Abstract
Transforming growth factor β (TGF-β) is the prototype of a large superfamily of signaling molecules involved in the regulation of cell growth and differentiation. In certain patients infected with human immunodeficiency virus type 1 (HIV-1), increased levels of TGF-β promoted the production of virus and also impaired the host immune system. In an effort to understand the signaling events linking TGF-β action and HIV production, we show here that TGF-β can stimulate transcription from the HIV-1 long terminal repeat (LTR) promoter through NF-κB binding sites in both HaCaT and 300.19 pre-B cells. When introduced into a minimal promoter, NF-κB binding sites supported nearly 30-fold activation from the luciferase reporter upon TGF-β treatment. Electrophoretic mobility shift assay indicated that a major factor binding to the NF-κB site is the p50-p65 heterodimeric NF-κB in HaCaT cells. Coexpression of Gal4-p65 chimeric proteins supported TGF-β ligand-dependent gene expression from a luciferase reporter gene driven by Gal4 DNA binding sites. NF-κB activity present in HaCaT cells was not affected by TGF-β treatment as judged by the unchanged DNA binding activity and concentrations of p50 and p65 proteins. Consistently, steady-state levels of IκBα and IκBβ proteins were not changed by TGF-β treatment. Our results demonstrate that TGF-β is able to stimulate transcription from the HIV-1 LTR promoter by activating NF-κB through a mechanism distinct from the classic NF-κB activation mechanism involving the degradation of IκB proteins.
Transforming growth factor β (TGF-β) families of cytokines regulate many aspects of cellular functions, including growth, differentiation, morphogenesis, and apoptosis (reviewed in references 26, 27, 35, and 42). It inhibits proliferation of epithelial, endothelial, and fibroblast cells (26, 35). In cells of hematopoietic and lymphoid origins, TGF-β inhibits proliferation of T lymphocytes (17), B lymphocytes (16), and thymocytes (34), as well as differentiation of natural killer cells (36), lymphocyte-activated killer cells (11), and macrophages (43). Thus, TGF-β displays immunosuppressive activities both in vivo and in vitro and is a potent endogenous immunosuppressor (10, 47). TGF-β plays vital functions in inflammatory responses, as targeted disruption of the TGF-β gene in mice caused multifocal inflammatory responses resembling autoimmune diseases and the eventual death of TGF-β1 null mice 2 to 3 weeks after birth (21, 41). The apparently normal embryonic development and perinatal survival of TGF-β1 null mice were likely due to the maternal rescue of TGF-β1 proteins crossing the placenta and from milk through breast feeding (24).
Several lines of evidence suggest that TGF-β is also involved in the regulation and pathogenesis of human immunodeficiency virus type 1 (HIV-1), which is the etiological agent causing AIDS. Increased expression of TGF-β was found in peripheral blood mononuclear cells (18), primary mononuclear phagocytes (22, 23), and brain tissues (45) from HIV-1-infected patients. The elevated expression of TGF-β has been partially attributed to the activation of the TGF-β1 promoter by the virally encoded Tat protein (7). Augmented production of TGF-β resulted in decreased T-cell counts and general immunodeficiency in patients infected with HIV-1, probably through the potent immunosuppressive activities of TGF-β. On the other hand, increased levels of TGF-β can enhance HIV production in certain cell types, such as primary macrophages and premonocytic U-937 cells (22). Therefore, an enhanced level of TGF-β dually benefits HIV-1 viral propagation: TGF-β suppresses the host antiviral immune responses through inhibition of growth of hematopoietic and lymphoid cells and, at the same time, may also increase viral production in certain cell types.
The mechanism by which TGF-β enhances HIV-1 production in specific cell types is unknown. Although TGF-β has been observed to enhance transcription from the transfected HIV-1 long terminal repeat (LTR) promoter in human mesangial cells (40), it was not clear how this activation was achieved. In this study, we used HaCaT, a spontaneously immortalized human keratinocyte cell line which is highly responsive to TGF-β (5), as a model system to show that TGF-β could activate the HIV-1 LTR promoter and that the transactivation requires NF-κB binding activity. NF-κB sites alone were sufficient to mediate gene activation by TGF-β. Our results establish a direct link between the TGF-β signal and NF-κB-mediated transcriptional activation, which may provide a better understanding of the molecular basis underlying the diverse biological effects of TGF-β.
MATERIALS AND METHODS
Materials.
Oligonucleotides representing the HIV-1 NF-κB binding sites have the sequence 5′ CAA GGG ACT TTC CGC TGG GGA CTT TCC AGG 3′ and were synthesized in the Oligo Core Facility at Duke University. Human TGF-β1 was a generous gift from Amgen, Inc. Polyclonal antibodies against human p50 (sc-114), p65 (sc-732), c-Rel (sc-070x), Rel-B (sc-226), IκBα (sc-371), and IκBβ (sc-945) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Antiserum against p100 was a generous gift from J. Nevins (Duke University Medical Center). Antiserum against MBP2 was a generous gift from R. Bernard (The Netherlands Cancer Institute). Recombinant p65 and p50 proteins were generous gifts from D. Baltimore (Massachusetts Institute of Technology). Cyclosporin A (CsA) was a generous gift from J. Heitman (Duke University Medical Center).
Plasmids.
The HIV-chloramphenicol acetyltransferase reporter plasmid and cytomegalovirus-Tat (CMV-Tat) were generous gifts from B. Cullen (Duke University Medical Center). All constructs are diagrammed in Fig. 2 and 3. The KpnI- to HindIII fragment of the HIV-chloramphenicol acetyltransferase plasmid was isolated and inserted between the KpnI and HindIII restriction sites on the pGL2 Basic luciferase reporter plasmid (Promega) to create HIVKH-luc. HIVRH-luc and HIVPH-luc were created by isolating the EcoRV-to-HindIII and the PvuII-to-HindIII fragments (the PvuII site was first blunt ended with Klenow fragment in the presence of 2 mM deoxynucleoside triphosphates), respectively, and they were then inserted between the SmaI and HindIII sites on the pGL2 Basic luciferase reporter plasmid. A SmaI site was created between the NF-κB and Sp1 sites to facilitate the isolation of the newly created SmaI-to-HindIII fragment. The fragment was then inserted between the SmaI and HindIII sites on the pGL2 Basic luciferase reporter plasmid to create HIVdκB-luc, which has all sequences upstream of the Sp1 sites, including both NF-κB sites, deleted. A second, similar SmaI site was introduced immediately downstream of the Sp1 sites on the HIVKH-luc construct by site-directed mutagenesis to create HIVdSp1-luc, which has both the NF-κB and Sp1 sites deleted. A construct containing both SmaI sites on HIVKH-luc was also created and then used to obtain HIVKHDSp1-luc, which has all three Sp1 sites deleted internally while retaining the rest of the sequences. HIVKHmκB1-luc and HIVKHmκB2-luc were obtained by mutating the upstream and downstream NF-κB sites, respectively, by site-directed mutagenesis. Luciferase reporter plasmid TI-luc, which contains the TATA box and initiator element (Inr) on its promoter (14), was a generous gift from S. Smale (UCLA). Double-stranded oligonucleotides representing both of the HIV-1 NF-κB sites were synthesized and inserted upstream of TI-luc to create TI+1×κB-luc. Two or three copies of the same oligonucleotides were first concatemerized and then introduced similarly to create TI+2×κB-luc and TI+3×κB-luc, respectively. The dominant-negative type I (TβRI-DN) and type II (TβRII-DN) TGF-β receptors contained lysine-to-arginine (K-to-R) mutations at amino acid 230 or 277 (3), respectively. The activated type I receptor (TβRI-act) (same as TβRI-T204D) was a generous gift from J. Massague. The IκBα(S32/36A) construct was a generous gift from A. S. Baldwin (University of North Carolina, Chapel Hill). Gal4-p65 and Gal4-p65TA1, which have the Gal4 DNA binding domain fused to either the full-length p65 cDNA sequences or the p65 transactivation domain 1 (TA1, amino acids 520 to 550), were generous gifts from M. L. Schmitz (Institute of Biochemistry and Molecular Biology, Heidelberg, Germany). The reporter construct containing the dihydrofolate reductase (DHFR) promoter, DHFR-luc, was a generous gift from J. Nevins (Duke University Medical Center).
