Abstract
We report a significantly improved system for studying single-copy lacZ operon fusions in Yersinia enterocolitica: a simple procedure for the stable integration of lacZ operon fusions into the ara locus and a strain with a deletion mutation that abolishes the low level of endogenous β-galactosidase activity.
Yersinia enterocolitica is a cause of foodborne human gastroenteritis and a favored model organism for studying bacterial virulence (2). Y. enterocolitica virulence gene regulation is studied in many laboratories. This is especially true for the genes encoding the Ysc type III secretion system, which Y. enterocolitica shares in common with the other pathogenic Yersinia species, Y. pestis and Y. pseudotuberculosis (5).
In Escherichia coli K-12, the ability to construct single-copy lacZ operon fusions greatly facilitates regulatory studies. Single-copy lacZ fusions overcome the problems of multicopy fusions, such as copy number variation and titration of transcription factors. The only existing method for constructing single-copy lacZ fusions in Y. enterocolitica relies on complete chromosomal integration of a suicide plasmid by homologous recombination (see, e.g., reference 6). This approach has many limitations, including the following. (i) The integrated plasmid encodes an antibiotic resistance no longer available for subsequent manipulations. (ii) Chromosomal integration is driven by the promoter fragment, which limits the minimum size of the fragment that can be studied. (iii) Studying promoters internal to an operon results in disruption of the operon after suicide plasmid integration. (iv) Integration of fragments with promoter mutations may be tedious to verify (homologous recombination can occur either upstream or downstream of the mutation, and only the former places the mutation upstream of lacZ). The use of lacZ fusions in Y. enterocolitica is further compromised because, although it is known as a Lac− species, there is a trace level of endogenous β-galactosidase activity that causes the formation of pale blue colonies on agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). This limits the use of X-Gal media for phenotypic screens and the analysis of weakly expressed promoters in enzyme assays. These limitations motivated our development of an improved system for the construction and analysis of single-copy lacZ operon fusions in Y. enterocolitica.
Construction of ara integration plasmid pAJD905.
We followed a strategy similar to that employed by Wu et al., who targeted the rha locus of Klebsiella oxytoca for lacZ operon fusion integration (21). They used a suicide plasmid with an rpsL+ (streptomycin sensitivity) allele for counter selection, which requires the use of a strain with an rpsL mutation conferring streptomycin resistance. Streptomycin resistance is a widely used marker in Y. enterocolitica, which is naturally ampicillin resistant. Therefore, we chose a different approach, based on the established technology of a suicide plasmid with a π protein-dependent R6K replication origin and the use of the sacB gene for selection of plasmid-free segregants as sucrose-resistant colonies. We extended this technology to develop a Y. enterocolitica site-specific integration lacZY operon fusion vector, with several features that increase its utility.
For the chromosomal integration site we chose the araFGHC-araBA locus of a Y. enterocolitica strain 8081 derivative, which is the highly pathogenic strain used in the genome sequencing project. Disruption of this locus abolishes arabinose catabolism, which allows integrants to be easily identified by their Ara− phenotype (e.g., formation of white colonies on MacConkey-arabinose agar plates). It also protects the lacZY operon fusions from flanking promoters by placing them between divergently transcribed operons which have had their promoters deleted (10).
