Abstract
Benzene is a highly toxic industrial compound that is essential to the production of various chemicals, drugs, and fuel oils. Due to its toxicity and carcinogenicity, much recent attention has been focused on benzene biodegradation, especially in the absence of molecular oxygen. However, the mechanism by which anaerobic benzene biodegradation occurs is still unclear. This is because until the recent isolation of Dechloromonas strains JJ and RCB no organism that anaerobically degraded benzene was available with which to elucidate the pathway. Although many microorganisms use an initial fumarate addition reaction for hydrocarbon biodegradation, the large activation energy required argues against this mechanism for benzene. Other possible mechanisms include hydroxylation, carboxylation, biomethylation, or reduction of the benzene ring, but previous studies performed with undefined benzene-degrading cultures were unable to clearly distinguish which, if any, of these alternatives is used. Here we demonstrate that anaerobic nitrate-dependent benzene degradation by Dechloromonas strain RCB involves an initial hydroxylation, subsequent carboxylation, and loss of the hydroxyl group to form benzoate. These studies provide the first pure-culture evidence of the pathway of anaerobic benzene degradation. The outcome of these studies also suggests that all anaerobic benzene-degrading microorganisms, regardless of their terminal electron acceptor, may use this pathway.
Benzene has a broad range of industrial uses and is one of the top 20 production volume chemicals in the United States, with an annual production of almost 9 million metric tons (16). In addition to its presence in petroleum-based fuels, benzene is often used as a raw material for the manufacture of other chemicals, rubbers, lubricants, dyes, detergents, drugs, and pesticides. Other sources, including volcanoes, forest fires, and cigarette smoke, also contribute to the benzene in the environment. Benzene is among the most prevalent organic contaminants in groundwater and is a major concern due to its toxicity and relatively high solubility (16). It is currently ranked sixth on the U.S. National Priorities List, and has been found in at least 813 of the 1,430 current or former National Priorities List sites (http://www.atsdr.cdc.gov/cxcx3.html). Benzene is biodegradable (16, 26), especially in the presence of oxygen; however, when soils and sediments are contaminated with benzene, extensive anaerobic zones frequently develop due to stimulation of the indigenous aerobic microbial population, resulting in rapid depletion of the dissolved oxygen content of the groundwater (2, 11, 12, 27). As a result, a lot of attention has been focused on anaerobic benzene degradation over the past two decades, and anaerobic benzene degradation has been demonstrated under nitrate-reducing (9), Fe(III)-reducing (29, 30), sulfate-reducing (14, 18, 28), and methanogenic (21, 37) conditions. However, until recently, no organism existed in pure culture that was capable of this metabolism (16, 26). Microbial community analysis of benzene-degrading sediments and enrichment cultures using molecular techniques has suggested that a functional role in anaerobic benzene degradation is played by members of the family Geobacteraceae under Fe(III)-reducing conditions (33) and by members of the family Desulfobacteriaceae under both sulfate-reducing and methanogenic conditions (31, 35). However, there is no direct evidence which shows that members of either of these families of organisms are capable of benzene degradation. Recently, Dechloromonas strains RCB and JJ, the first two organisms capable of anaerobic benzene degradation, were isolated from completely different environments and described (15). Strains RCB and JJ belong to the beta subclass of the Proteobacteria, exhibit 98.1% 16S rRNA gene sequence similarity, and metabolize benzene aerobically or anaerobically with nitrate as the sole electron acceptor (15). Anaerobically, Dechloromonas strain RCB completely degrades benzene to CO2, and concentrations as high as 160 μM are removed within 5 days (15).
The biochemical pathway of anaerobic benzene degradation by any organism is currently unknown, but several possibilities exist (16). These include initial carboxylation, hydroxylation, and methylation with subsequent transformation to the central aromatic intermediate benzoate or to its coenzyme A (CoA) thioester form, benzoyl-CoA (16). This compound then undergoes further enzymatic ring reduction and ring cleavage, which forms 3-acetyl-CoA and CO2. Previous studies of methanogenic benzene-degrading enrichments have indicated that phenol, cyclohexanone, propionate, and acetate are possible metabolites (21, 36, 37). Furthermore, phenol and benzoate were both detected as intermediates of benzene degradation with sulfate-reducing and Fe(III)-reducing enrichments (10, 32). However, all of these studies were performed with undefined sediments or enrichment cultures, and whether the compounds were formed directly from benzene catabolism by a single organism or were formed as a result of several sequential metabolic steps involving several different organisms is unknown.