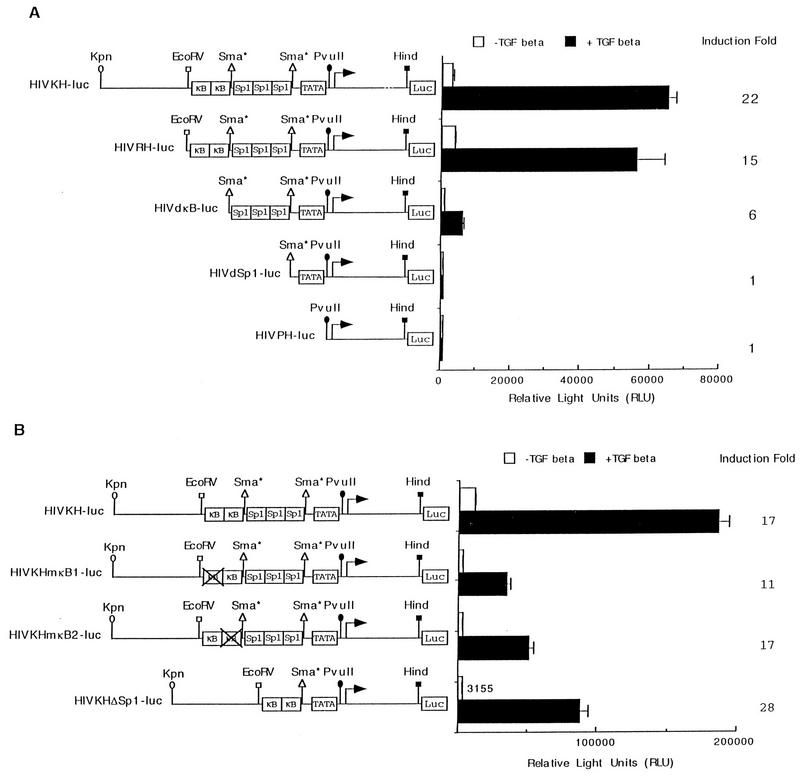
FIG. 2.
NF-κB binding sites are necessary for TGF-β-mediated gene expression from the HIV-1 LTR promoter. (A) Serial promoter deletion constructs of the HIV-1 LTR are shown. Also indicated are the restriction sites used to make the deletion constructs. These constructs were transfected transiently into HaCaT cells, and their relative luciferase activities were assayed after cells were treated or not with TGF-β1 for 24 h. The fold induction by TGF-β is indicated for each construct. Error bars represent standard deviations from duplicate determinations. (B) Either the upstream or the downstream NF-κB site was mutated by site-directed mutagenesis to create HIVKHmκB1-luc and HIVKHmκB2-luc. HIVKHDSp1-luc has all three Sp1 binding sites removed but retains the rest of the promoter sequences. These three constructs, as well as the wild-type HIVKH-luc, were transiently transfected into HaCaT cells, and their relative luciferase activities were assayed after cells were treated or not with TGF-β1 for 24 h. The fold induction by TGF-β is indicated. Error bars represent standard deviations from duplicate determinations.
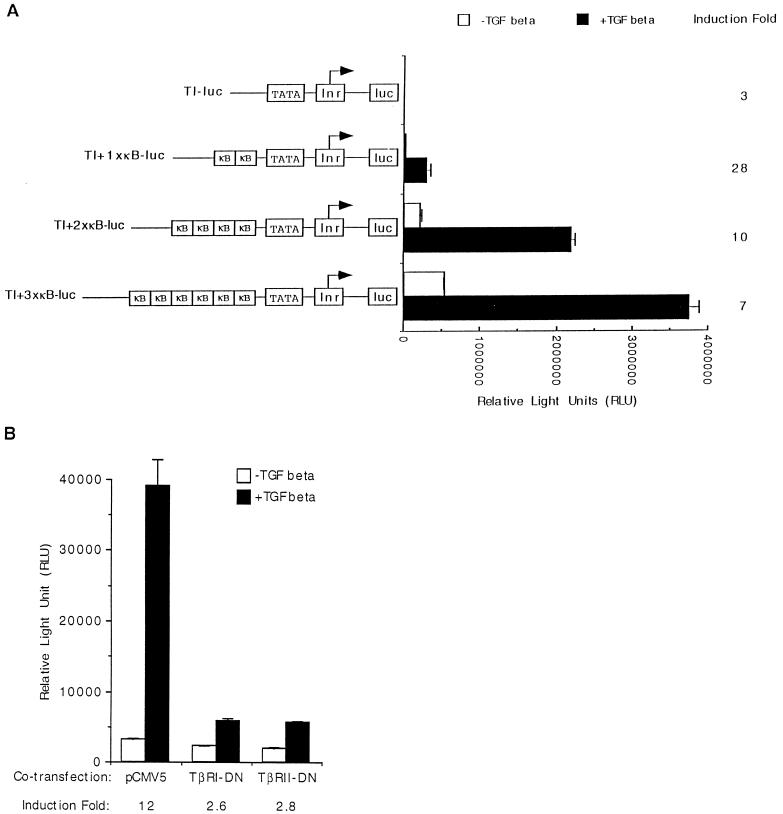
FIG. 3.
NF-κB sites are sufficient for TGF-β-mediated gene expression, which requires functional TGF-β receptors. (A) HIV-1 LTR NF-κB sites were synthesized as double-stranded oligonucleotides and inserted upstream of a minimal luciferase reporter (TI-luc) which contains only the TATA box and the initiator element to create TI+1×κB-luc. Two or three copies of the HIV-1 LTR NF-κB sites were similarly introduced to create TI+2×κB-luc and TI+3×κB-luc. All four constructs were transfected transiently into HaCaT cells, and their relative luciferase activities were assayed after cells were treated or not with TGF-β1 for 24 h. The fold induction by TGF-β is indicated. Error bars represent standard deviations from duplicate determinations. (B) The vector plasmid (pCMV5), a dominant-negative type I TGF-β receptor (TβRI-DN), or a dominant-negative type II TGF-β receptor (TβRII-DN) was cotransfected transiently with TI+1×κB-luc into HaCaT cells, and their relative luciferase activities were assayed after cells were treated or not with TGF-β1 for 24 h. The fold induction by TGF-β is indicated. Error bars represent standard deviations from duplicate determinations.
Cell culture and DNA transfection.
Human HaCaT cells, a generous gift from P. Baukamp and N. Fusenig (Institute of Biochemistry and Molecular Biology) were grown in minimal essential medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine (Gibco, BRL). Transient transfection into HaCaT cells was carried out by the standard DEAE-dextran method. Briefly, 105 cells were plated onto each well of a six-well plate and grown overnight. The cells were then washed once with phosphate-buffered saline (PBS) and submerged in 1 ml of serum-free minimal essential medium containing 100 mM chloroquine. Six micrograms of reporter plasmid, plus 6 μg of a cotransfection plasmid (or 6 μg of vector plasmid as filler DNA to keep the total amount of transfection plasmids constant) where applicable, was resuspended in 340 μl of PBS containing 85 μg of DEAE-dextran per ml and added dropwise onto each well of cells. Cells were cultured for 3 h before being treated with 10% glycerol for 2 min. The cells were then washed with PBS, and 2 ml of growth medium was added. Cells were grown for 24 h before the addition of 100 pM human TGF-β1. Cells were then grown for an additional 24 h, and the luciferase activities were measured according to standard protocols (37).
Mouse pre-B-cell line 300.19 was obtained from T. Tedder (Duke University Medical Center) and grown in RPMI 1640 supplemented with 10% fetal bovine serum and 2 mM β-mercaptoethanol (Gibco, BRL). For transient transfection into 300.19 cells, cells were collected, washed once with PBS, and resuspended in growth medium to obtain a cell density of 4 × 106 cells per ml. Ten micrograms of the luciferase reporter and 1 μg of CMV–β-galactosidase plasmid were mixed with 0.4 ml of cells and transferred into a 0.4-cm-gap-width electroporation cuvette. Cells were electroporated with a BTX electroporator (ECM600) which was set at a capacity of 1,700 mF and a charging voltage of 250 V, with a typical pulse time of 35 ms. The cells were then left for 20 min at room temperature to recover before being added to 12 ml of growth medium in a 10-cm-diameter plate. Cells were grown for 24 h and split in equal volumes (6 ml) into two 10-cm-diameter plates. TGF-β1 (100 pM) was added to one of the plates. Cells were grown for an additional 24 h, and luciferase activity was measured as described for HaCaT cells. β-Galactosidase activity was then measured to monitor transfection efficiency and ensure equal expression from both the TGF-β-treated and untreated cells.
Electrophoretic mobility shift assay (EMSA).
Nuclear and cytoplasmic extracts were prepared as described previously (20). Gel mobility shift assay was carried out with a 10-ml reaction mixture which contained 20 mM HEPES (pH 7.9), 40 mM KCl, 6 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.15% bovine serum albumin, 10% (wt/vol) Ficoll, and 0.1 mg of sonicated salmon sperm DNA (Sigma) per ml. Specific competitors or antisera were added to each reaction mixture as indicated in Fig. 4 before the addition of 5 μg of total proteins. One nanogram of probe, which was labeled with T4 polynucleotide kinase according to the standard method (37), was added last. DNA-protein complexes were separated from the free DNA probe on a 4% nondenaturing polyacrylamide gel containing 10% glycerol at 4°C. The gel was then dried and exposed to X-ray film with an intensifying screen.
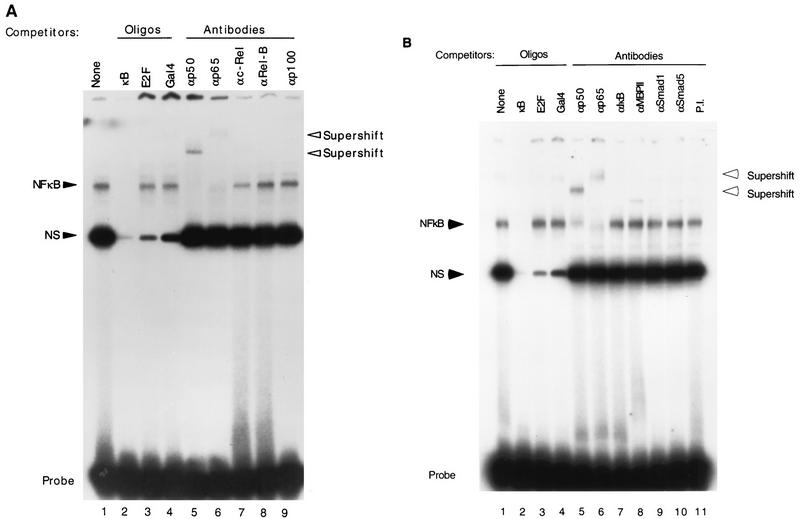
FIG. 4.