The allelic exchange vector pRE112 (Table 1) has a R6K replication origin and the sacB1 gene, which confers sucrose sensitivity (9). Two 1.5-kb fragments (′araGH′ and ′araBA′) were amplified by PCR and ligated into the pRE112 polylinker with a unique NotI site between them (pAJD898). Next, plasmid pAJD768 was constructed by joining the ∼7-kb BamHI lacZYA-cat fragment of pFUSE to the ∼3.1-kb BglII pSC101 ori fragment of pWSK29 and then inserting the StuI-SmaI pSL1180 polylinker upstream of lacZ. Finally, the polylinker-lacZY region of pAJD768 was amplified with primers that incorporate E. coli rrnBT1 and rrnBT2 terminators upstream and downstream, respectively. This fragment was cloned into the NotI site of plasmid pAJD898. Derivatives with the lacZY operon in either orientation were isolated. However, one of these orientations produced somewhat higher basal lacZY expression than the other (data not shown). Therefore, a plasmid with the lacZY operon in the orientation that gave the lowest basal lacZY expression was chosen. This was pAJD905, the ara-integration lacZY operon fusion vector (Fig. 1A). The rrnB terminators further insulate lacZY fusions from the influence of external chromosomal regions.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or feature(s) | Reference or source |
|---|---|---|
| Y. enterocolitica strain 8081 derivatives | ||
| JB580v | ΔyenR (R− M+) | 12 |
| AJD935 | ΔyenR (R− M+) ΔaraGFB::[Φ(−lacZY)] | This study |
| AJD957 | ΔyenR (R− M+) ΔaraGFB::[Φ(pspA−lacZY)] | This study |
| AJD1022 | ΔyenR (R− M+) ΔaraGFB::[Φ(yopH−lacZY)] | This study |
| AJD1024 | ΔyenR (R− M+) ΔYE2592 | This study |
| AJD1025 | ΔyenR (R− M+) ΔYE2592 ΔaraGFB::[Φ(−lacZY)] | This study |
| Y. enterocolitica CDCa reference strains and clinical isolates | ||
| 634-83 | Serogroup O:4,32, American strain | 17 |
| 637-83 | Serogroup O:5,27, non-American strain | 17 |
| MC8 | Bio. 1, serogroup O:9, non-American strain | 17 |
| MC17 | Bio. 1, serogroup O:3, non-American strain | 17 |
| Plasmids | ||
| pFUSE | Cmr, mob+ (RP4), lacZYA+, R6K ori | 1 |
| pRE112 | Cmr, mob+ (RP4), sacB1+, R6K ori | 9 |
| pSL1180 | Apr, super polylinker, colE1 ori | Amersham |
| pSR47S | Kmr, mob+ (RP4), sacB+, R6K ori | 16 |
| pVLT33 | Kmr, tacp expression vector, RSF1010 ori | 7 |
| pWSK29 | Apr, pSC101 ori | 20 |
| pAJD509 | tacp-yscC+ in pVLT33 | This study |
| pAJD554 | tacp-ysaC+ in pVLT33 | This study |
| pAJD768 | Cmr, lacZYA+, pSC101 ori | This study |
| pAJD898 | pRE112 with ′araHG′-NotI-′araBA′ insert | This study |
| pAJD905 | pAJD898 with polylinker and lacZY from pAJD768 | This study |
| pAJD930 | pAJD905 with pspAp fragment | This study |
| pAJD952 | pAJD905 with yopHp fragment | This study |
| pAJD990 | pSR47S with ΔYE2592 insert | This study |
CDC, Centers for Disease Control and Prevention.
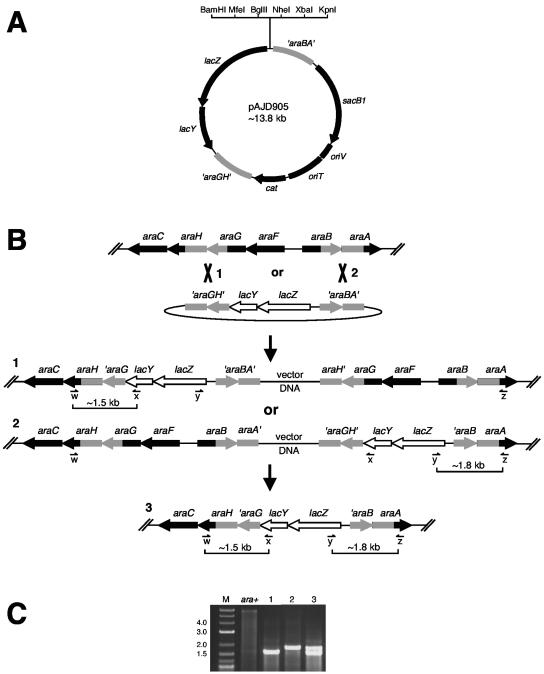
FIG. 1.