As part of our ongoing studies on the anaerobic degradation of benzene by Dechloromonas strain RCB, we investigated the metabolic intermediates formed with nitrate as the electron acceptor. The results of these studies provide the first documentation of the mechanism of anaerobic nitrate-dependent metabolism of benzene in pure culture and also provide significant insight into the initial process of oxidation of this compound in the absence of oxygen.
MATERIALS AND METHODS
Media and stock preparation.
All media and solutions were prepared using strictly anaerobic techniques, as previously described (8, 15). All anaerobic culturing was performed in sealed serum bottles under an N2-CO2 (80:20, vol/vol) headspace. Unless otherwise stated, all chemicals were acquired from Sigma Chemicals, Missouri. Anoxic aqueous stock solutions of propyl iodide (100 mM), sodium iodide (100 mM), 5,5-dimethyl-1-pyrroline-N-oxide (1 M), mannitol (1 M), sodium dithionite (100 mM), sodium nitrate (1 M), phenol (10 mM), benzoic acid (10 mM), and benzene (3.5 mM) were prepared under an N2 headspace. Amendments were made anaerobically as needed into experimental bottles from these anoxic stock solutions. Strain RCB was routinely cultured on anoxic minimal media (8) using 10 mM acetate and 16 mM nitrate as the electron donor and acceptor, respectively, in 1-liter bottles sealed with thick butyl rubber stoppers under an N2-CO2 (80:20, vol/vol) headspace. Small amounts of benzene (10 μM) were added to the culture media during growth and incubation of the cells to ensure induction of the appropriate biochemical pathway.
Sample preparation for analysis of benzene and its metabolites.
Samples (15 to 20 ml) of culture broth for analysis of volatile metabolites of benzene were collected at regular time intervals from active benzene-degrading cultures and extracted with diethyl ether (5 ml) by vigorous shaking for 90 s. The ether layer was then collected in Supelco screw-top clear vials with polytetrafluoroethylene/Neoprene-lined caps and dried over anhydrous sodium sulfate to remove any traces of water prior to injection using a Hamilton gas-tight syringe into a gas chromatograph-mass spectrometer (GC-MS) for analysis.
Samples (25 to 30 ml) of culture broth for analysis of water-soluble nonvolatile metabolites were first alkalized using 1 M NaOH to pH 12 for 30 min to cleave thioesters. This was followed by acidification of the samples for 30 min using concentrated HCl to approximately pH 2. Acidified samples were then extracted twice using 10 to 15 ml ethyl acetate each time. The ethyl acetate extracts were combined and filtered over anhydrous sodium sulfate prior to concentration to approximately 1 ml by rotary evaporation at 80°C. The concentrated ethyl acetate extracts were subsequently evaporated to dryness under a steady stream of nitrogen gas and then redissolved using 0.5 ml of ethyl acetate before derivatization with 0.1 ml/sample of N,O-bis[trimethylsilyl]trifluoroacetamide and 1% trimethylchlorosilane (Pierce Chemicals, Rockford, Ill.) according to the manufacturer's instructions.
Analytical methods.
14CO2 production in the headspace of active cultures amended with [14C]acetate (1 μCi) or [14C]benzene (1 μCi) was determined with a gas chromatograph equipped with gas proportional counting detection, as previously described (15). Both [14C]acetate (43 mCi/mmol) and [UL-14C]benzene (55mCi/mmol) were purchased from Sigma.
Chemicals.
Analysis of ether extracts for benzene and phenol detection was performed using a gas chromatograph equipped with a mass spectrometry detector (Shimadzu GCMS-QP2010) with electron ionization. The injector port temperature was set at 110°C with a 1:20 split ratio, and the carrier gas used was helium flowing through the column at a rate of 1.48 ml · min−1. Chromatography of each sample was performed using an XTI-5 column (Restek Corp., Pennsylvania). The column temperature was initially set at 40°C for 1 min and was subsequently ramped at a rate of 4°C min−1 to a final temperature of 125°C.
Analysis of aqueous-phase extracts for metabolite detection was performed similarly by GC-MS of 3-μl sample extracts. The injection port temperature was set at 110°C with a 1:50 split ratio, and helium was used as the carrier gas at a flow rate of 1.46 ml · min−1. Chromatography of each sample was performed using an XTI-5 column which was temperature ramped at a rate of 4°C · min−1 to a final temperature of 230°C after an initial 2-min hold at 80°C. The hold time at the final temperature was 5 min. In all cases, mass spectra data were characterized by comparison with the standard National Institute of Standards and Technology (NIST) library, and compound identification was confirmed by comparison with chemical standards freshly prepared and derivatized using the sample preparation protocols outlined above.
