Abstract
The anaerobic metabolism of acetate was studied in sediments and groundwater from a gas condensate-contaminated aquifer in an aquifer where geochemical evidence implicated sulfate reduction and methanogenesis as the predominant terminal electron-accepting processes. Most-probable-number tubes containing acetate and microcosms containing either [2-14C]acetate or [U-14C]acetate produced higher quantities of CH4 compared to CO2 in the presence or absence of sulfate.14CH4 accounted for 70 to 100% of the total labeled gas in the [14C]acetate microcosms regardless of whether sulfate was present or not. Denaturing gradient gel electrophoresis of the acetate enrichments both with and without sulfate using Archaea-specific primers showed identical predominant bands that had 99% sequence similarity to members of Methanosaetaceae. Clone libraries containing archaeal 16S rRNA gene sequences amplified from sediment from the contaminated portion of the aquifer showed that 180 of the 190 clones sequenced belonged to the Methanosaetaceae. The production of methane and the high frequency of sequences from the Methanosaetaceae in acetate enrichments with and without sulfate indicate that aceticlastic methanogenesis was the predominant fate of acetate at this site even though sulfate-reducing bacteria would be expected to consume acetate in the presence of sulfate.
Redox reactions in anaerobic environments play a pivotal role in the fate of organic compounds in contaminated aquifers (17, 19). The populations of microorganisms that are involved in the degradation of acetate, as well as other organic compounds in contaminated anaerobic sediments, depend on many factors, but the availability of an electron acceptor at the particular site is an important governing factor (17, 19). Typically, electron acceptors with a higher redox potential, such as NO3−, will be utilized first, followed by Fe(III), SO42−, and CO2 (19).
Relatively few studies have focused on the fate of acetate in hydrocarbon-contaminated environments. The majority of these studies that have been conducted in hydrocarbon-contaminated environments have used culture-independent molecular approaches to describe the microbial community and infer the putative function of the different phylotypes present (7, 8, 14, 39, 41). One recent study showed that sulfate reduction accounted for the degradation of petroleum hydrocarbon constituents in approximately 70% of all sites studied in a survey of 38 petroleum-impacted sites (14, 42). Kinetic studies have demonstrated that sulfate-reducing bacteria have a lower km value for acetate (34, 40, 44) and are capable of acetate utilization at lower threshold concentrations than aceticlastic methanogens (34, 40, 44). These findings along with the ability of the sulfate-reducing bacteria to completely mineralize a wide variety of hydrocarbon contaminants, including alkanes, aromatic hydrocarbons, and a variety of fatty acids, including acetate (44), suggest that sulfate-reducing bacteria should be responsible for acetate utilization in hydrocarbon-contaminated sites.
We used a combination of cultivation and molecular approaches to test the hypothesis that aceticlastic methanogenesis was the predominant fate of acetate in a hydrocarbon-contaminated aquifer where both methanogenesis and sulfate reduction have been implicated as the predominant electron-accepting processes (TEAP) (12). In the 1970s, the site was contaminated with gas condensate which contains a mixture of C5 to C15 hydrocarbons (20% of the gas condensate is composed of a mixture of benzene, toluene, ethylbenzene, and the xylene isomers) that was coproduced with the natural gas (12). Dissolved oxygen and nitrate were depleted in the contaminated portion of the aquifer with respect to uncontaminated sediments (12). Fe(III) was undetectable in the contaminated portion of the aquifer but was present at significantly higher levels in uncontaminated sediment (12). Geochemical data along with microcosm studies, which showed that the degradation of several compounds, including benzene, toluene, o-xylene, m-xylene, p-xylene, and ethylbenzene, was accompanied by sulfate loss, suggested that sulfate reduction is the predominant TEAP in the contaminated portion of this aquifer (12). However, dissolved methane within the contaminated portion of the aquifer ranged from 5 to 17 mg/liter (12), which suggested a role for methanogenesis within this site. Our study suggests that acetate is an important intermediate in hydrocarbon-contaminated aquifers where sulfate reduction occurs, even though one would predict that sulfate-reducing bacteria should completely mineralize the hydrocarbons.
MATERIALS AND METHODS
Sample collection.
Sediments and groundwater were collected from a shallow aquifer that lies just above a natural gas field that is located approximately 40 miles northeast of Denver, Colorado. Samples were collected in October 1998 and August 2000. Contaminated sediments and groundwater from well number 37 (12) were collected at a distance of approximately 10 m downgradient from a sump that leaked hydrocarbons in the 1970s (12). Contaminated sediments were collected by hand, boring to a depth of 1.5 m below the surface and placed in sterile 1-liter Mason jars. Samples were kept anaerobic by filling the jars to capacity with sediment and groundwater. Groundwater and sediments from an uncontaminated portion of the aquifer were collected as above from well 18 (12), which was located approximately 10 m upgradient from the original source of contamination. All sediment and groundwater samples were stored on ice until they were delivered to the laboratory. The samples were stored at 4°C upon arrival at the laboratory.