Integration of lacZY operon fusions into the ara locus. (A) Partial restriction map of plasmid pAJD905. Restriction sites shown in the polylinker are unique (note that MfeI is compatible with EcoRI). (B) Summary of lacZY fusion integration procedure (see text for experimental details). Plasmid pAJD905 is integrated into the ara locus by homologous recombination between either the ′araGH′ sequences or the ′araBA′ sequences. The structures of both possible integrants are shown as structures 1 and 2. Next, sucrose-resistant segregants lose the integrated pAJD905 backbone by homologous recombination between the tandemly duplicated ara sequences. This either regenerates the ara+ locus or leaves the lacZY fusion on the chromosome, shown as structure 3. (C) Colony PCR analysis of the ara+ parental strain, integrants with structure 1 or 2, and segregant 3 is shown. In this case the lacZY operon from pAJD905 without a promoter cloned upstream was used. Binding sites of each primer are shown in panel B as w, x, y, and z (see also Table 2). For the ara+ strain, primers w and z generate a single product of ∼6 kb that is only weakly amplified (primers x and y do not anneal). Integrants 1 and 2 give only one PCR product, as indicated in panel B (the second pair of primers are too far apart for amplification). The desired segregant gives two PCR products as shown. Note that a promoter fragment inserted upstream of the lacZY operon would increase the size of the 1.8-kb product. M, DNA marker (kilobase sizes of some marker bands are indicated). The gel lane images have been rearranged from the original order, but the data are from the same gel and a single experiment.
Chromosomal integration of lacZY operon fusions.
The method used to exchange lacZY operon fusions from pAJD905 into the ara locus is summarized in Fig. 1B. First, pAJD905 (or a derivative with a promoter insert) was transferred from E. coli S17-1 λpir (18) to Y. enterocolitica by conjugation. Plasmid integrants were identified by their Cmr Ara− phenotype on MacConkey-arabinose agar (the 1.5-kb regions flanking lacZY ensure that most integrations occur at ara). Segregants then lost the integrated plasmid by homologous recombination between the tandemly duplicated ara sequences. They were selected by streaking integrants on LB agar containing 10% sucrose (loss of the plasmid sacB1 gene restores a sucrose-resistant phenotype). The sucrose-resistant colonies were replica plated onto MacConkey-arabinose agar, which identified both Ara+ and Ara− segregants as expected. The authenticity of the desired Ara− segregants was confirmed by their Cms phenotype. Finally, the structure of segregants was confirmed by colony PCR analysis using an Expand PCR system (Roche; see Fig. 1C).
Application of the pAJD905 system.
We tested the new lacZY fusion system with the pspA promoter (studied in our laboratory) and the yopH promoter (studied in various laboratories). Approximately 500-bp control region fragments were amplified by PCR, confirmed by DNA sequencing, and ligated between the XbaI and BglII sites of pAJD905. The Φ(pspA-lacZ) and Φ(yopH-lacZ) fusions were integrated into the ara locus as described above (data not shown). In both cases, the initial integration step occurred in the ara locus rather than the yopH or pspA locus in over 99% of cases (initially identified by an Ara− phenotype). We have also done other studies with an approximately 2.2-kb fragment cloned into pAJD905. In this case the frequency of integration into the ara locus did decrease somewhat. However, the majority of integrations were still in the ara locus (data not shown).
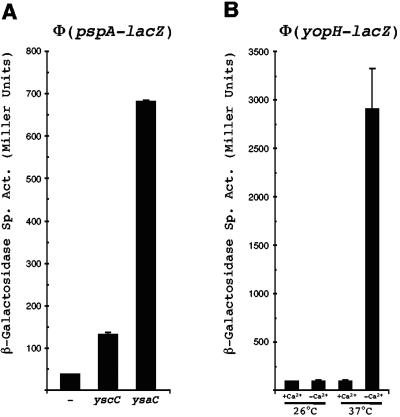
Previous studies have shown that pspA promoter expression is induced by the YscC and YsaC secretins, with YsaC being the most potent (6, 11, 15). The ΔaraGFB::[Φ(pspA-lacZY)] fusion demonstrated exactly the same pattern of regulation (Fig. 2A). The virulence plasmid (pYV)-encoded yopH gene is induced by a low calcium concentration at 37°C, and its expression requires the VirF transcription activator (4, 13). Consistent with this, expression of the ΔaraGFB::[Φ(yopH-lacZY)] fusion was induced approximately 30-fold by these growth conditions (Fig. 2B). Furthermore, in a strain lacking pYV (and, therefore, VirF), Φ(yopH-lacZY) expression was essentially abolished (7 Miller units; data not shown).
FIG. 2.