Iodide determinations were performed by ion chromatography by using a DX 500 ion chromatograph with suppressed conductivity detection and an IonPac AS16 anion-exchange column (Dionex Corp., Sunnyvale, CA). A mobile phase of 35 mM NaOH was used at a flow rate of 1 ml · min−1.
Optical density at 600 nm was determined using a Cary 50 Bio UV-visible spectrophotometer manufactured by Varian Analytical Instruments, Palo Alto, CA.
RESULTS AND DISCUSSION
Phenol as a metabolite of benzene degradation.
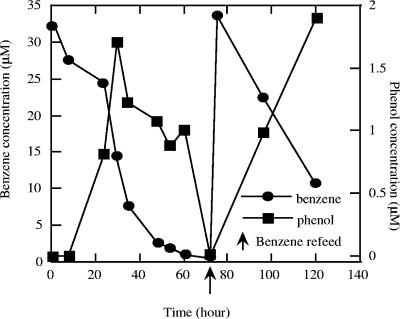
GC-MS of ether extracts of an active benzene-degrading anaerobic culture revealed the presence of a single peak in addition to benzene with a mass spectrum identical to that of phenol (data not shown). The peak identification was confirmed by comparison of mass spectra data with data from the NIST library and freshly prepared chemical standards. The phenol peak appeared in samples collected shortly after the onset of benzene degradation amended with 5 mM nitrate as the electron acceptor (Fig. 1). No benzene degradation or phenol formation was observed in the absence of the electron acceptor. The phenol concentration peaked at almost 2 μM within 30 h of inoculation, at which point almost 19 μM benzene had been utilized by the active culture (Fig. 1). Once benzene was completely depleted, the phenol concentration rapidly dropped to levels below detection (Fig. 1). When the culture was refed with 35 μM benzene, rapid benzene degradation with concomitant phenol formation was again observed (Fig. 1).
FIG. 1.
Transient phenol formation resulting from anaerobic benzene degradation by strain RCB using nitrate as the sole electron acceptor. The data are averages of duplicate determinations.
Benzoate as a metabolite of benzene degradation.
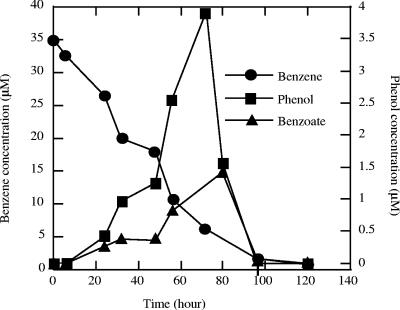
In order to investigate the production of nonvolatile water-soluble intermediates formed during anaerobic nitrate-dependent benzene catabolism by strain RCB, samples were collected and extracted with ethyl acetate prior to derivatization by silylation for chromatographic analysis. GC-MS analysis revealed the presence of a monoaromatic compound which was identified as benzoate by comparison of the mass spectra with data from the NIST library and authentic benzoate standards (data not shown). Benzoate accumulated gradually to a concentration of almost 1.5 μM after 70 h of incubation (Fig. 2). As observed with phenol, benzoate was formed concomitant with benzene degradation. The transient nature of benzoate observed correlated well with the transient formation and removal of phenol (Fig. 2). Benzoate formation was not observed in cultures that were not amended with the electron acceptor nitrate.
FIG. 2.
Transient formation of benzoate and phenol during anaerobic degradation of benzene by active cultures of strain RCB using nitrate as the sole electron acceptor. The data are averages of duplicate determinations.
Sequence of catabolite production.
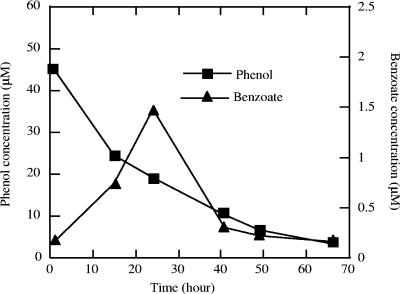
To determine the sequence of phenol and benzoate formation during benzene catabolism by strain RCB, anaerobic active cultures were grown with 45 μM phenol as the sole electron donor and 5 mM nitrate as the electron acceptor. At regular intervals throughout the incubation, samples were collected and analyzed by GC-MS. Analysis revealed that benzoate was transiently formed by strain RCB concomitant with phenol degradation (Fig. 3). No benzoate was formed in cultures that were not amended with either phenol or nitrate. After 24 h of incubation, almost 1.5 μM benzoate was detected in the culture broth.
