Abstract
Binding studies using 125I-Cry1Ac and biotinylated Cry1Fa toxins indicate the occurrence of a common receptor for Cry1Ac, Cry1Fa, and Cry1Ja in Helicoverpa armigera, Helicoverpa zea, and Spodoptera exigua. Our results, along with previous binding data and the observed cases of cross-resistance, suggest that this pattern seems to be widespread among lepidopteran species.
Transgenic plants expressing Bacillus thuringiensis insecticidal protein genes offer long-term and preventive measures against several species of insect pests, including tunneling insects. One of the main threats of the wide adoption of these crops is the evolution of insect resistance as a response to the strong selection pressure that will be imposed on the insect populations (7). Some strategies to delay or minimize the appearance of resistance are based on the use of more than one Cry toxin in either mixtures or rotations or combined in the same plant (6). For these strategies to be effective while at the same time avoiding the development of cross-resistance, the toxins to be considered have to have differing modes of action. Binding site modification is the most frequent mechanism of resistance to B. thuringiensis toxins and has been shown to be the basis of cross-resistance among Cry1A toxins (7). Therefore, from a resistance management perspective, toxins that use the same binding sites to exert their toxic actions cannot be used as replacements for or complements of each other.
Common binding sites for Cry1A and Cry1Ja have already been shown to occur in several insect species, which seems to be a general pattern in Lepidoptera (8). In the present study we have used Helicoverpa armigera, Helicoverpa zea, and Spodoptera exigua to integrate Cry1Fa into the binding model of Cry1Ja and Cry1A.
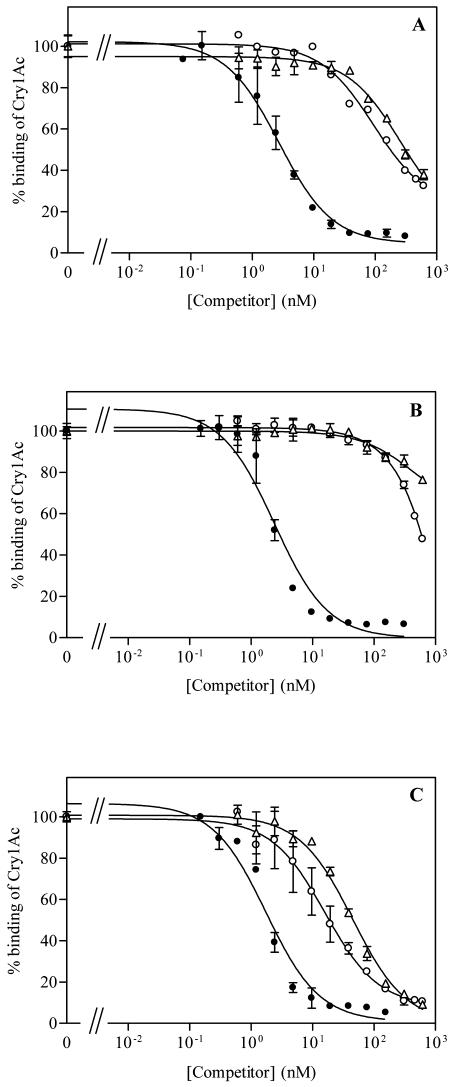
Cry1Ac, Cry1Fa, and Cry1Ja toxins were obtained from B. thuringiensis recombinant strains (EG11070, EG11069, and EG7279) and prepared as trypsin-activated and chromatography-purified toxins (5). Using 125I-Cry1Ac toxin (21) and unlabeled Cry1Ac, Cry1Fa, and Cry1Ja toxins as competitors, we performed binding competition experiments by incubating 25 μg/ml of brush border membrane vesicle (BBMV) proteins of H. armigera or H. zea or 50 μg/ml of S. exigua BBMV proteins following the protocol previously described (5). Cry1Fa and Cry1Ja toxins competed for the Cry1Ac binding site in the three species tested (Fig. 1). Quantitative estimates of the binding affinity of the three toxins indicate higher dissociation constant (KD) values for Cry1Fa and Cry1Ja than for Cry1Ac in the three insect species (Table 1). In S. exigua, Cry1Fa and Cry1Ja bound with moderate affinities (29 and 22 nM, respectively), whereas in H. armigera and H. zea, they bound with very low affinities (from 150 to 640 nM). In spite of the fact that the toxicities of Cry toxins do not always correlate with their binding affinities (4, 13, 22), the low affinity of Cry1Fa in H. armigera and H. zea agrees with its lack of toxicity against these species (12). Furthermore, Cry1Fa is toxic to S. exigua (3), which agrees with its higher affinity for the binding sites in this species.