Microorganisms and media.
The hydrogen-using organisms Methanospirillum hungatei strain JF-1 (DSM 864) and Desulfovibrio vulgaris strain G11 (DSM 7057) were grown with an 80% H2-20% CO2 gas phase (69 kPa) under strictly anaerobic conditions (2) using a previously described basal medium (21). The basal medium was amended with 10 mM acetate when growing JF-1 and with 10 mM acetate and 30 mM sulfate when growing G11.
The basal medium (21) with 10 mM acetate was used for most-probable-number (MPN) analysis and contained 10 mM sulfate for the growth of sulfate reducers. The MPN medium was prepared anaerobically (2), and each tube contained either 6 ml of medium in MPN tubes that contained either JF-1 or G11 medium or 9 ml of medium in MPN tubes that did not contain a hydrogen-using organism. The headspace of all the MPN tubes was replaced with an atmosphere containing 80% N2 and 20% CO2 (34 kPa) gas phase (2). MPN tubes were incubated at room temperature without shaking.
MPN analysis.
To test for the presence of different metabolic groups involved in acetate degradation, a three-tube MPN analysis was conducted using sediments from the contaminated and uncontaminated portions of the aquifer. Sterile sodium pyrophosphate solution (pH 7) was prepared by adding 1 g/liter sodium pyrophosphate to the basal medium (21) without rumen fluid. The sterile, anaerobically prepared (2) sodium pyrophosphate solution was taken into an anaerobic chamber where the stoppers and seals were removed. Three tubes of sodium pyrophosphate per MPN set were each amended with 1 g (wet weight) of sediment from the appropriate location, stoppered, sealed, removed from the anaerobic chamber, and used to inoculate the appropriate MPN set. Each of the three tubes was mixed by hand for 30 s, and 1 ml of each solution was removed aseptically and transferred into 9 ml of the appropriate MPN medium using needles and syringes flushed with 100% N2. This procedure was repeated using these first three tubes of inoculated MPN medium and continued until each tube of the dilution series was inoculated.
Three different series of MPN analyses were conducted using 10 mM acetate as the substrate. The first series contained no additional sulfate and 3 ml of a hydrogen-using methanogen, M. hungatei strain JF-1. The second series contained an additional 10 mM sulfate. The final series contained an additional 10 mM sulfate and 3 ml of a hydrogen-using sulfate-reducing bacterium, D. vulgaris strain G11. The hydrogen users were added to each tube of the dilution series to enrich for syntrophic bacteria capable of degrading acetate. Individual MPN tubes were scored positive if more than 50% of the acetate was metabolized after 120 days. As controls, MPN analysis was conducted using the basal medium without added acetate.
Preparation of [14C]acetate-amended microcosms.
Microcosms were prepared in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) using sterile, 40-ml serum bottles, which were left in the chamber overnight prior to inoculation. Thirteen grams of sediment from the contaminated portion of the aquifer was added to each serum bottle. Groundwater from a well located upgradient of the contaminated area was added to bring the final volume of each microcosm to 20 ml. The microcosms were stoppered and sealed inside the chamber, and the gas phase was exchanged three times by evacuation with vacuum and repressurization with 100% N2 (2). One set of microcosms received between 1.7 × 104 and 3.3 × 104 Bq of [2-14C]acetate; another set of microcosms received a similar amount of [U-14C]acetate. The labeled acetate was added by injecting 0.2 ml of either [2-14C]acetate or [U-14C]acetate stock solutions, which contained 1.13 × 105 Bq/ml and 9.93 × 104 Bq/ml, respectively. Unlabeled acetate was added to all of the microcosms to bring the final acetate concentration to approximately 500 μM. Each set of microcosms contained three replicates of each of the following treatments: sediment and acetate alone, acetate and an inhibitor of methanogenesis (7.5 mM 2-bromoethanesulfonic acid [BESA]), acetate with 7.5 mM sulfate, and acetate and an inhibitor of sulfate reduction (5 mM sodium molybdate). Heat-killed controls were run in duplicate for each of the above treatments. The heat-killed controls were autoclaved at 121°C for 20 min. All of the microcosms were incubated for 18 days at room temperature.
Analytical methods.