FIG. 3.
Transient formation of benzoate during anaerobic degradation of phenol by active cultures of strain RCB using nitrate as the sole electron acceptor. The data are averages of duplicate determinations.
In contrast, no phenol formation occurred in similar experiments performed with active cultures of strain RCB growing anaerobically on 50 μM benzoate with nitrate as the electron acceptor. These results indicate that phenol formation precedes formation of benzoate as an intermediate of anaerobic nitrate-dependent benzene oxidation by strain RCB.
Location of intermediate formation.
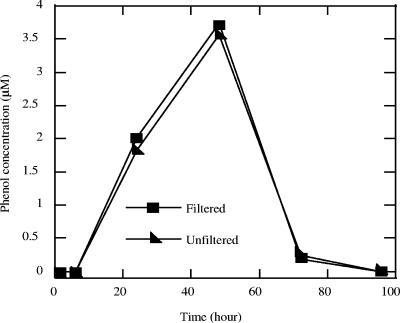
In order to determine if the intermediates were formed extracellularly or were extracted from the intracellular cytoplasmic pools of whole cells of strain RCB during sample preparation, subsamples from active benzene-oxidizing culture broths were prefiltered through 0.2-μm-pore-size filters to remove the cells prior to extraction and analysis. GC-MS analysis indicated that the phenol levels in extracts of filtered and unfiltered culture broths were identical throughout the incubation (Fig. 4), suggesting that the hydroxylation reaction occurs on the outer membrane or in the periplasm of the cell, from which the phenol readily diffuses into the culture milieu. Like the phenol levels, the benzoate levels were identical in extracts of both the filtered (0.2-μm-pore-size filters) and unfiltered culture broths throughout the incubation (data not shown), implying that benzoate formation also occurs on the outer membrane or in the periplasm of strain RCB.
FIG. 4.
Concentrations of phenol and benzoate in samples prepared from filtered and unfiltered culture broths of an active benzene-degrading culture of strain RCB. The data are averages of duplicate determinations.
Role of molecular oxygen.
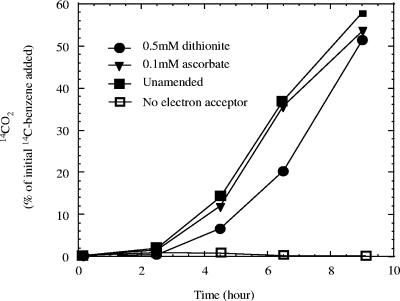
Hydroxylation of benzene to phenol is the accepted primary route of benzene metabolism by mammalian systems (25). However, this reaction is catalyzed by a monooxygenase P450 cytochrome isozyme and is dependent on the availability of molecular oxygen. To ensure that the benzene degradation in the active cultures of strain RCB was not due to a similar mechanism involving molecular oxygen, experiments were performed in anoxic culture media amended with 0.5 mM sodium dithionite or 0.1 mM sodium ascorbate as an alternative chemical reductant to remove any traces of dissolved O2 (Fig. 5). In spite of the presence of the chemical reductants at the concentrations tested, [14C]benzene was rapidly oxidized to14CO2 in these experiments only in the presence of nitrate (5 mM) (Fig. 5), discounting any possible role for molecular oxygen in the metabolism. GC-MS analysis revealed the formation of phenol concomitant with benzene degradation in culture broths amended with sodium dithionite (data not shown). In addition, no phenol was detected in benzene-degrading cultures of strain RCB incubated with oxygen as the electron acceptor (data not shown). This result is consistent with previous reports of studies on the metabolites of aerobic microbial benzene degradation, in which phenol was not detected as an intermediate (19).
FIG. 5.
Effect of the presence and absence of chemical reducing agents on [14C]benzene oxidation by active benzene-degrading cultures of strain RCB with nitrate as the electron acceptor. The data are averages of triplicate determinations.