FIG. 1.
Binding competition between 125I-Cry1Ac and increasing concentrations of unlabeled Cry1Ac (•), Cry1Fa (○), or Cry1Ja (▵) to BBMV proteins from H. armigera (A), H. zea (B), and S. exigua (C). Each data point is the mean of two independent replications.
TABLE 1.
KD values and concentrations of binding sites (Rt) for Cry1 toxins binding to BBMV proteins from H. armigera, H. zea, and S. exigua, determined using Cry1Ac as the labeled liganda
| Toxin |
H. armigera
|
H. zea
|
S. exigua
|
|||
|---|---|---|---|---|---|---|
| KD (nM) ± SD | Rt (pmol/ mg protein) ± SD | KD (nM) ± SD | Rt (pmol/ mg protein) ± SD | KD (nM) ± SD | Rt (pmol/ mg protein) ± SD | |
| Cry1Ac | 1.6 ± 0.2 | 16.4 ± 0.2 | 0.34 ± 0.04 | 29.6 ± 0.5 | 0.7 ± 0.5 | 4.6 ± 0.5 |
| Cry1Fa | 150 ± 40 | 220 ± 10 | 29 ± 7 | |||
| Cry1Ja | 250 ± 20 | 640 ± 2 | 22 ± 2 | |||
KD values of heterologous ligands are estimated assuming an Rt the same as that for the labeled ligand.
In the range of concentrations tested, complete competition of the 125I-Cry1Ac was observed only in S. exigua, indicating that Cry1Ac does not have binding sites other than those shared with the heterologous toxins. In the Helicoverpa species, neither complete competition with Cry1Fa and Cry1Ja nor a plateau which would have indicated the occurrence of unique Cry1Ac binding sites was achieved. The analysis of the heterologous curves gave a good fit to a single-site model, assuming a concentration of receptors the same as that for Cry1Ac. In a previous paper from our laboratory, Cry1Fa did not compete for Cry1Ac binding sites in H. armigera (5), but subsequent experiments carried out by us with the same batch of Cry1Fa toxin showed that the toxin had deteriorated during storage.
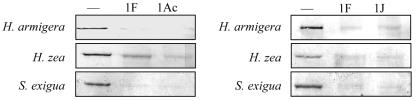
To determine whether Cry1Fa could have binding sites in addition to the one shared with Cry1Ac and Cry1Ja, Cry1Fa was biotinylated and its binding was tested in competition experiments using 200 μg/ml of BBMV proteins. Toxin biotinylation, competition assays, sample transference, and detection were done as previously described (8). The results showed that both Cry1Ac and Cry1Ja competed for the Cry1Fa binding site in the three species (Fig. 2). Furthermore, since competition by unlabeled Cry1Fa was not more effective than that by the heterologous toxins, there is no evidence of unshared sites for Cry1Fa. To our knowledge, this is the first time that reciprocal competition assays with labeled Cry1Fa have ever been performed.
FIG. 2.
Binding of biotinylated Cry1Fa to BBMV proteins from H. armigera, H. zea, and S. exigua in the absence of competitor (lanes labeled with horizontal lines) or in the presence of a 120-fold excess of competitor (lanes labeled 1F, 1Ac, and 1J).
For Plutella xylostella, an autosomal recessive gene conferring high resistance to Cry1Aa, Cry1Ab, Cry1Ac, Cry1Fa, and Cry1Ja has been reported (16, 17), suggesting that resistance to these five toxins has a common physiological basis. Since in this insect these five toxins share the same binding site in the midgut epithelial membrane (1) and reduced binding of Cry1A toxins is responsible for resistance (18), it is sensible to assume that reported cross-resistance to Cry1Fa and Cry1Ja (16, 17) might also be due to the alteration of the common receptor. In Heliothis virescens, a common binding site for Cry1Aa, Cry1Ab, Cry1Ac, Cry1Fa, and Cry1Ja has also been shown (10), and reduced binding of Cry1A toxins and Cry1Fa has been proposed as the mechanism responsible for resistance to these toxins (11). In Ostrinia nubilalis, low levels of cross-resistance to Cry1Fa were observed among Cry1Ab-selected strains (15), and inhibition of Cry1Ab binding by Cry1Fa has also been shown (9).