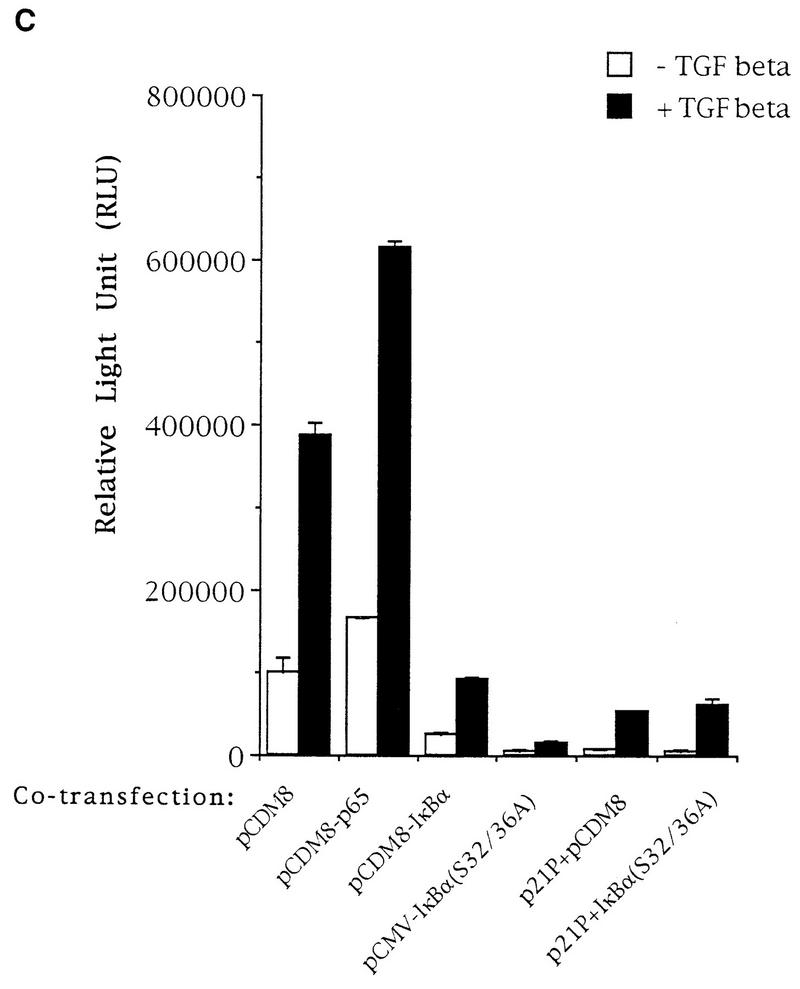
p50-p65 binds to NF-κB sites and is required for TGF-β-mediated gene expression. (A) Oligonucleotides representing the HIV-1 LTR NF-κB sites were labeled with T4 polynucleotide kinase and used as a probe in EMSA with nuclear extract prepared from TGF-β-treated HaCaT cells. In lanes 2 to 4, 20-fold excesses of various oligonucleotides (Oligos) were included in the EMSA as competitors as indicated above each lane. In lanes 5 to 9, antibodies against the indicated human proteins were included in the EMSA. The specific complex (NF-κB) is indicated. NS, nonspecific complex; Probe, free probe. Supershifted complexes are also indicated. (B) Same type of experiment as in panel A, except that the nuclear extract was prepared from HaCaT cells with no TGF-β treatment. Lanes 2 to 4 contained oligonucleotides in the EMSAs as indicated. Lanes 5 to 11 contained specific antibodies against proteins as indicated. (C). Expression vector alone (pCDM8) or expression plasmids for p65 (pCDM8-p65), IκBα (pCDM8-IκBα), or IκBα(S32/36A) [pCMV4-IκBα(S32/36A)] were cotransfected transiently with TI+1×κB-luc or p21P into HaCaT cells, and their relative luciferase activities were assayed 24 h after cells were treated or not with TGF-β. The fold induction by TGF-β is indicated. Error bars represent standard deviations from duplicate determinations.
Western blot analysis.
Equal amounts of total protein from the nuclear and cytoplasmic extracts were loaded onto a sodium dodecyl sulfate–10% polyacrylamide gel. After electrophoresis, proteins were electroblotted onto a polyvinylidene difluoride membrane by using a Bio-Rad Trans-Blot cell. The membrane was then blocked in 5% nonfat milk–0.1% Tween 20 before being probed with a 1:1,000 dilution of specific antibodies as indicated in the figures. Proteins detected by specific antibodies were visualized by ECL (Amersham) as described in the manufacturer’s manual and exposed to X-ray film.
RNase protection assay.
To isolate cytoplasmic RNA, HaCaT cells were first washed with ice-cold PBS three times and collected into a 15-ml conical tube in a small volume of PBS. Cells were then pelleted by centrifugation at 300 × g for 5 min, and the supernatant was discarded. The cell pellet was lysed with 375 μl of ice-cold lysis buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, and 0.5% Nonidet P-40. The lysate was incubated for 5 min on ice and centrifuged for 2 min in a cold room to pellet nuclei. The cytoplasmic supernatant was then extracted twice with water-saturated phenol and once with chloroform. The cytoplasmic RNA was precipitated with ethanol and dissolved in water. For the RNase protection assay, we used an RPA II RNase Protection Assay Kit (Ambion) and followed the protocol supplied by the manufacturer.
RESULTS
TGF-β stimulates the HIV-1 LTR promoter.
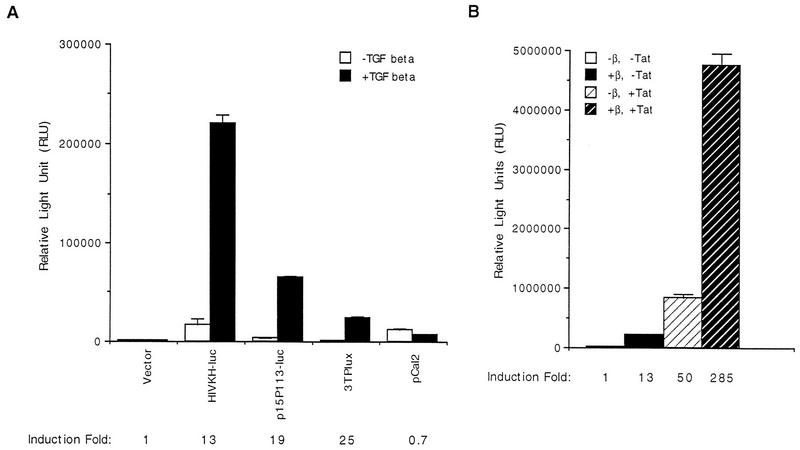
To investigate the effect of TGF-β on the expression of the HIV-1 LTR promoter, the construct HIVKH-luc was transiently transfected into HaCaT cells, and its relative luciferase activity was assayed after cells were treated with human TGF-β1 for 24 h or untreated. As shown in Fig. 1A, HIVKH-luc was activated 13-fold upon TGF-β treatment. As positive controls, the cyclin-dependent kinase inhibitor (CKI) p15INK4B gene promoter (25) and an artificial reporter construct, 3TPlux, which contains a portion of the plasminogen activator inhibitor-1 gene promoter linked to TRE sequences from the collagenase promoter (6), were induced similarly following TGF-β treatment (Fig. 1A, p15P113-luc and 3TPlux). On the other hand, the expression from the cyclin A promoter was reduced 30% upon TGF-β treatment (Fig. 1A, pCAL2), as previously reported (12). Therefore, TGF-β is able to potently activate the HIV-1 LTR promoter in HaCaT cells.
FIG. 1.
TGF-β stimulates the HIV-1 LTR promoter. (A) The KpnI-to-HindIII fragment of the HIV-1 LTR was placed upstream of a luciferase reporter plasmid (pGL2-Basic) to create HIVKH-luc. HIVKH-luc was transfected transiently into HaCaT cells, and its relative luciferase activity was assayed after cells were treated or not with human TGF-β1 for 24 h. p15P113-luc (25), 3TPlux, and the cyclin A promoter pCAL2-luc (12) were also transfected and assayed similarly. The fold induction is indicated for each construct. Error bars represent standard deviations from duplicate determinations. (B) HIVKH-luc was transiently transfected into HaCaT cells, and the relative luciferase activities were assayed after cells were treated (+β, −Tat) or not (−β, −Tat) human TGF-β1 for 24 h. CMV-Tat was cotransfected with HIVKH-luc into HaCaT cells, and the relative luciferase activities were assayed after cells were treated (−β, +Tat) or not (−β, +Tat) with TGF-β for 24 h. The fold induction by TGF-β is indicated. Error bars represent standard deviations from duplicate determinations.