It is interesting that although the metabolism was not inhibited, addition of dithionite as a reducing agent (0.5 mM) retarded the rate of benzene degradation and the subsequent phenol formation compared to unamended controls. At elevated concentrations (>1 mM) sodium dithionite completely inhibited benzene degradation. However, even at a dithionite concentration of 0.5 mM, the phenol degradation rates remained unaffected compared to those of unamended controls (data not shown). These results suggest that dithionite played an inhibitory role in the initial hydroxylation step of benzene degradation rather than the subsequent oxidation of phenol to CO2 by strain RCB.
H2O as a potential source of the hydroxyl group.
Previous studies performed using an undefined benzene-degrading methanogenic enrichment culture also showed that phenol is a metabolite of benzene degradation (36). In these studies, the undefined culture was incubated in media amended with 9% H218O, and a small fraction (2.5%) of the observed phenol pool contained the 18O label, suggesting that the hydroxyl group originated from H2O (36). When a similar experiment was performed by incubating strain RCB for 24 h in anaerobic medium prepared using 25% H218O, GC-MS analysis of the ether extracts indicated that there was an increase of only 1.40% [18O]phenol in the phenol pool compared to an unlabeled control culture. A similar increase (1.13% [18O]phenol) was observed in the phenol pool if the experiment was repeated using medium prepared with 50% H218O. The low level of incorporation of the 18O label into the hydroxyl group of phenol produced from benzene under the conditions tested and the lack of a correlation with the initial H218O content suggest that H2O is not the source of the hydroxyl group during benzene catabolism by strain RCB. This suggestion is supported by the fact that an increase of 1.40% [18O]phenol in the phenol pool was observed in sterile culture medium containing 50% H218O and 1.5 μM unlabeled phenol after 24 h of incubation, demonstrating that the 18O atom of H218O could interchange to a small extent with the more abundant 16O atom of phenol. This may more likely account for the [18O]phenol produced in the biologically active samples. This result is unexpected and leaves the source of the hydroxyl group unknown. Final elucidation of the details of the mechanism involved in the initial hydroxylation process may shed further light on this.
Mechanism of hydroxylation.
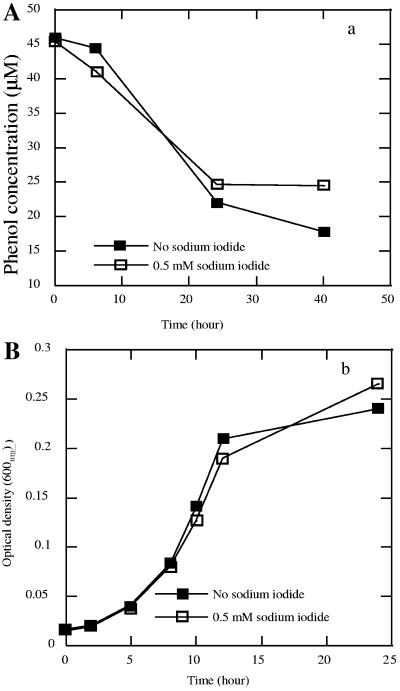
In other known microbial hydroxylation reactions involving substrates such as enoyl-CoA or fumarate, the double bond is generally adjacent to a carbonyl group that plays a critical role by delocalizing the negative charge of the reaction intermediate, making the reaction favorable (5). This cannot occur with benzene due to its molecular symmetry and lack of functional side groups. Thus, hydroxyl group addition for benzene under standard conditions is an endogonic reaction (ΔGo′ = 25.8 kJ/mol) (16). One possible mechanism for overcoming the unfavorable thermodynamics of benzene hydroxylation is through the formation of highly reactive hydroxyl free radicals (HO·), which subsequently attack the aromatic ring (1, 16, 38). When benzene-degrading cultures of strain RCB with nitrate as the electron acceptor were incubated in the presence of the hydroxyl free radical scavenger sodium iodide (0.5 mM), benzene degradation and subsequent phenol formation were inhibited compared to controls not amended with sodium iodide (Table 1). After 24 h of incubation, almost 19 μM of the initial benzene concentration (35 μM) was degraded by active cultures of strain RCB using nitrate as the electron acceptor, while only 1.75 μM of the initial benzene was degraded in the samples that were amended with sodium iodide (Table 1). In contrast, sodium iodide addition had no effect either on phenol degradation by strain RCB under nitrate-reducing conditions or on the growth of strain RCB on acetate and nitrate, demonstrating that the iodide was not generally toxic to the cell (Fig. 6). Phenol degradation by strain RCB proceeded at the same rate regardless of the presence of 0.5 mM sodium iodide, and almost 50% of the substrate was degraded within 24 h both in the presence and in the absence of sodium iodide (Fig. 6A). Ion chromatography analysis demonstrated that iodide concentrations remained constant throughout the incubation, indicating that the iodide was not used (oxidized to iodate) in place of benzene as an alternative, more favorable electron donor by strain RCB (data not shown).