Amino acid sequence similarity studies in domain II, the specificity-determining domain of Cry toxins and the one mainly involved in receptor binding (14), have shown that Cry1Fa and Cry1Ja are closer to the Cry1A cluster than the rest of the Cry toxins are (2, 19, 20). So far, all available information on binding site competition suggests that Cry1Aa, Cry1Ab, Cry1Ac, Cry1Fa, and Cry1Ja share a common binding site in most, if not all, members of the order Lepidoptera. Since it seems that Cry1Fa does not have binding sites other than those shared with Cry1Ac or Cry1Ja in the species tested, we propose that Cry1Fa and Cry1Ja exert their toxic actions in some Lepidoptera species by using the same target sites as those used by the Cry1A toxins.
That Cry1Aa, Cry1Ab, Cry1Ac, Cry1Fa, and Cry1Ja bind to common sites explains, in an elegant form, the biochemical basis of multiple resistances and cross-resistances among these toxins observed in some insect species. Insects that lack additional sites for Cry1Fa or Cry1Ja could become resistant simultaneously to the five toxins relatively easily. Nevertheless, this model does not preclude other outcomes, since alterations in the receptor molecule may not always render a reduction in binding involving all five toxins but may be more selective, affecting binding of just some of them, as has been described for P. xylostella strains lacking binding of Cry1Ab without affecting binding of Cry1Ac (1, 18, 23).
In Bt cotton, genes expressing B. thuringiensis Cry1Ac and Cry1Fa toxins have been combined in the same plant to confer a broader-spectrum resistance to cotton pests. With this approach, species which are nonsusceptible to Cry1Fa, such as H. armigera and H. zea, can be controlled with the Cry1Ac toxin, whereas the Cry1Fa toxin is effective against Spodoptera spp., which are little affected by Cry1Ac. However, from a resistance management standpoint, neither transgenic plants expressing pairwise combinations of Cry1Ac, Cry1Fa, and Cry1Ja nor rotations of Bt crops containing single genes of these three toxins will offer a good strategy for controlling those insects susceptible to more than one of these toxins. Populations of insects without alternative sites for Cry1Fa, previously exposed to first-generation Cry1A crops, could have already started to develop cross-resistance to Cry1Fa. In the case of cotton, populations exposed to the dual gene strategy would be under strong pressure to select for mutations affecting the common receptor. Since primary pests of this crop are not susceptible to both Cry1Ac and Cry1Fa, the risk for cross-resistance is not as great for them as it is for potential secondary pests susceptible to both toxins. In the case of corn, primary pests susceptible to Cry1Ab and Cry1Fa, such as O. nubilalis (9, 15) and Sesamia nonagrioides (F. Ortego and P. Castañera, personal communication), do exist. Therefore, establishing the binding site model in these species is of extreme importance for the appropriate design of resistance management strategies.
Acknowledgments
We thank J. Van Rie and J. González-Cabrera for critically reading the manuscript.
This work was conducted within a Bayer BioScience/Universitat de València Research Collaboration.
REFERENCES
- 1.Ballester, V., F. Granero, B. E. Tabashnik, T. Malvar, and J. Ferré. 1999. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 65:1413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo, A. 1997. Phylogenetic relationships of Bacillus thuringiensis δ-endotoxin family proteins and their functional domains. J. Bacteriol. 179:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers, J. A., A. Jelen, M. P. Gilbert, C. S. Jany, T. B. Johnson, and C. Gawron-Burke. 1991. Isolation and characterization of a novel insecticidal crystal protein gene from Bacillus thuringiensis subsp. aizawai. J. Bacteriol. 173:3966-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escriche, B., J. Ferré, and F. J. Silva. 1997. Occurrence of a common binding site in Mamestra brassicae, Phthorimaea operculella, and Spodoptera exigua for the insecticidal crystal proteins CryIA from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 27:651-656. [DOI] [PubMed] [Google Scholar]
- 5.Estela, A., B. Escriche, and J. Ferré. 2004. Interaction of Bacillus thuringiensis toxins with larval midgut binding sites of Helicoverpa armigera (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 70:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferré, J. 2003. Insect resistance to Bacillus thuringiensis toxins, p. 141-155. In T. Lelley, E. Balázs, and M. Tepfer (ed.), Ecological impact of GMO dissemination in agro-ecosystems. Facultas Universitätsverlag, Vienna, Austria.
