Abstract
The yeast protein Prp19p is essential for pre-mRNA splicing and is associated with the spliceosome concurrently with or just after dissociation of U4 small nuclear RNA. In splicing extracts, Prp19p is associated with several other proteins in a large protein complex of unknown function, but at least one of these proteins is also essential for splicing (W.-Y. Tarn, C.-H. Hsu, K.-T. Huang, H.-R. Chen, H.-Y. Kao, K.-R. Lee, and S.-C. Cheng, EMBO J. 13:2421–2431, 1994). To identify proteins in the Prp19p-associated complex, we have isolated trans-acting mutations that exacerbate the phenotypes of conditional alleles of prp19, using the ade2-ade3 sectoring system. A novel splicing factor, Snt309p, was identified through such a screen. Although the SNT309 gene was not essential for growth of Saccharomyces cerevisiae under normal conditions, yeast cells containing a null allele of the SNT309 gene were temperature sensitive and accumulated pre-mRNA at the nonpermissive temperature. Far-Western blot analysis revealed direct interaction between Prp19p and Snt309p. Snt309p was shown to be a component of the Prp19p-associated complex by Western blot analysis. Immunoprecipitation studies demonstrated that Snt309p was also a spliceosomal component and associated with the spliceosome in the same manner as Prp19p during spliceosome assembly. These results suggest that the functions of Prp19p and Snt309p in splicing may require coordinate action of these two proteins.
Splicing of pre-mRNA occurs on a large ribonucleoprotein particle (RNP) called the spliceosome, which consists of five small nuclear RNAs (snRNAs) and a number of protein factors (for reviews, see references 18, 38, 39, 47, and 49). The roles of snRNAs in spliceosome assembly have been extensively studied in both Saccharomyces cerevisiae and mammals. Base-pairing-mediated interactions between small nuclear RNPs (snRNPs), and between snRNPs and the pre-mRNA, appear to play important roles in assembly of the spliceosome (for reviews, see references 18, 47, and 49). Additional proteins are also required for proper assembly and function of the spliceosome (for reviews, see reference 4, 42, 44, and 47).
Assembly of the spliceosome is a multistep process that involves sequential binding of snRNAs to the pre-mRNA in the order U1, U2, and then U4-U6 plus U5 as a preformed tri-snRNP (8, 12, 28, 40). After all five snRNAs are associated with the pre-mRNA, U4 becomes only loosely associated with the spliceosome and does not participate in the subsequent splicing reaction (61). A large conformational rearrangement of the spliceosome occurs, accompanying U4 dissociation as the mode of interactions between pre-mRNA and snRNAs changes. New base-pairings between U5 and the pre-mRNA and between U6 and the 5′-splice site region of the pre-mRNA are detected (56). It is generally believed that an RNA helicase activity is involved in this step of the assembly process to unwind base-pairings between U4 and U6 snRNAs and between pre-mRNA and snRNAs. Nevertheless, no RNA helicase activity has been demonstrated. Factors mediating such conformational change have also not yet been identified.
Identification of protein factors involved in pre-mRNA splicing has been greatly facilitated by yeast genetics. A large number of PRP (precursor RNA processing) genes that encode protein splicing factors have been identified by screening temperature-sensitive mutants defective in pre-mRNA splicing (55). Other genes have been identified through genetic interactions with introns, PRP genes, or snRNA genes. They include suppressors of temperature-sensitive alleles of PRP genes (37), suppressors of snRNA mutations (46, 57), and mutants with a synthetic lethal phenotype for mutations in snRNA or protein factors (17, 32, 50). Biochemical and genetic studies reveal that many of these genes encode snRNP-associated proteins. The SNP1, MUD1, PRP39, and PRP40 genes encode protein components of the U1 snRNP (16, 25, 26, 32, 35). Snp1p is the yeast homolog of human U1-70K protein (16, 25), and Mud1p is a U1A-like protein (32). Prp8p and Prp18p are components of the U5 snRNP (21, 36), while Prp4p and Prp6p are part of the U4-U6 snRNP (1, 3, 9). Prp24p and Sdb23p were shown to be associated with both free U6 snRNA and U4-U6 base-paired snRNAs (15, 46). In addition to Msl1p being identified as the homolog of U2 snRNP B" protein (50), Cus1p was identified as homologous to human SAP145, which is a U2 snRNP component (57). The demonstration of genetic interactions between U2 snRNA and Prp5p, Prp9p, Prp11p, and Prp21p/Spp91p suggests functional associations of these proteins factors with U2 snRNA (43). In fact, a Prp9p-related splicing factor, SF3a, has consistently been identified in the mammalian system and has been demonstrated to interact with the U2 snRNP in the presence of another splicing factor, SF3b (10).