TABLE 1.
Degradation of benzene by strain RCB in the presence and absence of various hydroxyl free radical scavengers
| Compound | Initial benzene concn added (μM) | Residual benzene concn after 24 h (μM) | % of initial benzene degraded after 24 h |
|---|---|---|---|
| No amendment (positive control) | 35 | 15.7 ± 0.6 | 55 |
| 0.5 mM sodium iodide | 35 | 33.2 ± 0.9 | 5 |
| 0.5 mM propyl iodide | 35 | 24.5 ± 1.6 | 30 |
| 10 mM mannitol | 35 | 35 ± 0.4 | 0 |
| 10 mM DMPOa | 28.5 | 25 ± 1.1 | 12 |
DMPO, 5,5-dimethyl-1-pyrroline-N-oxide.
FIG. 6.
Effect of 0.5 mM sodium iodide on phenol degradation by strain RCB (A) and growth of strain RCB with acetate (B). The data are averages of triplicate determinations.
Similar inhibition of anaerobic benzene degradation and phenol formation by strain RCB with nitrate as the terminal electron acceptor was also observed in the presence of other hydroxyl free radical scavengers, such as 10 mM 5,5-dimethyl-1-pyrroline-N-oxide or 10 mM mannitol, which inhibited benzene degradation to different extents (Table 1). Phenol degradation and acetate oxidation under nitrate-reducing conditions by strain RCB remained unaffected in the presence of these scavengers after 24 to 48 h of incubation (data not shown). Although the results described above are not direct evidence, they strongly support the hypothesis that benzene ring hydroxylation is mediated through hydroxyl free radical attack.
Mechanism of benzoate formation from phenol.
Reports on anaerobic phenol degradation by the monoaromatic hydrocarbon degrader Thauera aromatica have demonstrated that phenol is initially phosphorylated in an ATP-dependent mechanism to form phenylphosphate, which is then carboxylated by phenylphosphate carboxylase, forming 4-hydroxybenzoate (34). The 4-hydroxybenzoate is further activated by a specific CoA ligase, and the hydroxyl group is reductively removed (22). Molybdenum-containing iron-sulfur proteins have been shown to catalyze the dehydroxylation of hydroxy-benzoyl-CoA thioesters (6, 7, 20, 23). Whether a similar mechanism is involved during benzene metabolism by strain RCB is currently unknown. Phenylphosphate or 4-hydroxybenzoate was not detected in culture extracts of strain RCB analyzed during nitrate-dependent benzene degradation. Furthermore, recent reports have demonstrated that 0.1 mM dithionite inhibits carboxylation of phenylphosphate to form 4-hydroxybenzoate (34). Whether this is a reversible reaction is not known. In our experiments, while the addition of 0.1 mM dithionite to active benzene-degrading cultures decreased the rate of degradation of benzene and the subsequent formation of phenol (data not shown), it did not completely inhibit the process and had no significant effect on phenol catabolism by strain RCB.
Conclusion.