Regulation of ΔaraGFB::[Φ(pspA-lacZY)] and ΔaraGFB::[Φ(yopH-lacZY)] fusions. (A) Φ(pspA-lacZY) expression. Plasmid pVLT33 (−), pAJD509 (yscC), or pAJD554 (ysaC) encoding the indicated genes expressed from the tac promoter were transferred into a pYV-negative derivative of strain AJD957. Strains were grown exactly as described previously (11). (B) Φ(yopH-lacZ) expression. Strain AJD1022 was grown at 26°C or 37°C in low- or high-calcium media as described previously (6). β-Galactosidase activities were determined as described previously (14) and are expressed in Miller units (each value is the average of measurements from three independent cultures). Sp. Act. = specific activity.
These results demonstrate the functionality of the new lacZY fusion system. The ability to construct stable lacZY fusions with pYV-encoded promoters is particularly important. The alternative approach of integrating suicide plasmids into the relatively small pYV could affect the stability or copy number of pYV or the expression of pYV-encoded genes.
Elimination of Y. enterocolitica endogenous β-galactosidase activity.
Y. enterocolitica is phenotypically Lac− on MacConkey-lactose agar but has a low level of β-galactosidase activity in enzyme assays and forms blue colonies on X-Gal media. The Y. enterocolitica genome encodes a protein (YE2592) approximately 60% identical to E. coli LacZ (data not shown). To determine whether this gene is responsible for endogenous β-galactosidase activity we constructed a strain with a YE2592 in-frame deletion mutation. The mutation was made using the sacB+ suicide plasmid pSR47S with a ΔYE2592 insert, which was generated by PCR. Plasmid construction and mutagenesis procedures were identical to those described by others (19).
In enzyme assays, the YE2592+ Y. enterocolitica strain produced 2 to 4 Miller units of β-galactosidase activity, depending on growth conditions, whereas the ΔYE2592 mutant had no detectable activity (data not shown). Furthermore, whereas the YE2592+ strain was blue on agar containing X-Gal, the ΔYE2592 mutant was white (data not shown). These results confirm that YE2592 is responsible for Y. enterocolitica endogenous β-galactosidase activity. The ΔYE2592 mutant strain provides a significant enhancement for the use of β-galactosidase enzyme assays and X-Gal media in screens and to analyze weakly expressed lacZ operon fusions.
Use of the pAJD905 system with different Y. enterocolitica strains.
There are different biotypes and serotypes of Y. enterocolitica (2). In particular, there are two groups of strains known as American and non-American, which differ in their virulence properties (American strains are more virulent) (3). We were interested to know whether plasmid pAJD905 could be used in different Y. enterocolitica strains, in addition to our strain 8081 derivative (an American strain of serotype O:8). We randomly chose one American strain (serotype O:4,32) and three non-American strains (serotypes O:5,27; O:3; and O:9), all of which are Ara+ (as were all of the Y. enterocolitica strains in our collection). First, we attempted PCR amplification of the ′araBA′ and ′araGH′ fragments with the same primers used to construct pAJD898. Both fragments were successfully amplified from the O:4,32; O:5,27; and O:3 strains (data not shown). However, only the ′araGH′ fragment could be amplified from the O:9 strain (therefore, this strain was not used further). Next, we attempted the pAJD905 integration and segregation procedure. For the American O:4,32 strain, the process proceeded with speed and efficiency similar to that seen with our strain 8081 derivative. The procedure was also successful for the non-American O:5,27 and O:3 strains. However, in these cases the segregation procedure heavily favored reversion to Ara+. Only a few desired Ara− segregants were isolated. Even so, colony PCR analysis identified segregants with the correct integration in both cases (data not shown). From these experiments we conclude that pAJD905 can be used in different Y. enterocolitica strains, although not all. However, the system is likely to be most easily used in the highly pathogenic American strains.
Summary.
The Y. enterocolitica single-copy lacZY operon fusion system described here provides many benefits, including the following. (i) Fusion strains are stable, are not marked with an antibiotic resistance, and do not contain any suicide vector DNA, all of which facilitate further manipulation. (ii) There is no lower limit to the size of promoter fragments that can be analyzed. (iii) There is low background (integration of lacZY without a promoter fragment upstream causes only approximately 2 to 4 Miller units of β-galactosidase activity; data not shown). (iv) The native locus of the promoter under study is not disrupted (polar effects are not a concern). (v) The integration procedure is simple and takes only a few days. (vi) Correct lacZY fusion integrants are easily and unambiguously confirmed by their Ara− phenotype and colony PCR analysis. Our identification of the Y. enterocolitica gene responsible for endogenous β-galactosidase activity, and construction of a deletion mutant, further improves the utility of lacZ fusions in this organism.