Proteins involved in cleavage-ligation reactions have also been demonstrated. Prp2p is required for the first step of the reaction and is dispensable for assembly of the spliceosome (33). The second step of the reaction requires Prp16p, Prp17p/Slu4p, Prp18p, and Slu7p, all of which can genetically interact with U5 snRNA (17). The U5 snRNP, required for early steps of the spliceosome assembly process, may play an additional role in coordinating a set of factors required for the second catalytic step of the splicing reaction (17). Both Prp2p and Prp16p contain the DEAD-DEAH box RNA helicase motif and have been demonstrated to possess RNA-dependent nucleoside triphosphatase (NTPase) activities (27, 45). Prp16p is further proposed to play a role in promoting the fidelity of mRNA splicing in an ATP hydrolysis-dependent manner (11), reminiscent of NTPases which are believed to enhance accuracy in other macromolecular biological processes (54). Other proteins, such as Prp5p, Prp22p, Prp28p, and Slt22p, also contain the DEAD-DEAH-DEIH box motif, although none of them have been demonstrated to have RNA helicase activity (43, 60). None of these proteins has been shown to be involved in the step of U4 dissociation during spliceosome assembly.
Besides base-pairing-mediated RNA-RNA interactions, protein-protein interactions may also play important roles in the splicing reaction. Interactions between yeast splicing factors Prp9p and Spp91p and between Spp91p and Prp11p have been demonstrated with the yeast two-hybrid system (30, 31). These three proteins together form a protein complex equivalent to the mammalian splicing factor SF3a (30), which is part of the 17S U2 snRNP (5, 6, 10, 29). A recent demonstration of interaction between Prp2p and Spp2p, also by the two-hybrid system, further attests to the important role of protein-protein interaction in splicing (41).
The yeast Prp19p protein is a spliceosomal component but is not tightly associated with snRNPs (13, 52). It has previously been shown that Prp19p becomes associated with the spliceosome concomitantly with or just after dissociation of U4 from the spliceosome (52, 53). Thus, Prp19p may play an important role in mediating the conformational rearrangement of the spliceosome during this transition. Attempts to purify the Prp19p protein led to the discovery that Prp19p is associated with a large protein complex consisting of at least seven other proteins in addition to Prp19p. Although it is not known whether all of the proteins in the complex are essential splicing factors, at least one of them in addition to Prp19p is required for splicing (51).
Examination of protein-protein interactions by far-Western blotting reveals that Prp19p can directly interact with three other proteins in the complex, namely, Ntc85p, Ntc40p, and Ntc25p (Ntc stands for PRP19 complex [51]), of which Ntc25p shows the strongest interaction. Prp19p can also interact with itself in far-Western blots and appears to form a homo-oligomer, possibly a tetramer, in solution (51). Deletion mutants incapable of forming oligomers are also unable to interact with the above-mentioned proteins in far-Western blots, suggesting a link between oligomerization of Prp19p and its interaction with other proteins.
In order to identify components in the Prp19p-associated complex, we have taken a genetic approach to isolate synthetic lethal mutants to prp19 mutations. Using the ade2-ade3 red-white colony sectoring system (7, 22), we have isolated several such mutants. Here, we report the identification of a novel splicing factor, Snt309p (Snt represents synthetic lethal to prp19 mutation), that can directly interact with Prp19p and is a component of the Prp19p-associated complex. We also show that, like Prp19p, Snt309p is associated with the spliceosome concurrently with or just after dissociation of U4 during spliceosome assembly.
MATERIALS AND METHODS
Yeast strains.