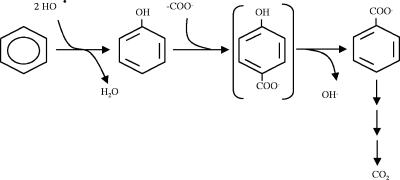
The results presented here provide the first elucidation of the initial steps involved in the anaerobic metabolic pathway for benzene degradation in pure culture (Fig. 7). These studies demonstrated that phenol is a metabolic intermediate of anaerobic benzene degradation by Dechloromonas strain RCB growing with nitrate as the terminal electron acceptor. Contrary to previous reports on methanogenic benzene degradation in which the source of the hydroxyl group of phenol was suggested to be water (36), in strain RCB under nitrate-reducing conditions, phenol formation appears to be mediated through reaction with a hydroxyl free radical that does not originate from H2O but is formed on the outer membrane or in the periplasm of the organism. Initial hydroxylation has been suggested to be the first step in degradation of other hydrocarbons, such as naphthalene under sulfate-reducing conditions (3) and recently ethylbenzene under nitrate-reducing conditions (24). The recently purified Mo-containing iron-sulfur protein that adds a hydroxyl group to ethylbenzene to form (S)-(−)-1-phenylethanol may provide a model for hydroxylation of benzene (24). The phenol formed from benzene by strain RCB is further carboxylated and dehydroxylated to form benzoate or its CoA ester, benzoyl-CoA. Both phenol and benzoate are formed outside the cytoplasm, and it appears that benzoate is transported into the cell for subsequent catabolism to CO2. The environmental implications of such extracellular processing of benzene to more readily degradable substrates and the availability of these substrates to other organisms in the natural environment are currently unknown. Although the concentrations of phenol and benzoate detected in our studies are very low, they are in agreement with previous reports on intermediates formed during anaerobic benzene and toluene degradation (4, 10). Prior studies detected phenol, cyclohexanone, and propionate as potential intermediates in undefined methanogenic enrichments incubated with benzene (21, 37). Similarly, phenol, benzoate, and acetate were also detected in benzene-degrading iron-reducing and sulfate-reducing sediments (10). As Dechloromonas species are not known to grow by Fe(III) reduction, by sulfate reduction, or through syntrophic interactions with methanogenic bacteria (13, 17), it is unlikely that organisms closely related to strain RCB or JJ are involved in the benzene metabolism observed in the previous studies. This, combined with the fact that both phenol and benzoate have now been identified during anaerobic benzene degradation in each of these disparate terminal electron-accepting processes, suggests that a single universal biochemical pathway may exist for anaerobic benzene degradation and that it is similar to that shown here for Dechloromonas strain RCB. In this regard, strain RCB serves as an ideal model organism to study the biochemistry and genetic regulation of anaerobic benzene oxidation in pure culture.
FIG. 7.
Model of proposed universal pathway of anaerobic benzene degradation. The transient formation of para-hydroxybenzoate (shown in parentheses) is hypothetical.
Acknowledgments
Studies on Dechloromonas aromatica strain RCB were supported by independent grants to J.D.C. from the U.S. Department of Energy NABIR program (DE-FG02-98-ER-62689) and the U.S. Department of Defense (DACA72-00-C-0016).
We acknowledge K. V. Kellaris for iodide analysis in this project.
REFERENCES
- 1.Acero, J. L., F. J. Benítez, F. J. Real, and A. I. Leal. 2001. Degradation of p-hydroxyphenylacetic acid by photoassisted Fenton reaction. Water Sci. Technol. 44:31-38. [PubMed] [Google Scholar]
- 2.Anderson, R. T., and D. R. Lovley. 1997. Ecology and biogeochemistry of in situ groundwater bioremediation. Adv. Microb. Ecol. 15:289-350. [Google Scholar]
- 3.Bedessem, M. E., N. G. Swoboda-Colberg, and P. J. S. Colberg. 1997. Naphthalene mineralization coupled to sulfate reduction in aquifer-derived enrichments. FEMS Microbiol. Lett. 152:213-218. [Google Scholar]
- 4.Beller, H. R., and E. A. Edwards. 2000. Anaerobic toluene activation by benzylsuccinate synthase in a highly enriched methanogenic culture. Appl. Environ. Microbiol. 66:5503-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benning, M. M., T. Haller, J. A. Gerlt, and H. M. Holden. 2000. New reactions in the crotonase superfamily: structure of methylmalonyl-CoA decarboxylase from Escherichia coli. Biochemistry 39:4630-4639. [DOI] [PubMed] [Google Scholar]
- 6.Brackmann, R., and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from a denitrifying Pseudomonas species. Eur. J. Biochem. 213:563-571. [DOI] [PubMed] [Google Scholar]
- 7.Breese, K., and G. Fuchs. 1998. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica. Prosthetic groups, electron donor, and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur. J. Biochem. 251:916-923. [DOI] [PubMed] [Google Scholar]
- 8.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 9.Burland, S. M., and E. A. Edwards. 1999. Anaerobic benzene biodegradation linked to nitrate reduction. Appl. Environ. Microbiol. 65:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldwell, M. E., and J. M. Suflita. 2000. Detection of phenol and benzoate as intermediates of anaerobic benzene biodegradation under different terminal electron-accepting conditions. Environ. Sci. Technol. 34:1216-1220. [Google Scholar]
- 11.Christensen, T., P. Kjeldsen, H. Albrechtsen, and G. Heron. 1994. Attenuation of pollutants in landfill leachate polluted aquifers. Crit. Rev. Environ. Sci. Technol. 24:119-202. [Google Scholar]
- 12.Coates, J. D., and L. A. Achenbach. 2001. The biogeochemistry of aquifer systems, p. 719-727. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. W. Walter (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, DC.