TABLE 2.
Primers
| Primer name and function | Sequence (5′ to 3′) |
|---|---|
| For amplification of ′araGH′ and ′araBA′ fragments | |
| ′araG NotI | GAAGCGGCCGCGGGAAATCGAGCAACTGTTCCG |
| araH′ SpeI | CGGACTAGTGGTTATCAGGGCATTTATCTTCAGC |
| ′araB NotI | GAAGCGGCCGCTGAACGCCAGGTTGGTCTG |
| araA′ SacI | CGGGAGCTCCTGTTTGGCTGCCGCAACG |
| For amplification of lacZY fragment with flanking rrnB terminators | |
| lacZ Up rrnBT1 | GAACGGCCGATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCGGTACCTCTAGAACTATAGC |
| lacY Dwn rrnBT2 | AGAATGCGGCCGCAAAAAGGCCATCCGTCAGGATGGCCTTCTGCCGACATTGATTGCTTAAGCG |
| For colony PCR confirmation of lacZY operon fusion integrants | |
| araH check (w) | GAAATGATGTAAGCCAGACC |
| lacY check (x) | CGTCAGGTGAATGAAGTCGC |
| M13-40 (y) | GTTTTCCCAGTCACGAC |
| araA check (z) | GCATATTATCGCCGAAACGG |
| For amplification of pspA and yopH control regions | |
| pspAp Up | AGGCGATGGGCTATCAGCTC |
| pspAp Dwn | GGATCCGCACTAACTTCTGTGGAT |
| yopHp Up | GGCTCTAGATATTCGTTGCTATCACTGG |
| yopHp Dwn | GCGAGATCTAAGTTCATGCTTCCCTCC |
| For amplification of fragments flanking YE2592 | |
| ΔYE2592 Up Fwd | CGCGTCGACCCCATTTAGCGCCATCTTG |
| ΔYE2592 Up Rev | CCGGGATCCCTGCGGTTTAACTTCTTGC |
| ΔYE2592 Dwn Fwd | CCGGGATCCTGGCTAAATATCGATGGT |
| ΔYE2592 Dwn Rev | GAAGCGGCCGCTACAGGGAGCTTCACCA |
Acknowledgments
We thank Joe Vogel and Kimberly Walker for providing plasmid pSR47S and its DNA sequence. Y. enterocolitica genome sequence data were produced by the Y. enterocolitica Sequencing Group at the Sanger Institute and can be obtained online at http://www.sanger.ac.uk/Projects/Y_enterocolitica/.
This work was supported by Public Health Service grant AI-052148 from the National Institute of Allergy and Infectious Diseases and by a grant from the Speaker's Fund for Biomedical Research: Toward the Science of Patient Care, awarded by the City of New York.
REFERENCES
- 1.Bäulmer, A. J., R. M. Tsolis, A. W. M. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 2.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelis, G., Y. Laroche, G. Balligand, M.-P. Sory, and G. Wauters. 1987. Y. enterocolitica, a primary model for bacterial invasiveness. Rev. Infect. Dis. 9:64-87. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis, G., C. Sluiters, C. Lambert de Rouvroit, and T. Michiels. 1989. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis, G. R. 2002. The Yersinia Ysc-Yop “Type III” weaponry. Nat. Rev. Mol. Cell Biol. 3:742-754. [DOI] [PubMed] [Google Scholar]
- 6.Darwin, A. J., and V. L. Miller. 2001. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39:429-444. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, R. C., and A. J. Darwin. 2004. PspG, a new member of the Yersinia enterocolitica phage shock protein regulon. J. Bacteriol. 186:4910-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 13.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6:395-409. [PubMed] [Google Scholar]
- 14.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 15.Maxson, M. E., and A. J. Darwin. 2004. Identification of inducers of the Yersinia enterocolitica phage shock protein system and comparison to the regulation of the RpoE and Cpx extracytoplasmic stress responses. J. Bacteriol. 186:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, V. L., J. J. Farmer III, W. E. Hill, and S. Falkow. 1989. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect. Immun. 57:121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker, K. A., and V. L. Miller. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 21.Wu, S., W. Chai, J. T. Lin, and V. Stewart. 1999. General nitrogen regulation of nitrate assimilation regulatory gene nasR expression in Klebsiella oxytoca M5al. J. Bacteriol. 181:7274-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]