Virally encoded Tat protein is a potent transactivator of the HIV-1 LTR promoter (reviewed in reference 15). We next examined the relationship between TGF-β-mediated activation of the HIV-1 LTR promoter and Tat-mediated transactivation. The Tat expression plasmid CMV-Tat was cotransfected with HIVKH-luc, and the luciferase activity was assayed after cells were treated with TGF-β or untreated. As shown in Fig. 1B, TGF-β treatment by itself caused 13-fold induction of the reporter expression, whereas coexpression of CMV-Tat alone stimulated the expression of HIVKH-luc by about 50-fold. Combination of the CMV-Tat coexpression and TGF-β treatment, however, synergistically increased the promoter activity to 285-fold. These results indicate that TGF-β stimulates gene expression from the HIV-1 LTR promoter through a mechanism distinct from that of Tat transactivation.
NF-κB binding sites are required for TGF-β-mediated gene expression from the HIV-1 LTR promoter.
The HIV-1 LTR promoter contains three copies of Sp1 binding sites in addition to two NF-κB binding sites (reviewed in reference 15). To determine the promoter elements required for TGF-β induction of the HIV-1 LTR promoter, we generated serial deletion constructs of the HIV-1 LTR promoter with HIVKH-luc as the parental construct (Fig. 2A) and tested for their induction by TGF-β treatment. Deletion of sequences immediately upstream of the NF-κB sites did not change TGF-β induction of the promoter (Fig. 2A, HIVRH-luc). However, deletion of both NF-κB sites significantly decreased TGF-β-induced expression as well as the uninduced expression (Fig. 2A, HIVdκB-luc), suggesting that NF-κB sites were needed for the promoter induction by TGF-β. The remaining sixfold induction by TGF-β, after NF-κB sites were deleted, resulted from the presence of three Sp1 binding sites, since deletion of both the NF-κB and Sp1 binding sites completely abolished TGF-β induction of the promoter (Fig. 2A, HIVdSp1-luc). Deletion of the TATA box did not further affect the promoter expression (Fig. 2A, HIVPH-luc). Previously we have shown that Sp1 binding sites are capable of supporting TGF-β-induced gene activation from the promoters of the CKIs p15INK4B and p21(CIP1/WAF1) (8, 25). Results from the current promoter deletion analysis demonstrated that both NF-κB and Sp1 binding sites contribute to TGF-β induction of the HIV-1 LTR promoter, with NF-κB sites playing a more prominent role.
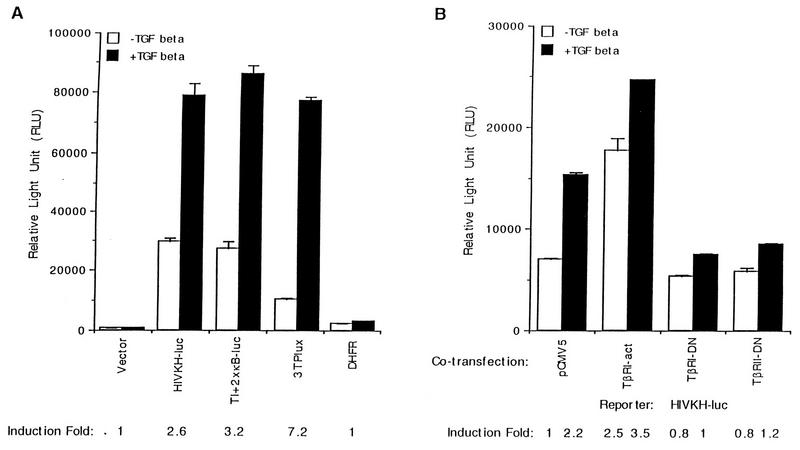
NF-κB binding sites are sufficient for TGF-β-mediated gene activation.
To further define promoter elements required for the induction of the HIV-1 LTR promoter by TGF-β treatment, we mutated either copy of the NF-κB binding sites by site-directed mutagenesis to create HIVKHmκB1-luc and HIVKHmκB2-luc (Fig. 2B). We also created HIVKHDSp1-luc, which had all three Sp1 binding sites deleted but retained rest of the sequences (Fig. 2B). Mutating either copy of the NF-κB binding sites significantly reduced the basal expression but not the fold induction by TGF-β treatment, indicating that either of the two NF-κB binding sites was sufficient for TGF-β-mediated induction of the HIV-1 LTR promoter (Fig. 2B, HIVKHmκB1-luc and HIVKHmκB2-luc). Deleting all three Sp1 binding sites while retaining the rest of the promoter sequences dramatically reduced the uninduced expression but, surprisingly, resulted in an even higher fold induction after TGF-β treatment (Fig. 2B, HIVDSp1-luc). These results indicate that NF-κB sites alone were sufficient to support TGF-β-mediated gene activation from the HIV-1 LTR promoter. To generate further support for this hypothesis, we synthesized oligonucleotides comprising NF-κB binding sites from the HIV-1 LTR promoter and inserted them upstream of a minimal-promoter luciferase reporter gene containing only the TATA box (TATA) and the initiator element (Inr, as defined in reference 14), which by itself is induced only slightly by TGF-β treatment (Fig. 3A, TI-luc). To our satisfaction, introduction of one copy of the NF-κB oligonucleotides (which contains both HIV-1 NF-κB binding sites) caused the minimal promoter to be induced 28-fold upon TGF-β treatment (Fig. 3A, TI+1×κB-luc). Introduction of two or three copies of the NF-κB oligonucleotides caused a dosage-dependent increase in the overall levels of TGF-β-induced expression, although the induction decreased from 28- to 7-fold due to the more significant increases in uninduced expression (Fig. 3A, TI+2×κB-luc and TI+3×κB-luc). Induction from NF-κB sites specifically required signals initiated by a functional TGF-β receptor complex, since coexpression of a dominant-negative type I (TβRI-DN) or a dominant-negative type II (TβRII-DN) receptor abolished the induction by TGF-β without affecting the uninduced expression (Fig. 3B). Taken together, these results suggest that NF-κB binding sites alone are sufficient to support TGF-β-mediated gene activation.
The p50-p65 complex binds to NF-κB sites and is required for TGF-β-mediated gene expression.
Many cellular transcription factors bind to NF-κB sites, including Rel/NF-κB family members and the zinc finger-containing proteins MBP-1/PRDII-BP-1/HIV-EP1 and MBP-2 (reviewed in references 2 and 30). To determine the identity of factors binding to the HIV-1 NF-κB sites, we used EMSA and detected a specific complex formed on the HIV-1 NF-κB binding sites in both TGF-β-induced and uninduced HaCaT cells (Fig. 4A and B). This complex was specific to the NF-κB probe used, since it was readily competed away by a 20-fold excess of the unlabeled NF-κB oligonucleotides (Fig. 4A and B, lanes 2) but not by the same excess amount of E2F or Gal4 binding oligonucleotides (Fig. 4A and B, lanes 3 and 4). Addition of antiserum against either the human p50 or p65 abolished the complex formation and caused formation of supershifted complexes, indicating that the complex contained both p50 and p65 proteins (Fig. 4A and B, lanes 5 and 6). Previous studies have demonstrated that endogenous p50-p65 heterodimers are much more stable and capable of binding to DNA with higher affinity than p65 homodimers (38). In addition, the p50-p65 heterodimer complex has been shown to run typically at a position between those of the p50 and p65 homodimers in EMSA (38). Consistent with those findings, the p50-p65-containing complex from HaCaT cells runs at precisely the same position as the complex consisting of the recombinant p50-p65 heterodimer, and its mobility lies between those of the recombinant p50 and p65 homodimers (data not shown). Addition of antisera against human c-Rel, Rel-B, and p100 (Fig. 4A, lanes 7 to 9) and against IκBα, MBP2, Smad1, and Smad5 (Fig. 4B, lanes 7 to 10), or preimmune antiserum (Fig. 4B, lane 11), did not affect the complex formation. These results suggest that the major factor binding to the HIV-1 NF-κB sites in HaCaT cells is the p50-p65 heterodimer.
To investigate whether NF-κB activities are required for promoter induction by TGF-β through NF-κB binding sites, we cotransfected p65, IκBα, or a mutant form of IκBα, IκBα(S32/36A), into HaCaT cells and assayed their effects on the expression of the TI+1×κB-luc reporter construct (Fig. 3A) in the absence or presence of TGF-β treatment. The IκBα(S32/36A) mutant represents a supersuppressive form of IκBα, as it contains serine-to-alanine substitutions at residues 32 and 36 which when phosphorylated can lead to ubiquitin-mediated degradation of IκBα (4, 44, 46). Coexpression of p65 stimulated slightly both the uninduced and TGF-β-induced expression from the TI+1×κB-luc reporter gene (Fig. 4C), while coexpression of the wild-type IκBα significantly reduced both the uninduced and TGF-β-induced expression (Fig. 4C). In these cotransfection experiments, the fold induction by TGF-β remained the same even though the uninduced expression was either higher (pCDM8-p65) or lower (pCDM8-IκBα) than that with the vector cotransfection control. Identical results were obtained when the construct containing the HIV-1 LTR promoter, HIVKH-luc, was used in the assay (data not shown). A negative control, the p21(CIP1/WAF1) promoter, which contains only Sp1 binding sites as its TGF-β-responsive elements (9), was not affected by the coexpression of either p65 or IκBα(S32/36A) (Fig. 4C). These results suggest that TGF-β induction from NF-κB sites depends on the activity of NF-κB. Indeed, when the NF-κB activity was further reduced by coexpression of the ubiquitin-mediated-degradation-resistant super suppressive form of IκBα, IκBα(S32/36A), both the uninduced and TGF-β-induced expression from NF-κB sites was further reduced (Fig. 4C). The remaining induction by TGF-β probably reflects the presence of residual NF-κB activities. Taken together, these results suggest that NF-κB activity is required for TGF-β-induced gene expression from NF-κB binding sites.