- 13.Coates, J. D., and L. A. Achenbach. 2004. Microbial perchlorate reduction: rocket fuelled metabolism. Nat. Rev. Microbiol. 2:569-580. [DOI] [PubMed] [Google Scholar]
- 14.Coates, J. D., R. T. Anderson, and D. R. Lovely. 1996. Oxidation of polycyclic aromatic hydrocarbons under sulfate-reducing conditions. Appl. Environ. Microbiol. 62:1099-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039-1043. [DOI] [PubMed] [Google Scholar]
- 16.Coates, J. D., R. Chakraborty, and M. J. McInerney. 2002. Anaerobic benzene biodegradation—a new era. Res. Microbiol. 153:621-628. [DOI] [PubMed] [Google Scholar]
- 17.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coates, J. D., J. Woodward, J. Allen, P. Philp, and D. R. Lovley. 1997. Anaerobic degradation of polycyclic aromatic hydrocarbons and alkanes in petroleum-contaminated marine harbor sediments. Appl. Environ. Microbiol. 63:3589-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, J., M. Dispensa, and C. S. Harwood. 1997. 4-Hydroxybenzoyl coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J. Bacteriol. 179:634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grbic-Galic, D., and T. Vogel. 1987. Transformation of toluene and benzene by mixed methanogenic cultures. Appl. Environ. Microbiol. 53:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heider, J., and G. Fuchs. 1997. Anaerobic metabolism of aromatic compounds. Eur. J. Biochem. 243:577-596. [DOI] [PubMed] [Google Scholar]
- 23.Heider, J., and G. Fuchs. 1997. Microbial anaerobic aromatic metabolism. Anaerobe 3:1-22. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, H. A., D. A. Pelletier, and A. M. Spormann. 2001. Isolation and characterization of anaerobic ethylbenzene dehydrogenase, a novel Mo-Fe-S enzyme. J. Bacteriol. 183:4536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstrom, A. B., K. Yeowell-O'Connell, S. Waidyanatha, B. T. Golding, R. Tornero-Velez, and S. M. Rappaport. 1997. Measurement of benzene oxide in the blood of rats following administration of benzene. Carcinogenesis 18:1637-1641. [DOI] [PubMed] [Google Scholar]
- 26.Lovley, D. R. 2000. Anaerobic benzene degradation. Biodegradation 11:107-116. [DOI] [PubMed] [Google Scholar]
- 27.Lovley, D. R. 1997. Potential for anaerobic bioremediation of BTEX in petroleum-contaminated aquifers. J. Ind. Microbiol. Biotechnol. 18:75-81. [Google Scholar]
- 28.Lovley, D. R., J. D. Coates, J. C. Woodward, and E. J. P. Phillips. 1995. Benzene oxidation coupled to sulfate reduction. Appl. Environ. Microbiol. 61:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1996. Rapid anaerobic benzene degradation with a variety of chelated Fe(III) forms. Appl. Environ. Microbiol. 62:288-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1994. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature 370:128-131. [DOI] [PubMed] [Google Scholar]
- 31.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 32.Phelps, C. D., X. Zhang, and L. Y. Young. 2001. Use of stable isotopes to identify benzoate as a metabolite of benzene degradation in a sulphidogenic consortium. Environ. Microbiol. 3:600-603. [DOI] [PubMed] [Google Scholar]
- 33.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schühle, K., and G. Fuchs. 2004. Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186:4556-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulrich, A. C., and E. A. Edwards. 2003. Physiological and molecular characterization of anaerobic benzene-degrading mixed cultures. Environ. Microbiol. 5:92-102. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, T. M., and D. Grbic-Galic. 1986. Incorporation of oxygen from water into toluene and benzene during anaerobic fermentative transformation. Appl. Environ. Microbiol. 52:200-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiner, J. M., and D. R. Lovley. 1998. Rapid benzene degradation in methanogenic sediments from a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 64:1937-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon, J., Y. Lee, and S. Kim. 2001. Investigation of the reaction pathway of OH radicals produced by Fenton oxidation in the conditions of wastewater treatment. Water Sci. Technol. 44:15-21. [PubMed] [Google Scholar]