Nonlabeled acetate loss was measured by high-pressure liquid chromatography (16). The mobile phase was 25 mM KH2PO4 (pH 2.5) at a flow rate of 1 ml min−1. Labeled acetate was measured by using a radioisotope detector. The radioisotope detector was calibrated by comparing its response to that of a scintillation counter. Standards of both [2-14C]acetate and [U-14C]acetate were prepared from the same stock solutions that were used to amend the microcosms. One-hundred-microliter aliquots of standard solutions ranging from 1.67 × 103 Bq to 1.67 × 104 Bq were run on the radioisotope detector. The same volume of each standard was also placed into 5 ml of scintillation cocktail and counted using a scintillation counter. Each standard and unamended scintillation cocktail was counted by using a scintillation counter. Quenching of standards during liquid scintillation counting was corrected by autocalibration using an unquenched 14C standard and through the use of both an H-number monitor and a random coincidence monitor.
CH4 and CO2 production were measured by using a gas chromatograph (GC) equipped with a thermal conductivity detector. The GC had a 3.05-m by 0.004-m Carbosphere 80/100 column (Altech Inc., Deerfield, IL). Helium was the carrier gas at 2 ml min−1. The injector and the column were set at 175°C, and the detector was set at 81°C. The gas chromatograph was connected to a gas proportional counter (Insus Systems Incorporated, Fairfield, NJ). Standards containing 14CO2 were prepared from a stock solution containing 6.03 × 102 Bq/ml of H14CO3. This solution was then diluted to concentrations ranging from 3.01 × 102 to 3.02 × 102 Bq/ml by adding the appropriate volume of stock solution to enough 0.1 N NaOH to bring the final volume of each standard to 20 ml. Each standard was then acidified with 1 ml of 12 N HCl. A 0.2-ml aliquot of the headspace of each standard was then injected into the GC. Also, 0.2-ml aliquots of each standard were slowly bubbled into 0.8 ml of 0.1 N NaOH, and 0.45 ml of the solution was added to 5 ml of scintillation cocktail and counted using the same procedure described for the [14C]acetate standards. The retention time of CH4 was determined through the use of nonlabeled methane standards, which were detected with the thermal conductivity detector. An enrichment culture that degraded [U-14C]methyl tert-butyl ether (MTBE) was provided by the laboratory of Joseph M. Suflita. This enrichment culture, which was known to produce 14CH4, was used to verify the retention time of 14CH4.
The pH of the individual microcosms was measured with Color pHast indicator strips (EM Science, Gibbstown, NJ) at the end of the 18-day incubation period. The final pH of each microcosm and the amount of 14CO2 (obtained by GC) were used to calculate the amount of H14CO3 that was dissolved in the liquid phase of the microcosms by using the Henderson-Hasselbach equation: final pH of microcosm = 6.35 + log([H14CO3]/[14CO2]). The amount of H14CO3 determined with this equation was added to the amount of 14CO2 measured by gas chromatography to obtain the total amount of 14CO2 produced in each microcosm.
Sulfate concentrations were determined by ion chromatography (16), and methane was quantified by gas chromatography (13).
Molecular analysis.
DNA was extracted from enrichments that were prepared by inoculating MPN medium with 1-ml aliquots of sediment and groundwater from microcosms containing [2-14C]acetate, [2-14C]acetate with sulfate, [U-14C]acetate, and [U-14C]acetate with sulfate. These enrichments were transferred three times prior to being used as a source of material for DNA extraction. Two milliliters of each enrichment was added to sterile 2-ml polypropylene screw-cap tubes that contained 1 g of 0.1-mm zirconia-silica beads (Biospec Products, Bartlesville, OK). Samples were centrifuged for 5 min at 14,000 × g to pellet the cells, and any remaining supernatant was discarded. DNA was also extracted directly from sediments by using approximately 1 g of sediment (wet weight) that was added directly to a 2-ml polypropylene screw-cap tube containing zirconia-silica beads. DNA was extracted from enrichments and sediments using a bead beating protocol as previously described (28).
DNA extracted from contaminated aquifer sediments was used as a template to screen for the presence of different groups of sulfate-reducing bacteria. Five sets of group-specific 16S rRNA gene primers were used to screen for members of the Desulfobulbus, Desulfobacterium, Desulfovibrio, Desulfobacter, and Desulfotomaculum genera (5). DNAs from Desulfobulbus propionicus, Desulfobacterium autotrophicum, Desulfovibrio vulgaris strain G11, Desulfobacter curvatis, and Desulfotomaculum nigrificans were used as positive controls to ensure that each set of primers amplified the 16S rRNA gene of the appropriate group. PCRs and cycling conditions were set up and carried out as previously described (5).