TGF-β treatment does not change NF-κB and IκB activities.
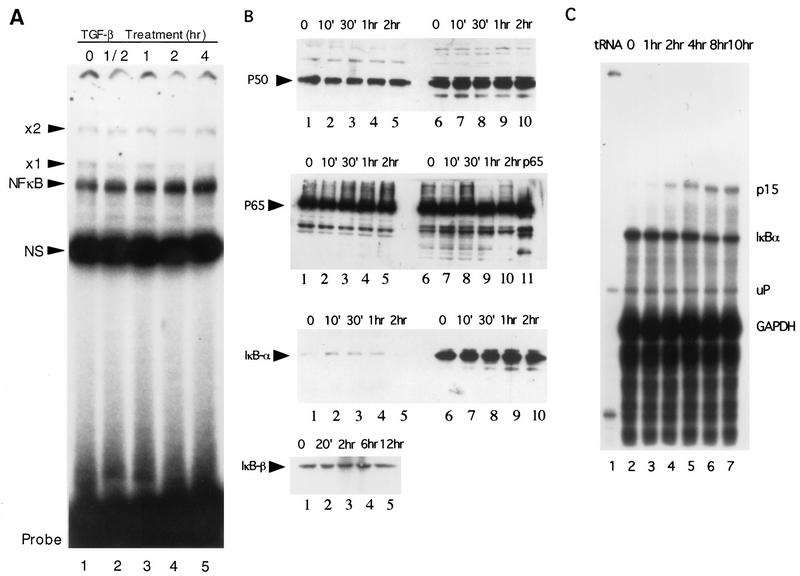
NF-κB activity has been shown to be readily inducible upon activation of most premature lymphoid cells, thus providing a rapid response to various external stimuli (reviewed in references 2 and 30). We investigated if TGF-β treatment caused similar changes in NF-κB activity by measuring its DNA binding activity with extracts prepared from cells treated with TGF-β for various length of time as well as with extracts from untreated cells. As shown in Fig. 5A, TGF-β treatment did not cause significant changes in the NF-κB binding activity over a 4-h period. Similar experiments showed no changes in the DNA binding activity from 10 min up to 24 h (data not shown). We also measured levels of p50, p65, and IκBα proteins in the same nuclear and cytoplasmic extracts by Western blot analysis. As shown in Fig. 5B, relatively equal amounts of p50 and p65 were present in the nuclear and cytoplasmic extracts. Neither p50 nor p65 protein levels changed significantly upon TGF-β treatment (Fig. 5B). IκBα was located mostly in the cytoplasmic fraction, and its protein levels did not change after treatment with TGF-β. Likewise, total IκBβ protein levels did not change with TGF-β treatment, consistent with unchanged DNA binding activity of NF-κB (Fig. 5B). Furthermore, RNase protection analysis showed that the IκBα mRNA levels remained unchanged with TGF-β treatment from 1 to 10 h, whereas the p15INK4B mRNA level was induced more than 20-fold with the same treatment (Fig. 5C).
FIG. 5.
TGF-β does not change NF-κB DNA binding activities or NF-κB levels in HaCaT cells. (A) Nuclear extracts were prepared from HaCaT cells treated with human TGF-β1 for 0 h (lane 1), 1/2 h (lane 2), 1 h (lane 3), 2 h (lane 4), or 4 h (lane 5) and used in an EMSA. The NF-κB complex is indicated. Also indicated are the nonspecific complex (NS) and two uncharacterized complexes, ×1 and ×2. Probe, free probe. (B) Western blot analyses were performed with specific antibodies against human p50, p65, and IκBα as indicated. Nuclear (lanes 1 to 5) and cytoplasmic (lanes 6 to 10) extracts were prepared from HaCaT cells treated with TGF-β for 0 min (lanes 1 and 6), 10 min (lanes 2 and 7), 30 min (lanes 3 and 8), 1 h (lanes 4 and 9) and 2 h (lanes 5 and 10). For Western analysis of IκBβ, total cell lysates treated with TGF-β for 20 min to 12 h were used. Specific bands for p50, p65, IκBα, and IκBβ are indicated by arrows. Recombinant p65 was loaded as a control in lane 11 of the p65 panel. (C) Cytoplasmic RNA was isolated from HaCaT cells treated with TGF-β for 1 h (lane 3), 2 h (lane 4), 4 h (lane 5), 8 h (lane 6), and 10 h (lane 7), as well as from untreated cells (lane 2), and used in an RNase protection assay. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and p15 mRNAs were included in the same experiment as controls. tRNA was used as negative control (lane 1). mRNA bands for p15, IκBα, and GAPDH are indicated. uP, undigested GAPDH riboprobe.
In addition, we used two pharmacological inhibitors, calpain inhibitor I, which inhibits the degradation of phosphorylated IκB (29), and PDTC, which inhibits IκB phosphorylation (39), to test if they could affect the TGF-β-mediated promoter transactivation through NF-κB sites. No significant effect on the TGF-β-induced promoter activity was observed when calpain inhibitor I was present in concentrations of between 0.1 and 2 mM (data not shown). We were unable to use higher concentrations of the inhibitor because of the significant cytotoxic effect of the drug on HaCaT cells (data not shown). Interestingly, addition of PDTC in concentrations of between 10 and 40 μM to HaCaT cells led to a more-than-10-fold activation of both the uninduced and TGF-β-induced activities from the TI+1×κB-luc reporter construct (data not shown), suggesting that PDTC may have some unknown activities, other than inhibiting specifically the phosphorylation of IκB (39). Taken together, these results suggest that TGF-β affects the transactivation activity of NF-κB in HaCaT cells through a yet-unidentified mechanism which is different from the classic mechanism involving IκB degradation observed in premature lymphoid cells.
Gal4-p65 can mediate TGF-β-induced promoter activation through Gal4 binding sites.
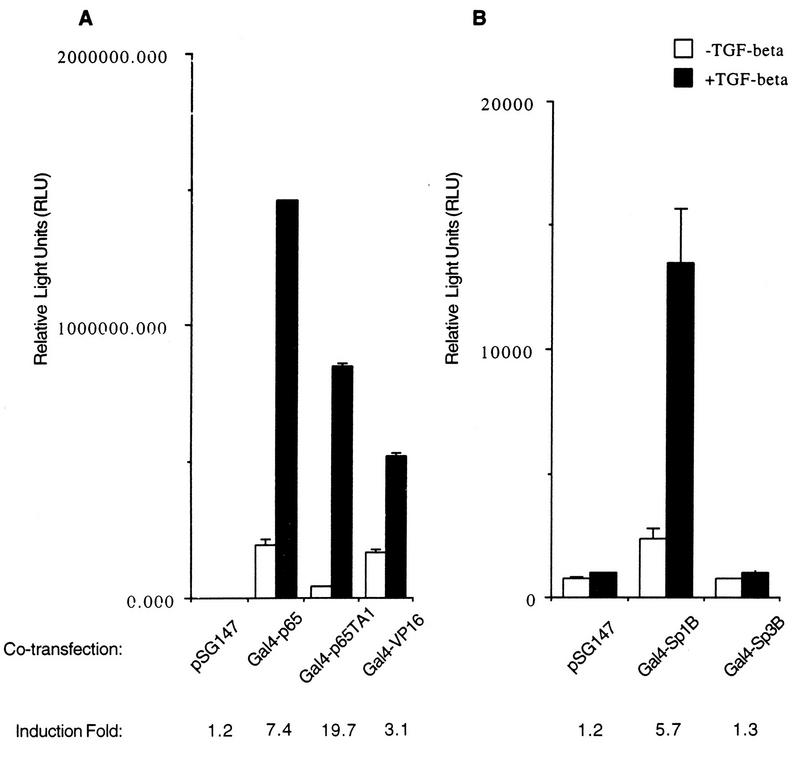
If NF-κB is indeed required for TGF-β-mediated transcription activation through NF-κB binding sites and yet its protein levels and DNA binding activities are not affected by TGF-β treatment, we would expect Gal4-p65 fusion proteins to support TGF-β-mediated expression from a promoter containing Gal4 DNA binding sites. Chimeric constructs fusing the first 147 amino acids of the Gal4 DNA binding domain to either the full-length p65 or the TA1 of p65 alone (38) were cotransfected with a luciferase reporter which contains five copies of Gal4 DNA binding sites in its promoter in addition to the TATA box and initiator element. As shown in Fig. 6A, the Gal4 fusion protein containing the full-length p65 supported significantly elevated expression from the luciferase reporter which was further induced strongly upon TGF-β treatment (Fig. 6A). Interestingly, 30 amino acids (amino acids 520 to 550) of the p65 TA1 alone also supported transcription from Gal4 binding sites, which was induced by TGF-β almost 20-fold (Fig. 6A). The weaker TGF-β-induced transactivating activity of the Gal4–full-length p65 construct is possibly the result of a combinational effect derived from both the presence of the DNA binding domain of p65 in the fusion construct and a higher noninduced transactivating activity. A control, the Gal4-VP16 construct, elicited a similarly strong transactivation response from the reporter which was also further induced by TGF-β treatment (Fig. 6A). This apparently nonspecific two- to threefold TGF-β-induced promoter activity is observed frequently in our assay systems with certain other types of luciferase constructs, and we attribute the source of the nonspecific promoter activity to the potential change in the stability of luciferases in cells treated with TGF-β, as has been reported in other circumstances (33). In any event, we have clearly demonstrated that the Gal4-p65 constructs could specifically mediate the TGF-β-induced transactivating activity, even when the nonspecific induced activity is taken into account. It is also important to point out that the noninduced transactivating activity of the Gal4-p65 full-length construct is at least comparable to, if not stronger than, that of the Gal4-VP16 construct (Fig. 6A), a known potent transactivator of transcription.
