIH 5 T32 GM08349, and NIH GM65891.
We thank all members of the University of Wisconsin—Madison Laboratory for Molecular and Computational Genomics.
REFERENCES
- 1.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1997. Genomics via optical mapping. II. Ordered restriction maps. J. Comput. Biol. 4:91-118. [DOI] [PubMed] [Google Scholar]
- 2.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1999. Genomics via optical mapping. III. Contiging genomic DNA and variations. Seventh International Conference on Intelligent Systems for Molecular Biology, vol. 7, p. 18-27. [PubMed]
- 3.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1998. Genomics via optical mapping. III. Contiging genomic DNA and variations. Courant Technical Report 760. Courant Institute, New York University, New York, N.Y.
- 4.Armbrust, E. V., J. A. Berges, C. Bowler, B. R. Green, D. Martinez, N. H. Putnam, S. Zhou, A. E. Allen, K. E. Apt, M. Bechner, M. A. Brzezinski, B. K. Chaal, A. Chiovitti, A. K. Davis, M. S. Demarest, J. C. Detter, T. Glavina, D. Goodstein, M. Z. Hadi, U. Hellsten, M. Hildebrand, B. D. Jenkins, J. Jurka, V. V. Kapitonov, N. Kroger, W. W. Y. Lau, T. W. Lane, F. W. Larimer, J. C. Lippmeier, S. Lucas, M. Medina, A. Montsant, M. Obornik, M. S. Parker, B. Palenik, G. J. Pazour, P. M. Richardson, T. A. Rynearson, M. A. Saito, D. C. Schwartz, K. Thamatrakoln, K. Valentin, A. Vardi, F. P. Wilkerson, and D. S. Rokhsar. 2004. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306:79-86. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, M., A. Scheffer, A. Ben-Dor, N. Sampas, D. Lipson, R. Kincaid, P. Tsang, B. Curry, K. Baird, P. Meltzer, Z. Yakhini, L. Bruhn, and S. Laderman. 2004. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl. Acad. Sci. USA 101:17765-17770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonam, D., L. Lehman, G. P. Roberts, and P. W. Ludden. 1989. Regulation of carbon monoxide dehydrogenase and hydrogenase in Rhodospirillum rubrum: effects of CO and oxygen on synthesis and activity. J. Bacteriol. 171:3102-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandl, H., E. J. Knee, Jr., R. C. Fuller, R. A. Gross, and R. W. Lenz. 1989. Ability of the phototrophic bacterium Rhodospirillum rubrum to produce various poly (β-hydroxyalkanoates): potential sources for biodegradable polyesters. Int. J. Biol. Macromol. 11:49-55. [DOI] [PubMed] [Google Scholar]
- 8.Dimalanta, E. T., A. Lim, R. Runnheim, C. Lamers, C. Churas, D. K. Forrest, J. J. de Pablo, M. D. Graham, S. N. Coppersmith, S. Goldstein, and D. C. Schwartz. 2004. A microfluidic system for large DNA molecule arrays. Anal. Chem. 76:5293-5301. [DOI] [PubMed] [Google Scholar]
- 9.Fitzmaurice, W. P., L. L. Saari, R. G. Lowery, P. W. Ludden, and G. P. Roberts. 1989. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol. Gen. Genet. 218:340-347. [DOI] [PubMed] [Google Scholar]
- 10.Fuller, R. C. 1995. Polyesters and photosynthetic bacteria: from lipid cellular inclusions to microbial thermoplastics, p. 991-1003. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with Autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handrick, R., S. Reinhardt, D. Schultheiss, T. Reichart, D. Schuler, V. Jendrossek, and D. Jendrossek. 2004. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB granule protein (phasin). J. Bacteriol. 186:2466-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imhoff, J. F. 2000. The anoxygenic phototrophic purple bacteria, p. 631-637. In D. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, NY. [Google Scholar]
- 14.Imhoff, J. F. 2001, posting date. The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag. [Online.]