PCR amplification of the archaeal 16S rRNA gene sequences used a previously described touchdown PCR protocol (28), with 5 pmol of GM5F (5′ CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCG TAC GGG AGG CAG CAG 3′) (22) and 20 pmol of the archaea-specific primer Arc 958R (5′ YCC GGC GTT GAM TCC ATT T 3′) (27). Denaturing gradient gel electrophoresis (DGGE) was performed with PCR-amplified products of DNA from enrichments using GM5F and Arc 958R (22). Predominant bands were excised from the gel, reamplified by the above touchdown PCR protocol, checked for purity by DGGE, and then sequenced.
For constructing archaeal 16S rRNA gene clone libraries from aquifer sediments, 16S rRNA genes were amplified from the DNA extracted from the sediments by using the GM5F and Arc 958r primers. The PCR product obtained was cloned into the TOPO 2.1 cloning vector (Invitrogen Corp., Carlsbad, CA) according to the instructions of the manufacturer. Randomly picked clones (190 total) were sequenced at the Advanced Center for Genome Technology at the University of Oklahoma. Details of the sequencing protocols applied were described previously (10) and can be found at http://www.genome.ou.edu/ds_seq_template_isol_hydra.html.
The 16S rRNA gene sequences were initially screened with the Basic Local Alignment Search Tool (1) to determine their rough phylogenetic affiliations. Sequences with greater than 98% similarity were grouped into the same operational taxonomic unit (OTU) using Sequencher (Gene Codes Corp., Ann Arbor, MI). Sequences from this study and GenBank downloaded sequences were aligned using the Clustal X program version 1.83 (37). The alignment obtained with Clustal X was also manually checked for errors. The aligned sequences were exported from Clustal X and loaded into Phylogenetic Analysis Using Parsimony (PAUP) version 4.0 beta 10 (Sinauer Associates, Sunderland, MA). Evolutionary distance trees were constructed using the neighbor-joining algorithm with Jukes-Cantor corrections. Bootstrap support values are based on 1,000 replicates.
Nucleotide sequence accession numbers.
The 16S rRNA sequences of the excised DGGE band and the OTUs from the sediment clone libraries have been assigned the following GenBank accession numbers: AY894806 for DGGE band 1, AY894807 for OTU1, and AY894808 for OTU2.
RESULTS
Most-probable-number analysis.
There was no significant (P < 0.05) difference in the number of acetate degraders (as defined by acetate depletion) in MPN samples amended with either Methanospirillum hungatei JF-1 or Desulfovibrio vulgaris G11 relative to MPN samples containing only acetate and sulfate (Table 1). Acetate consumption was coupled to methane production, and no sulfate loss was observed in any of the acetate MPN samples inoculated with sediments from the contaminated portion of the aquifer. Substrate-unamended MPN samples that contained sulfate and were inoculated with contaminated sediments produced up to 50 μmol of methane in the 10−5 dilutions after 120 days. Acetate MPN samples inoculated with uncontaminated sediments showed no acetate loss, no methane production, and no sulfate loss after 120 days.
TABLE 1.
Summary of MPN results obtained using acetate as a substratea
| Treatment | MPN contaminated | 95% confidence limitb
|
MPN uncontaminated | |
|---|---|---|---|---|
| Upper | Lower | |||
| Acetate + JF-1d | 2.4 × 106c | 5.8 × 106 | 9.9 × 107 | BDLd |
| Acetate + sulfate | 1.1 × 106 | 3.6 × 105 | 3.5 × 106 | BDL |
| Acetate + sulfate + G11d | 2.1 × 106 | 7.9 × 105 | 5.6 × 106 | BDL |
The samples used for MPN analysis were collected in 1998.
MPN values are per gram of sediment.
The 95% upper and lower confidence values used for three tube MPN analysis (3).
Abbreviations: JF-1, Methanospirillum hungatei strain JF-1; G11, Desulfovibrio vulgaris strain G11; BDL, below detection limit.
[14C]acetate-amended microcosms.
The quantities of both 14CH4 and 14CO2 produced as a result of the degradation of either [U-14C]acetate or [2-14C]acetate are shown in Table 2. The majority of the label (between 70 and 90%) from [U-14C]acetate was recovered as 14CH4 rather than 14CO2 in the active bottles regardless of whether or not sulfate was present. The addition of BESA caused this ratio to shift such that approximately 80% of the label appeared as 14CO2 in bottles with BESA compared to bottles without BESA. The addition of molybdate did not affect the fate of carbon from [U-14C]acetate compared to active bottles with acetate alone or with both acetate and sulfate (Table 2). The heat-killed controls containing [U-14C]acetate showed only a minor decrease in the quantities of labeled acetate with respect to the active bottles. However, no accumulation of either 14CH4 or 14CO2 was observed. The addition of [2-14C]acetate produced similar results as in microcosms containing [U-14C]acetate (Table 2).