FIG. 6.
p65 can mediate TGF-β-induced transcriptional activation. (A) Gal4 vector plasmid (pSG147), Gal4-p65 chimeric constructs containing either the full-length p65 (Gal4-p65) or TA1 of p65 (Gal4-p65TA1), and Gal4-VP16 were cotransfected transiently with the TI+5×Gal4-luc reporter plasmid into HaCaT cells by the standard DEAE-dextran method, and their relative luciferase activities were measured after cells were treated or not with TGF-β1 for 24 h. The fold induction by TGF-β is indicated. Error bars represent standard deviations from duplicate determinations. (B) In the same experiment as shown in panel A, the Gal4-Sp1B and Gal4-Sp3B constructs were used as controls in cotransfection with the reporter plasmid. The only difference between the two panels is the scale of relative light units as a reflection of differences in the overall transactivating activities of those Gal4 fusion constructs.
To further demonstrate the specificity of this TGF-β-induced transcriptional activation through the Gal4-p65 fusion proteins, we used two more control Gal4 fusion constructs in the same experiments. As shown in Fig. 6B, the Gal4-Sp1B construct, which contains the transcription factor Sp1 transactivation subdomain B fused to the Gal4 DNA binding domain, responded to TGF-β similarly to the Gal4-p65 constructs, a result consistent with our previous findings (references 9 and 25 and unpublished results). In contrast, the Gal4-Sp3B construct, which contains the corresponding transactivation subdomain B of transcription factor Sp3 fused to the Gal4 DNA binding domain, failed to respond to the TGF-β signal (Fig. 6B). The striking difference between these two Gal4 fusion proteins, which belong to the same Sp1 family of transcription factors, was primarily the result of their differential responses to a specific signal, in this case TGF-β, as the expression levels and DNA binding capabilities of the two fusion proteins are essentially the same (9a). Of interest are the reproducible significant differences in the overall activities of the NF-κB p65 constructs versus the Sp1 and Sp3 constructs, which display transactivating activities that are almost 2 orders of magnitude lower in the same assay. While the exact reason underlying the differences is not known, we suspect that the expression levels of the two types of fusion proteins may contribute to the differences. Taken together, these results demonstrated that p65 was sufficient to support TGF-β-mediated promoter activation. Possibly, TGF-β treatment enhanced gene expression by increasing the transactivation potential of p65, as in the case of the TA1 domain of p65, or the ability of p65 to interact with other coactivators. It is also possible that TGF-β modifies specific coactivators which could then be recruited to the promoter region more readily through interaction with p65 or one of its functional domains.
Activation of the HIV-1 LTR by TGF-β does not require the activity of transcription factor NF-AT.
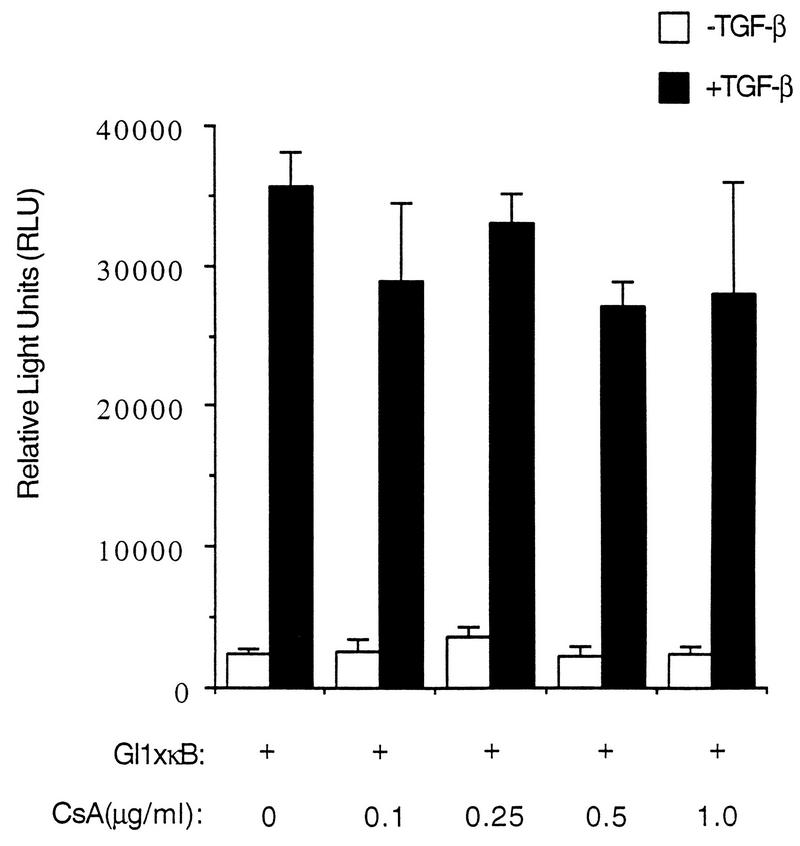
A recent study demonstrated that transcription factor NF-AT, which interacts specifically with the NF-κB sites in the HIV-1 LTR, can synergize with NF-κB and Tat in the transcription activation of HIV-1 and can enhance HIV-1 replication in T cells (19). To investigate if TGF-β can activate transcription through the activity of NF-AT, we measured the TGF-β-mediated transactivation of the construct TI+1×κB-luc in the presence of various concentrations of CsA, a specific pharmacological inhibitor of NF-AT (13). As shown in Fig. 7, CsA has no effect on TGF-β-induced promoter activity, suggesting that NF-AT is probably not involved in the mediation of the TGF-β transactivation signal in HaCaT cells.
FIG. 7.
TGF-β-induced activation of NF-κB transcriptional activity does not require NF-AT activity. TI+1×κB was transiently transfected into HaCaT cells, and different concentrations of CsA were added 12 h after transfection, right before addition of TGF-β. Relative luciferase activities were assayed after cells were treated or not with TGF-β for 24 h. Error bars represent standard deviations from duplicate determinations.
TGF-β activates the HIV-1 LTR in 300.19 pre-B cells.
To investigate if TGF-β could activate transcription from the HIV-1 LTR promoter in cell types other than HaCaT cells, we screened more than a half-dozen cell lines of lymphoid and myeloid lineages for TGF-β responsiveness. Only one cell line, the mouse pre-B cell line 300.19, tested positive for TGF-β-mediated growth arrest as determined by a thymidine incorporation assay (data not shown). When HIVKH-luc was transiently transfected into 300.19 cells by electroporation, two- to threefold induction of relative luciferase activity was consistently observed after cells were treated with TGF-β (Fig. 8A). The luciferase reporter construct driven by two copies of the HIV-1 NF-κB sites alone was similarly activated by TGF-β treatment (Fig. 8A, TI+2×κB-luc). In the same experiment, the 3TPlux luciferase reporter was also activated about sevenfold (Fig. 8A). In contrast, the expression from the negative-control DHFR luciferase reporter was not affected by TGF-β treatment (Fig. 8A). Coexpression of either the dominant-negative type I or the dominant-negative type II receptor significantly reduced TGF-β induction of the HIV-1 LTR promoter (Fig. 8B, TβRI-DN and TβRII-DN), while a constitutively active type I receptor induced HIVKH-luc expression in a ligand-independent manner (Fig. 8B, TβRI-act). These results indicate that the observed induction of the HIV-1 LTR promoter by TGF-β treatment is indeed mediated by a specific TGF-β signaling pathway. Thus, TGF-β-mediated transcriptional activation through NF-κB is not limited to HaCaT cells but also exists in other cell types such as 300.19 pre-B cells of lymphoid origin.
FIG. 8.