- 7.Ferré, J., and J. Van Rie. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501-533. [DOI] [PubMed] [Google Scholar]
- 8.Herrero, S., J. González-Cabrera, B. E. Tabashnik, and J. Ferré. 2001. Shared binding sites in Lepidoptera for Bacillus thuringiensis Cry1Ja and Cry1A toxins. Appl. Environ. Microbiol. 67:5729-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hua, G., L. Masson, J. L. Jurat-Fuentes, G. Schwab, and M. J. Adang. 2001. Binding analyses of Bacillus thuringiensis Cry δ-endotoxins using brush border membrane vesicles of Ostrinia nubilalis. Appl. Environ. Microbiol. 67:872-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurat-Fuentes, J. L., and M. J. Adang. 2001. Importance of Cry1 δ-endotoxin domain II loops for binding specificity in Heliothis virescens (L.). Appl. Environ. Microbiol. 67:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurat-Fuentes, J. L., F. L. Gould, and M. J. Adang. 2002. Altered glycosylation of 63- and 68-kilodalton microvillar proteins in Heliothis virescens correlates with reduced Cry1 toxin binding, decreased pore formation, and increased resistance to Bacillus thuringiensis Cry1 toxins. Appl. Environ. Microbiol. 68:5711-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao, C., D. Heckel, and R. Akhurst. 2002. Toxicity of Bacillus thuringiensis insecticidal proteins for Helicoverpa armigera and Helicoverpa punctigera (Lepidoptera: Noctuidae), major pests of cotton. J. Invertebr. Pathol. 80:55. [DOI] [PubMed] [Google Scholar]
- 13.Luo, K., D. Banks, and M. J. Adang. 1999. Toxicity, binding, and permeability analyses of four Bacillus thuringiensis Cry1 δ-endotoxins using brush border membrane vesicles of Spodoptera exigua and Spodoptera frugiperda. Appl. Environ. Microbiol. 65:457-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siqueira, H. A., D. Moellenbeck, T. Spencer, and B. D. Siegfried. 2004. Cross-resistance of Cry1Ab-selected Ostrinia nubilalis (Lepidoptera: Crambidae) to Bacillus thuringiensis delta-endotoxins. J. Econ. Entomol. 97:1049-1057. [DOI] [PubMed] [Google Scholar]
- 16.Tabashnik, B. E., K. W. Johnson, J. T. Engleman, and J. A. Baum. 2000. Cross-resistance to Bacillus thuringiensis toxin Cry1Ja in a strain of diamondback moth adapted to artificial diet. J. Invertebr. Pathol. 76:81-83. [DOI] [PubMed] [Google Scholar]
- 17.Tabashnik, B. E., Y. B. Liu, N. Finson, L. Masson, and D. G. Heckel. 1997. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc. Natl. Acad. Sci. USA 94:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabashnik, B. E., Y. B. Liu, T. Malvar, D. G. Heckel, L. Masson, V. Ballester, F. Granero, J. L. Ménsua, and J. Ferré. 1997. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 94:12780-12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabashnik, B. E., T. Malvar, Y. B. Liu, N. Finson, D. Borthakur, B. S. Shin, S. H. Park, L. Masson, R. A. de Maagd, and D. Bosch. 1996. Cross-resistance of the diamondback moth indicates altered interactions with domain II of Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 62:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson, M. A., H. E. Schnepf, and J. S. Feitelson. 1995. Structure, function and engineering of Bacillus thuringiensis toxins. Genet. Eng. (N.Y.) 17:99-117. [PubMed] [Google Scholar]
- 21.Van Rie, J., S. Jansens, H. Hofte, D. Degheele, and H. Van Mellaert. 1989. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur. J. Biochem. 186:239-247. [DOI] [PubMed] [Google Scholar]
- 22.Wolfersberger, M. G. 1990. The toxicity of two Bacillus thuringiensis delta-endotoxins to gypsy moth larvae is inversely related to the affinity of binding sites on midgut brush border membranes for the toxins. Experientia 46:475-477. [DOI] [PubMed] [Google Scholar]
- 23.Wright, D. J., M. Iqbal, F. Granero, and J. Ferré. 1997. A change in a single midgut receptor in the diamondback moth (Plutella xylostella) is only in part responsible for field resistance to Bacillus thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai. Appl. Environ. Microbiol. 63:1814-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]