TABLE 2.
Fate of acetate carbon in microcosms from the contaminated site
| Treatment | Acetate lost (Bq [104]) | 14CH4 produced (Bq [104]) | 14CO2 produced (Bq [104]) |
|---|---|---|---|
| [2-14C]acetate | 1.7 ± 0.28a | 1.4 ± 0.05 | 0.15 ± 0.1 |
| [2-14C]acetate + HKb | 0 | 0 | 0 |
| [2-14C]acetate + BESA | 1.1 ± 0.11 | 0.06 ± 0.01 | 0.4 ± 0.03 |
| [2-14C]acetate + BESA HK | 0.03-0.4c | 0 | 0 |
| [2-14C]acetate + sulfate | 1.3 ± 0.27 | 1.2 ± 0.08 | 0.11 ± 0.02 |
| [2-14C]acetate + sulfate HK | 0 | 0 | 0 |
| [2-14C]acetate + sulfate + molybdate | 1.8 ± 0.27 | 1.9 ± 0.01 | 0.17 ± 0.04 |
| [2-14C]acetate + sulfate + molybdate HK | 0.53-0.92 | 0 | 0 |
| [U-14C]acetate | 1.4 ± 0.23 | 0.78 ± 0.1 | 0.15 ± 0.07 |
| [U-14C]acetate + HK | 0.013-0.24 | 0 | 0 |
| [U-14C]acetate + BESA | 0.77 ± 0.09 | 0.05 ± 0.02 | 0.4 ± 0.03 |
| [U-14C]acetate + BESA HK | 0.025-0.4 | 0 | 0 |
| [U-14C]acetate + sulfate | 0.93 ± 0.15 | 0.82 ± 0.08 | 0.1 ± 0.02 |
| [U-14C]acetate + sulfate HK | 0 | 0 | 0 |
| [U-14C]acetate + sulfate + molybdate | 1.7 ± 0.28 | 1.1 ± 0.1 | 0.17 ± 0.03 |
| [U-14C]acetate + sulfate + molybdate HK | 0-0.23 | 0 | 0 |
Mean ± standard deviation.
HK indicates heat-killed samples.
Range of values obtained for duplicate heat-killed samples.
Molecular analyses.
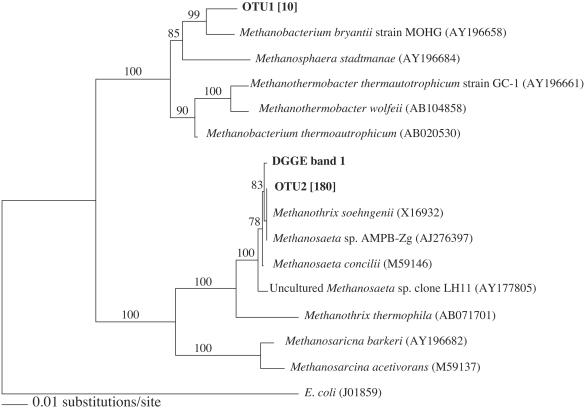
Clone libraries of 16S rRNA genes amplified from DNA extracted from contaminated sediments contained only two OTUs (Fig. 1). Sequences from OTU 2 (180 out of 190 clones) were most similar to clone SSADM_AG7 (100% similar based on a total of 564 bases sequenced). The sequence of OTU 2 was also 99.8% similar (563/564 bases were identical) to the sequence of the most intense DGGE band (Fig. 1), which was common to all microcosm enrichments regardless of whether sulfate was present or not (data not shown). OTU 1 (10 out of 190 clones) grouped with hydrogen-using methanogens from the genus Methanobacterium.
FIG. 1.
Evolutionary distance tree showing the relationship of each OTU found in contaminated sediments and DGGE band 1 from acetate enrichments with other members of the Archaea. The phylogenetic tree was constructed using the neighbor-joining algorithm. Bootstrap values that are greater than 50% are shown at each clade and are based on 1,000 replicates. Accession numbers are listed in parentheses. The frequencies of occurrence of specific OTUs in clone libraries from contaminated sediments are listed in brackets.