TGF-β stimulates expression from the HIV-1 LTR promoter in 300.19 pre-B cells. (A) HIVKH-luc was transiently transfected into 300.19 pre-B cells by electroporation as detailed in Methods and Materials, and its relative luciferase activity was assayed after cells were treated or not with human TGF-β1 for 24 h. Similarly transfected and assayed were the vector plasmid, 3TPlux, TI+2×κB-luc (Fig. 3A), and DHFR-luc. The fold induction is indicated. Error bars represent standard deviations from duplicate determinations. (B) HIVKH-luc was cotransfected transiently with the pCMV5 expression vector, a constitutively active TGF-β receptor expression plasmid (TβRI-act), a dominant-negative TGF-β receptor I (TβRI-DN), or a dominant-negative TGF-β receptor II (TβRII-DN), and luciferase activities were assayed as described for panel A. The fold induction relative to the uninduced (pCMV5) expression is indicated. Error bars represent standard deviations from duplicate determinations.
DISCUSSION
TGF-β has been shown to promote HIV-1 viral production in primary macrophages and U937 cells (22), however, the mechanism by which TGF-β increases HIV-1 viral production in these cells is not known. We demonstrated here that TGF-β activated the HIV-1 LTR promoter in HaCaT cells as well as in 300.19 pre-B cells, primarily through NF-κB binding sites. TGF-β can synergize with the virally encoded Tat transactivator to induce greater induction from the HIV-1 LTR. Tat has previously been shown to be a strong inducer of TGF-β1 gene expression, which is correlated with its potent immunosuppressive activity (7). Taken together, these findings indicate that both Tat and TGF-β can be employed by HIV-1 to perform similar functions as immunosuppressors of the host immune system and at the same time as transcription activators of the HIV-1 LTR promoter to promote viral production. This hypothesis is consistent with reports of increased TGF-β production or expression levels in peripheral blood mononuclear cells (18), primary mononuclear phagocytes (22, 23), and brain tissues (45) from HIV-1-infected patients. It will be important to investigate if TGF-β displays the same transactivating effect of the HIV-1 LTR in those cell types, which are natural hosts of HIV. On the other hand, it will be equally important to identify endogenous target genes in immune cells regulated by TGF-β through this novel signaling pathway, thus providing a better understanding of the molecular basis for the diverse biological effects of TGF-β on the immune system.
In this study, we have demonstrated that NF-κB activity, specifically p65, is required for the mediation of the TGF-β transactivating effect in HaCaT cells. NF-κB is the prototype of a group of transcription factors which bind to the NF-κB DNA binding sites. NF-κB controls many genes of biological importance, such as those encoding the immunoglobulin κ light chain, inflammatory cytokines, chemokines, interferons, major histocompatibility complex proteins, growth factors, cell adhesion molecules, and viruses (reviewed in references 2 and 30). Most premature lymphocytes contain NF-κB in an inactive form through its association with the inhibitory IκB family of proteins, and the inactive NF-κB–IκBα complex resides in the cytoplasm. Upon treatment with specific cytokines such as interleukin-2 and tumor necrosis factor alpha or stress signals such as UV irradiation, IκBα is rapidly phosphorylated and degraded through the ubiquitin-proteasome degradation pathway (31). Dissociation from IκBα enables NF-κB to translocate into the nucleus, where it elicits various gene responses (reviewed in references 2 and 30). Unlike this classic NF-κB activation mechanism, we have demonstrated that HaCaT cells contain constitutively active NF-κB activities which are apparently not changed upon TGF-β treatment (Fig. 5A). Furthermore, HaCaT cells contain abundant IκBα mRNA and proteins which are located primarily in the cytoplasm (Fig. 7B). This is reminiscent of mature B cells, which express constitutively active NF-κB as well as large amounts of IκBα proteins and mRNAs in the cytoplasm (28). Therefore, at least in HaCaT cells, TGF-β-mediated transcription activation through NF-κB sites does not involve changes in the NF-κB DNA binding activity. Rather, there is a constitutive fraction of active NF-κB present in the nucleus, probably due to rapid IκBα dissociation and degradation as proposed for mature B cells (28), a notion consistent with a recent finding that NF-κB is present in significant quantities in keratinocyte nuclei (48). The TGF-β signal appears to reach the NF-κB molecules in the nucleus and activate their transactivating activity through a yet-unidentified pathway. This is consistent with our results that Gal4-p65 chimeric constructs could support ligand-dependent activation of a promoter driven by Gal4 DNA binding sites.
In WEHI231 immature B cells, TGF-β was shown to cause apoptosis by increasing the IκBα mRNA level and consequently decreasing NF-κB binding activity (1). However, we did not see significant changes in the protein levels of either IκBα, IκBβ, p50, or p65 after TGF-β treatment for up to 24 h in HaCaT cells (Fig. 5B and data not shown), nor did we observe changes in IκBα mRNA levels after TGF-β treatment (Fig. 5C). In fact, we routinely observed slightly increased NF-κB DNA binding activities upon TGF-β treatment (Fig. 5A). This discrepancy most likely reflects the difference in the cell types employed. Perhaps both mechanisms exist, depending on cell types and growth conditions. Thus, TGF-β treatment could down-regulate NF-κB activity and cause apoptosis in WEHI231 immature B cells (1), whereas the same treatment causes G1 growth arrest in HaCaT cells which contain constitutive NF-κB activity. It is possible that the constitutively active NF-κB activity, which was unchanged upon TGF-β treatment, might protect certain cell types from undergoing apoptosis, since NF-κB has recently been shown to exert antiapoptotic activity (4, 44, 46).
The notion that there are alternative mechanisms for gene activation by NF-κB which do not involve changes in the NF-κB DNA binding activity is also supported by other studies. For example, it was shown that overexpression of p21(CIP1/WAF1) could stimulate HIV-1 gene expression without inducing changes in NF-κB DNA binding activities in Jurkat cells (32). Indeed, coexpression of p21(CIP1/WAF1) and p65 (RelA) caused synergistic induction from the HIV-1 promoter, suggesting that p21(CIP1/WAF1) induces expression from NF-κB sites by a mechanism distinct from that of p65 (RelA) (32). TGF-β causes growth arrest in certain cell types by inducing the genes for CKI p15INK4B and/or p21(CIP1/WAF1) (8, 25). It is possible that the TGF-β-induced p21(CIP1/WAF1) could in turn stimulate HIV-1 gene expression, presumably through the p300/CBP coactivator proteins as proposed by Perkins et al. (32). Our preliminary results support the possibility of such a mechanism since coexpression of p21(CIP1/WAF1) stimulated HIV-1 gene expression about two- to threefold in HaCaT cells. Moreover, Gal4-p300/CBP chimeric proteins could support gene induction by TGF-β from Gal4 DNA binding sites, which is consistent with the involvement of p300/CBP coactivators in transcription activation mediated by the TGF-β signaling pathway (our unpublished results). However, additional mechanisms may be involved in the stimulation of HIV-1 gene expression by TGF-β through NF-κB sites, because the induction of the HIV-1 promoter through the coexpression of p21(CIP1/WAF1) is further enhanced significantly by TGF-β treatment (our unpublished results).
It is possible that TGF-β could induce phosphorylation of p65 through an unidentified kinase, leading to the activation of NF-κB, as phosphorylation of p65 by PKA has recently been demonstrated to play an important role in the regulation of NF-κB activity (49). Our attempts to address the question of whether PKA is involved in this specific TGF-β signaling pathway have so far generated mixed results. In those studies, we tested the ability of two reagents to abrogate the effect of TGF-β on NF-κB-mediated transactivation: a dominant-negative mutant form of PKA and a pharmacological inhibitor of PKA kinase activity, H89, both of which were used successfully in that recent study (49) to demonstrate the involvement of PKA in the activation of NF-κB activity. We found that coexpression of the dominant-negative mutant form of PKA had no effect on the fold induction mediated by TGF-β, even though the overall promoter activity was slightly reduced. In contrast, the presence of 20 μM H89 significantly reduced TGF-β-induced activity of the HIV-1 LTR, as well as that of several TGF-β-responsive control promoters, such as the Sp1 site-driven p15 and p21 promoters and the AP1-Smad-driven p3TP-lux promoter (data not shown), suggesting that H89 could have additional effects on other kinases. Based on those conflicting results, we are unable to reach a firm conclusion as to the exact role of PKA, if any, in the mediation of the TGF-β signal in activating transcription through the activity of NF-κB. Further experiments will certainly be needed to elucidate the precise mechanisms by which TGF-β regulates NF-κB activation, consequently helping us understand how TGF-β signals specific gene responses and the roles that TGF-β may play in HIV-1 viral gene regulation and pathogenesis.
ACKNOWLEDGMENTS
We thank Albert S. Baldwin, Jr., for critical reading of the manuscript and for many valuable suggestions related to potential involvement of IκBα, Lishan Su for critical reading of the manuscript and helpful discussions, and Yong Yu for excellent technical support. We also thank Brian Cullen for HIV-chloramphenicol acetyltransferase and CMV-Tat plasmids, M. Lienhard Schmitz for the Gal4-p65 and Gal4-p65TA1 constructs, Rik Derynck for the pCAL2 plasmid, Tony Koleske for the pCDM8-p50 and pCDM8-p65 plasmids, and Rene Bernards for antiserum against MBP2.
This work was supported by grants from NIH (DK45746) and the Council for Tobacco Research (3613A). X.-F.W. is a Leukemia Society Scholar.
The first two authors contributed equally to this work.