- 15.Kytola, S., J. Rummukainen, A. Nordgren, R. Karhu, F. Farnebo, J. Isola, and C. Larsson. 2000. Chromosomal alterations in 15 breast cancer cell lines by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer 28:308-317. [DOI] [PubMed] [Google Scholar]
- 16.Lai, Z., J. Jing, C. Aston, V. Clarke, J. Apodaca, E. T. Dimalanta, D. J. Carucci, M. J. Gardner, B. Mishra, T. S. Anantharaman, S. Paxia, S. L. Hoffman, J. C. V. J. Huff, and D. C. Schwartz. 1999. A shotgun optical map of the entire Plasmodium falciparum genome. Nat. Genet. 23:309-313. [DOI] [PubMed] [Google Scholar]
- 17.Lim, A., E. T. Dimalanta, K. D. Potamousis, G. Yen, J. Apodoca, C. Tao, J. Lin, R. Qi, J. Skiadas, A. Ramanathan, N. T. Perna, G. Plunkett, V. Burland, B. Mau, J. Hackett, F. R. Blattner, T. S. Anantharaman, B. Mishra, and D. C. Schwartz. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11:1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, J., R. Qi, C. Aston, J. Jing, T. S. Anantharaman, B. Mishra, O. White, M. J. Daly, K. W. Minton, J. C. Venter, and D. C. Schwartz. 1999. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science 285:1558-1562. [DOI] [PubMed] [Google Scholar]
- 19.Lucito, R., J. Healy, J. Alexander, A. Reiner, D. Esposito, M. Chi, L. Rogers, A. Brady, et al. 2003. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 13:2291-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marra, M. A., T. A. Kucaba, N. L. Dietrich, E. D. Green, B. Brownstein, R. K. Wilson, K. M. McDonald, L. W. Hillier, J. D. McPherson, and R. H. Waterston. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7:1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng, X., K. Benson, K. Chada, J. E. Huff, and D. C. Schwartz. 1995. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat. Genet. 9:432-438. [DOI] [PubMed] [Google Scholar]
- 22.Pfennig, N., H. Lünsdorf, J. Süling, and J. F. Imhoff. 1997. Rhodospira trueperi gen. nov., spec. nov., a new phototrophic Proteobacterium of the alpha group. Arch. Microbiol. 168:39-45. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki, S., and I. Karube. 1999. The development of microfabricated biocatalytic fuel cells. TIBTECH 17:50-52. [DOI] [PubMed] [Google Scholar]
- 24.Schick, H. J. 1971. Interrelationship of nitrogen fixation, hydrogen evolution and photoreduction in Rhodospirillum rubrum. Arch. Microbiol. 75:102-109. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz, D., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67-77. [DOI] [PubMed] [Google Scholar]
- 27.Tabita, F. R. 1995. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria, p. 885-914. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.von Felten, P., H. Zürrer, and R. Bachofen. 1985. Production of molecular hydrogen with immobilized cells of Rhodospirillum rubrum. Appl. Microbiol. Biotechnol. 23:15-20. [Google Scholar]
- 29.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1995. Comparison studies of dinitrogenase reductase ADP-ribosyl transferase/dinitrogenase reductase activating glycohydrolase regulatory systems in Rhodospirillum rubrum and Azospirillum brasilense. J. Bacteriol. 177:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou, S., W. Deng, T. S. Anantharaman, A. Lim, E. T. Dimalanta, J. Wang, T. Wu, T. Chunhong, R. Creighton, A. Kile, E. Kvikstad, M. Bechner, G. Yen, A. Garic-Stankovic, J. Severin, D. Forrest, R. Runnheim, C. Churas, C. Lamers, N. T. Perna, V. Burland, F. R. Blattner, B. Mishra, and D. C. Schwartz. 2002. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl. Environ. Microbiol. 68:6321-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, S., A. Kile, M. Bechner, M. Place, E. Kvikstad, W. Deng, J. Wei, J. Severin, R. Runnheim, C. Churas, D. Forrest, E. T. Dimalanta, C. Lamers, V. Burland, F. R. Blattner, and D. C. Schwartz. 2004. A single-molecule approach to bacterial genomic comparisons via optical mapping. J. Bacteriol. 186:7773-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, S., E. Kvikstad, A. Kile, J. Severin, D. Forrest, R. Runnheim, C. Churas, J. W. Hickman, C. Mackenzie, M. Choudhary, T. Donahue, S. Kaplan, and D. C. Schwartz. 2003. Whole-genome shotgun optical mapping of Rhodabacter sphaeroides strain 2.4.1 and its use for whole-genome shotgun sequence assembly. Genome Res. 13:2142-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]