PCR products were observed in reaction mixtures containing primers specific for members of the Desulfobulbus, Desulfovibrio, Desulfotomaculum, and Desulfobacter genera. PCR product was not observed in PCRs that contained primers specific for members of the Desulfobacterium.
DISCUSSION
This work showed that aceticlastic methanogenesis was the predominant fate of acetate in an aquifer where geochemical evidence implicated both sulfate reduction and methanogenesis as important terminal electron-accepting processes (12). Anaerobic acetate degradation can occur by iron reduction (4, 29), sulfate reduction (6, 18, 25, 26, 30, 32, 35), aceticlastic methanogenesis (6, 18, 23, 32, 33, 36), or syntrophic acetate degradation (31). Iron reduction did not appear to play an important role in contaminated sediments (12). The high quantities of methane observed in MPN samples, the high ratios of 14CH4/14CO2 in microcosms containing [14C]acetate, the lack of sulfate consumption in MPN samples amended with sulfate, and the high frequency of sequences from the family Methanosaetaceae in enrichments and clone libraries from contaminated sediments lead to the conclusion that aceticlastic methanogenesis rather than sulfate reduction is responsible for acetate consumption at this site. The possibility of syntrophic acetate metabolism seems unlikely, since one methane was produced per acetate consumed in all MPN tubes and all label from the microcosms amended with [2-14C]acetate was recovered as 14CH4, as expected for aceticlastic methanogenesis (11). The lack of sulfate consumption in MPN samples with sulfate and G11 would also suggest that syntrophic metabolism of acetate is not occurring at this site. The ratios of 14CH4/14CO2 observed in some of the microcosms amended with [U-14C]acetate were higher than the expected 1/1 ratio, which may indicate that hydrogenotrophic methanogenesis is also an important process in the contaminated sediments.
The results of this work raise the question of why aceticlastic methanogens control the fate of acetate in an aquifer where geochemical evidence indicates that sulfate reduction is an important TEAP. Several studies have shown that a number of different factors, including pH (38), sulfide toxicity (38), substrate specificity (26), sulfate limitation (15), and kinetic factors, including km, Vmax, and acetate threshold concentration (24, 26, 34, 40, 43), control whether acetate is utilized by methanogens or sulfate-reducing bacteria in anaerobic environments. The microcosms used in this study had a pH range from around 7.0 to 7.4 throughout the experiment, and sulfide concentrations ranged from 1 mg/liter to 10 mg/liter in the contaminated portion of the aquifer. These pH values and sulfide concentrations have been shown to be favorable for the growth of both aceticlastic methanogens and acetate-utilizing sulfate-reducing bacteria (38). The predominance of members of the family Methanosaetaceae at this site, which are only capable of using acetate (26, 33), is interesting since sulfate-reducing bacteria are known to completely mineralize the hydrocarbons in petroleum-contaminated environments where sulfate is present (14, 42, 44), and group-specific PCR indicates the presence of a potential acetate-using sulfate reducer, Desulfobacter sp. From the specific radioactivity of [14C]acetate and the bequerels present, we estimate that the [14C]acetate concentration after 18 days was approximately 0.3 μM in microcosms containing [14C]acetate, [14C]acetate with sulfate, and [14C]acetate with sulfate and molybdate. By HPLC analysis the final concentration of acetate was below the detection limit of 50 μM. These results show that the final acetate concentration in the microcosms was between 0.3 μM and 50 μM, which is consistent with previously described acetate threshold concentrations in Methanosaeta (20, 26). These findings, along with those describing similar kinetic properties (km, Vmax, and acetate threshold concentration) in two acetate-utilizing sulfate reducers and Methanosaeta soehngenii (26), suggest that some members of the Methanosaeta may be able to compete with sulfate reducers for acetate. Low levels of sulfate in the contaminated region of the aquifer may favor acetate degradation by methanogenesis rather than by sulfate reducers, since the acetate user Desulfobacter postgatei was a less successful competitor for limiting sulfate than two other hydrogen-using sulfate reducers (15).
While it is clear that acetate is an important intermediate in this hydrocarbon-contaminated site, the source of acetate in the contaminated portion of this aquifer is unclear. Bacterial clone libraries prepared with DNA from acetate enrichments and contaminated sediments contained a large number of both clostridial and Cytophaga-Flavobacter-Bacteroides sequences (data not shown), indicating that fermentative metabolism could be a source of acetate. Another possibility is the incomplete metabolism of the benzene, toluene, ethylbenzene, and xylene isomers (BTEX), which are major components of gas condensate. Dolfing (9) suggests that the incomplete metabolism of benzoate, which is known to be an important intermediate in the anaerobic biodegradation of BTEX, to acetate is more energetically favorable than its complete mineralization to CO2 under methanogenic conditions (PH2 > 2 Pa). Thus, under sulfate-limiting conditions, it is likely that incomplete BTEX hydrocarbon degradation is occurring, resulting in acetate excretion, which creates a niche for the aceticlastic methanogens to function in this hydrocarbon-contaminated environment.