REFERENCES
- 1.Arsura M, Wu M, Sonenshein G E. TGFβ1 inhibits NF-κB/Rel activity inducing apoptosis of B cells: transcriptional activation of IκB alpha. Immunity. 1996;5:31–40. doi: 10.1016/s1074-7613(00)80307-6. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P A, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 3.Bassing C H, Yingling J M, Howe D J, Wang T, He W W, Gustafson M L, Shah P, Donahoe P K, Wang X-F. A transforming growth factor β type 1 receptor that signals to activate gene expression. Science. 1994;263:87–89. doi: 10.1126/science.8272871. [DOI] [PubMed] [Google Scholar]
- 4.Beg A A, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 5.Boukamp P, Petrussevska R T, Breitkreutz D, Hornung J, Makham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carcamo J, Weis F M B, Ventura F, Wieser R, Wrana J L, Attisano L, Massague J. Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor β and activin. Mol Cell Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cupp C, Taylor J P, Khalili K, Amini S. Evidence for stimulation of the transforming growth factor β1 promoter by HIV-1 Tat in cells derived from CNS. Oncogene. 1993;8:2231–2236. [PubMed] [Google Scholar]
- 8.Datto M B, Li Y, Panus J, Howe D J, Xiong Y, Wang X-Y. TGF-β mediated growth inhibition is associated with induction of the cyclin-dependent kinase inhibitor, p21. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datto M B, Yu Y, Wang X-F. Functional analysis of the transforming growth factor β responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 9a.Datto, M. B., and X.-F. Wang. Unpublished data.
- 10.de Martin R, Haendler B, Hofer-Warbinek R, Gaugitsch H, Wrann M, Schlusener H, Seifert J M. Complementary DNA for human glioblastoma-derived T cell suppressor factor, a novel member of the transforming growth factor-β gene family. EMBO J. 1987;6:3673–3677. doi: 10.1002/j.1460-2075.1987.tb02700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espevik T, Figari I S, Ranges G E, Palladino M A., Jr Transforming growth factor-β1 (TGF-β1) and recombinant human tumor necrosis factor-alpha reciprocally regulate the generation of lymphokine-activated killer cell activity. Comparison between natural porcine platelet-derived TGF-β1 and TGF-β2, and recombinant human TGF-β1. J Immunology. 1988;140:2312–2316. [PubMed] [Google Scholar]
- 12.Feng X H, Derynck R. Ligand-independent activation of transforming growth factor (TGF) β signaling pathways by heteromeric cytoplasmic domains of TGF-β receptors. J Biol Chem. 1996;271:13123–13129. doi: 10.1074/jbc.271.22.13123. [DOI] [PubMed] [Google Scholar]
- 13.Flanagan W M, Corthesy B, Bram R J, Crabtree G R. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 14.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones K A. Tat and the HIV-1 promoter. Curr Opin Cell Biol. 1993;5:461–468. doi: 10.1016/0955-0674(93)90012-f. [DOI] [PubMed] [Google Scholar]
- 16.Kehrl J H, Roberts A B, Wakefield L M, Jakowlew S, Sporn M B, Fauci A S. Transforming growth factor β is an important immunomodulatory protein for human B lymphocytes. J Immunol. 1986;137:3855–3860. [PubMed] [Google Scholar]
- 17.Kehrl J H, Wakefield L M, Roberts A B, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn M B, Fauci A S. Production of transforming growth factor β by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med. 1986;163:1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kekow J, Wachsman W, McCutchan J A, Cronin M, Carson D A, Lotz M. Transforming growth factor β and noncytopathic mechanisms of immunodeficiency in human immunodeficiency virus infection. Proc Natl Acad Sci USA. 1990;87:8321–8325. doi: 10.1073/pnas.87.21.8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita S, Su L, Amano M, Timmerman L A, Kaneshima H, Nolan G P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 20.Koff A, Ohtsuki M, Polyak K, Roberts J M, Massague J. Negative regulation of G1 in mammalian cells: inhibition of cyclin E-dependent kinase by TGF-β. Science. 1993;260:536–539. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni A B, Huh C G, Becker D, Geiser A, Lyght M, Flanders K C, Roberts A B, Sporn M B, Ward J M, Karlsson S. Transforming growth factor β1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazdins J K, Klimkait T, Alteri E, Walker M, Woods-Cook K, Cox D, Bilbe G. TGF-β: upregulator of HIV replication in macrophages. Res Virol. 1991;142:239–242. doi: 10.1016/0923-2516(91)90063-9. [DOI] [PubMed] [Google Scholar]
- 23.Lazdins J K, Klimkait T, Woods-Cook K, Walker M, Alteri E, Cox D, Cerletti N. In vitro effect of transforming growth factor-β on pregression of HIV-1 infection in primary mononuclear phagocytes. J Immunol. 1991;147:1201–1207. [PubMed] [Google Scholar]
- 24.Letterio J J, Geiser A G, Kulkarni A B, Roche N S, Sporn M B, Roberts A B. Maternal rescue of transforming growth factor-β1 null mice. Science. 1994;264:1936–1938. doi: 10.1126/science.8009224. [DOI] [PubMed] [Google Scholar]
- 25.Li J-M, Nichols M A, Chandrasekharan S, Xiong Y, Wang X-F. Transforming growth factor β activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem. 1995;270:26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 26.Lyons R M, Moses H L. Transforming growth factors and the regulation of cell proliferation. Eur J Biochem. 1990;187:467–473. doi: 10.1111/j.1432-1033.1990.tb15327.x. [DOI] [PubMed] [Google Scholar]
- 27.Massague J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto S, Chiao P J, Verma I M. Enhanced IκB alpha degradation is responsible for constitutive NF-κB activity in mature murine B-cell lines. Mol Cell Biol. 1994;14:3276–3282. doi: 10.1128/mcb.14.5.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto S, Maki M, Schmitt M J, Hatanaka M, Verma I M. Tumor necrosis factorα-induced phosphorylation of IκBα is a signal for its degradation but not dissociation from NF-κB. Proc Natl Acad Sci USA. 1994;91:12740–12744. doi: 10.1073/pnas.91.26.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto S, Verma I M. Rel/NF-κB/IκB story. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 31.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 32.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 33.Plevy S E, Gemberling J H, Hsu S, Dorner A J, Smale S T. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ristow H J. BSC-1 growth inhibitor/type beta transforming growth factor is a strong inhibitor of thymocyte proliferation. Proc Natl Acad Sci USA. 1986;83:5531–5533. doi: 10.1073/pnas.83.15.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts A B, Sporn M B. The transforming growth factor-β. In: Sporn M B, Roberts A B, editors. Handbook of experimental pharmacology, peptide growth factors and their receptors. Heidelberg, Germany: Springer; 1990. pp. 419–472. [Google Scholar]
- 36.Rook A H, Kehrl J H, Wakefield L M, Roberts A B, Sporn M B, Burlington D B, Lane H C, Fauci A S. Effects of transforming growth factor-β on the functions of natural killer cells: depressed cytolytic activity and blunting of interferon responsiveness. J Immunol. 1986;136:3916–3920. [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Schmitz M L, Baeuerle P A. The p65 subunit is responsible for the strong transcription activating potential of NF-κB. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreck R, Meier B, Mannel D N, Droge W, Baeuerle P A. Dithiocarbamates as potent inhibitors of nuclear factor κB activation in intact cells. J Exp Med. 1992;175:1181–1194. doi: 10.1084/jem.175.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla R R, Kumar A, Kimmel P L. Transforming growth factor beta increases the expression of HIV-1 gene in transfected human mesangial cells. Kidney Int. 1993;44:1022–1029. doi: 10.1038/ki.1993.344. [DOI] [PubMed] [Google Scholar]
- 41.Shull M M, Ormbsy I, Kier A B, Pawlowski S, Diebold R J, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sporn M B, Roberts A B. Transforming growth factor-beta: recent progress and new challenges. J Cell Biol. 1992;119:1017–1021. doi: 10.1083/jcb.119.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsunawaki S, Sporn M, Ding A, Nathan C. Deactivation of macrophages by transforming growth factor-β. Nature. 1988;334:260–262. doi: 10.1038/334260a0. [DOI] [PubMed] [Google Scholar]
- 44.Van Antwerp D J, Martin S J, Kafri T, Green D R, Berma I M. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 45.Wahl S M, Allen J B, McCartney-Francis N, Morganti-Kossmann M C, Kossmann T, Ellingsworth L, Mai U E, Mergenhagen S E, Orenstein J M. Macrophage- and astrocyte-derived transforming growth factor beta as a mediator of central nervous system dysfunction in acquired immune deficiency syndrome. J Exp Med. 1991;173:981–991. doi: 10.1084/jem.173.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C Y, Mayo M W, Baldwin A S., Jr TNF-α and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 47.Wrann M, Bodmer S, de Martin R, Siepl C, Hofer-Warbinek R, Frei K, Hofer E. T cell suppressor factor from human glioblastoma cells is a 12.5 kd protein closely related to transforming growth factor-beta. EMBO J. 1987;6:1633–1636. doi: 10.1002/j.1460-2075.1987.tb02411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu R-L, Chen T-T, Sun T-T. Functional importance of an Sp1- and an NFκB-related nuclear protein in a keratinocyte-specific promoter of rabbit K3 deratin gene. J Biol Chem. 1994;269:28450–28459. [PubMed] [Google Scholar]
- 49.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]