Acknowledgments
We thank N. Wofford for technical assistance and J. M. Suflita and K. Kropp for providing the MTBE enrichment culture used in this study.
This work was supported by contract DE-FG03-96-ER-20212 from the U.S. Department of Energy.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banwart, G. J. 1981. Basic food microbiology. AVI Publishing Co., Westport, Conn.
- 4.Coates, J. D., E. J. Phillips, D. J. Lonergan, H. Jenter, and D. R. Lovley. 1996. Isolation of Geobacter species from diverse sedimentary environments. Appl. Environ. Microbiol. 62:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly, K., R. J. Sharp, and A. J. McCarthy. 2000. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146:1693-1705. [DOI] [PubMed] [Google Scholar]
- 6.de Graaf, W., P. Wellsbury, R. J. Parkes, and T. E. Cappenberg. 1996. Comparison of acetate turnover in methanogenic and sulfate-reducing sediments by radiolabeling and stable isotope labeling and by use of specific inhibitors: evidence for isotopic exchange. Appl. Environ. Microbiol. 62:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dojka, M. A., P. Hugenholz, S. K. Haak, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolfing, J. 2001. The microbial logic behind the prevalence of incomplete oxidation of organic compounds by acetogenic bacteria in methanogenic environments. Microb. Ecol. 41:83-89. [DOI] [PubMed] [Google Scholar]
- 10.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferry, J. G. 1992. Methane from acetate. J. Bacteriol. 174:5489-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gieg, L. M., R. V. Kolhatkar, M. J. McInerney, R. S. Tanner, S. H. Harris, K. L. Sublette, and J. M. Suflita. 1999. Intrinsic bioremediation of petroleum hydrocarbons in a gas condensate-contaminated aquifer. Environ. Sci. Technol. 33:2550-2560. [Google Scholar]
- 13.Jenneman, G. E., M. J. McInerney, and R. M. Knapp. 1986. Effect of nitrate on biogenic sulfide production. Appl. Environ. Microbiol. 51:1205-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleikemper, J., M. H. Schroth, W. V. Sigler, M. Schmucki, S. M. Bernasconi, and J. Zeyer. 2002. Activity and diversity of sulfate-reducing bacteria in a petroleum hydrocarbon-contaminated aquifer. Appl. Environ. Microbiol. 68:1516-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laanbroek, H. J., H. J. Geerligs, L. Sijtsma, and H. Veldkamp. 1984. Competition for sulfate and ethanol among Desulfobacter, Desulfobulbus, and Desulfovibrio species isolated from intertidal sediments. Appl. Environ. Microbiol. 47:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Londry, K. L., P. M. Fedorak, and J. M. Suflita. 1997. Anaerobic degradation of m-cresol by a sulfate-reducing bacterium. Appl. Environ. Microbiol. 63:3170-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovley, D. R., F. H. Chapelle, and J. C. Woodward. 1994. Use of dissolved H2 concentrations to determine distribution of microbially catalyzed redox reactions in anoxic groundwater. Environ. Sci. Technol. 28:1205-1210. [DOI] [PubMed] [Google Scholar]
- 18.Lovley, D. R., and M. J. Klug. 1982. Intermediary metabolism of organic matter in the sediments of a eutrophic lake. Appl. Environ. Microbiol. 43:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lueders, T., and M. W. Friedrich. 2002. Effects of amendment with ferrihydrite and gypsum on the structure and activity of methanogenic populations in rice field soil. Appl. Environ. Microbiol. 68:2484-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McInerney, M. J. 1999. Anaerobic metabolism and its regulation, p. 129-135. In H. J. Rehm and G. Reed (ed.), Biotechnology. Wiley-VCH, Weinheim, Germany.
- 21.McInerney, M. J., M. P. Bryant, and N. Pfennig. 1979. Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens. Arch. Microbiol. 122:129-135. [DOI] [PubMed] [Google Scholar]
- 22.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nusslein, B., W. Eckert, and R. Conrad. 2003. Stable isotope biogeochemistry of methane formation in profundal sediments of Lake Kinneret (Israel). Limnol. Oceanogr. 48:1439-1446. [Google Scholar]
- 24.Ohtsubo, S., K. Demizu, S. Kohno, I. Miura, T. Ogawa, and H. Fukuda. 1992. Comparison of acetate utilization among strains of an aceticlastic methanogen, Methanothrix soehngenii. Appl. Environ. Microbiol. 58:703-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oude Elferink, S. J. W. H., W. M. Akkermans-van Vliet, J. J. Bogte, and A. J. M. Stams. 1999. Desulfobacca acetoxidans gen. nov., sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Bacteriol. 49:345-350. [DOI] [PubMed] [Google Scholar]
- 26.Oude Elferink, S. J. W. H., S. B. I. Luppens, C. L. M. Marcelis, and A. J. M. Stams. 1998. Kinetics of acetate oxidation by two sulfate reducers isolated from anaerobic granular sludge. Appl. Environ. Microbiol. 64:2301-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purdy, K. J., M. A. Munson, D. B. Nedwell, and T. M. Embley. 2002. Comparison of the molecular diversity of the methanogenic community at the brackish and marine ends of a UK estuary. FEMS Microbiol. Ecol. 39:17-21. [DOI] [PubMed] [Google Scholar]
- 28.Rios-Hernandez, L. A., L. M. Gieg, and J. M. Suflita. 2003. Biodegradation of an alicyclic hydrocarbon by a sulfate-reducing enrichment from a gas condensate-contaminated aquifer. Appl. Environ. Microbiol. 69:434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roden, E. E., and R. G. Wetzel. 2003. Competition between Fe(III)-reducing and methanogenic bacteria for acetate in iron-rich freshwater sediments. Microb. Ecol. 45:252-258. [DOI] [PubMed] [Google Scholar]
- 30.Rooney-Varga, J. N., B. R. Genthner, R. Devereux, S. G. Willis, S. D. Friedman, and M. E. Hines. 1998. Phylogenetic and physiological diversity of sulphate-reducing bacteria isolated from a salt marsh sediment. Syst. Appl. Microbiol. 21:557-568. [DOI] [PubMed] [Google Scholar]
- 31.Schnürer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Appl. Environ. Microbiol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 32.Scholten, J. C., and A. J. Stams. 2002. Isolation and characterization of acetate-utilizing anaerobes from a freshwater sediment. Microb. Ecol. 40:292-299. [DOI] [PubMed] [Google Scholar]
- 33.Scholten, J. C., R. Conrad, and A. J. Stams. 2000. Effect of 2-bromo-ethane sulfonate, molybdate and chloroform on acetate consumption by methanogenic and sulfate-reducing populations in freshwater sediment. FEMS Microbiol. Ecol. 32:35-42. [DOI] [PubMed] [Google Scholar]
- 34.Schönheit, P., J. K. Kristjansson, and R. K. Thauer. 1982. Kinetic mechanism for the ability of sulfate reducers to out-compete methanogens for acetate. Arch. Microbiol. 132:285-288. [Google Scholar]
- 35.Sorenson, J., D. Christensen, and B. B. Jorgensen. 1981. Volatile fatty acids and hydrogen as substrates for sulfate-reducing bacteria in anaerobic marine sediment. Appl. Environ. Microbiol. 42:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stams, A. J. 1994. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Leeuwenhoek 66:271-294. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visser, A., L. W. Hulshoff Pol, and G. Lettinga. 1996. Competition of methanogenic and sulfidogenic bacteria. Water. Sci. Technol. 33:99-110. [Google Scholar]
- 39.Voordouw, G., S. M. Armstrong, M. F. Reimer, B. Fouts, A. J. Telang, Y. Shen, and D. Gevertz. 1996. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl. Environ. Microbiol. 62:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, D. M., and M. R. Winfrey. 1985. Interactions between methanogenic and sulfate-reducing bacteria in sediments, p. 219-286. In H. W. Jannasch and P. J. L. Williams (ed.), Advances in microbial ecology. Plenum Press, New York, N.Y.
- 41.Watanabe, K., Y. Kodama, N. Hamamura, and N. Kaku. 2002. Diversity, abundance, and activity of archaeal populations in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Appl. Environ. Microbiol. 68:3899-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weideneier, T. H., H. S. Rifai, C. J. Newell, and J. T. Wilson. 1999. Natural attenuation of fuels and chlorinated solvents in the subsurface. John Wiley and Sons Inc., New York, N.Y.
- 43.Westermann, P., B. K. Ahring, and R. A. Mah. 1989. Threshold acetate concentrations for acetate catabolism by aceticlastic methanogenic bacteria. Appl. Environ. Microbiol. 55:514-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Widdel, F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p. 469-483. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. Wiley-Liss, New York, N.Y.