The following yeast strains were used: HR112 [MATa ade2 ade3 leu2 ura3 his3 trp1 PRP19::LEU2(pPRP19-ADE3-URA, pprp19-1-pRS413)], HR412 [(MATa ade2 ade3 leu2 ura3 his3 trp1 PRP19::LEU2(pPRP19-ADE3-URA, pprp19-4-pRS413)], SEY6210 (MATα leu2 ura3 his3 trp1 lys2 suc2), SEY6210.5 (MATa/MATα leu2/leu2 ura3/ura3 his3/his3 trp1/trp1 suc2/suc2 lys2/LYS2 ADE2/ade2), CH1462 (MATα ade2 ade3 leu2 ura3 his3 can1r), BJ2168 (MATa prc1 prb1 pep4 leu2 trp1 ura3), and HR3091 (MATα leu2 ura3 his3 trp1 lys2 suc2 SNT309::LEU2).
Oligonucleotides.
The following oligonucleotides were used: N3, AGGATTTGGGTAAATCCTTC; N39, CACTATTGAGAACTACGGTTGCACT; N74, CATTTCTTTACACAATCAAGCGTAGTCTGGGACGTCGTATGGGTAA TTCCAGAGCTTGAT; N97, GGCCGGATCCGATTTGATGGACGGCC; N98, GGCCGGATCCGTTGGTAACTCCCA; and N117, TCCATTGCGACATTAAGCGTAGTCTGGGACGTCGTATGGGTATAACCTCTTTCTGTA.
Plasmids.
The plasmids used for the genetic screen and their relevant characteristics are as follows: pPRP19-YEp, 3.6-kb fragment of the PRP19 gene in YEp24; pPRP19-ADE3-URA3, 5.0-kb SalI-BamHI fragment of the ADE3 gene from pB166 cloned into the BamHI site of pPRP19-YEp; pPRP19-pRS414, 3.6-kb fragment of the PRP19 gene in pRS414; pprp19-pRS414, 3.6-kb fragment of the prp19 mutant allele in pRS414; pprp19-pRS413, 3.6-kb fragment of the prp19 mutant allele in pRS413; pPRP19-pRS424, 3.6-kb fragment of the PRP19 gene in pRS424; and pPRP19-ADE3-TRP1, 5.0-kb SalI-BamHI fragment of the ADE3 gene from pB166 cloned into the BamHI site of pPRP19-pRS424. The plasmid used for epitope tagging was pHR95 (4.5-kb SalI-ClaI fragment of SNT309 in pRS416). The plasmids used for disruption of the SNT309 gene and their descriptions are as follows: pHR9651, 3-kb ClaI-KpnI fragment of SNT309 in pBluescript; and pHR9652, 2-kb LEU2 fragment cloned into the AccI site of pHR9651.
Plasmid SNT309-pGEM1 was constructed as follows. An 824-bp fragment corresponding to positions −6 to 818 relative to the SNT309 open reading frame (ORF) with BamHI sites at both ends was generated by PCR using oligonucleotides N97 and N98 and cloned into the BamHI site of pGEM-1 in such an orientation that the SP6 promoter could be used for in vitro transcription of the SNT309 gene.
Plasmids were constructed as follows for expression in Escherichia coli. An EcoRI-SalI fragment containing the SNT309 ORF was excised from plasmid SNT309-pGEM1 and cloned into the EcoRI-SalI site of plasmid pET15b(DH4) (a gift from D.-H. Huang). The resulting plasmid was digested with HindIII, and then the ends were repaired with Klenow polymerase. After digestion with NdeI, the SNT309-containing fragment was subcloned into pAR2156 (a gift from C. Wang) and pET15b for expression of Snt309p and His-tagged Snt309p by using T7 RNA polymerase.
Mutagenesis and genetic screen.
Strains HR112 and HR412 were grown in liquid media to saturation. After being washed with water, the cells were resuspended in water at 107 per ml, transferred to petri dishes, and UV irradiated for 2 to 3 min (UVP Inc. transilluminator, model TM-36, 306 nm). This treatment gave 10 to 20% survival with these strains. The cells were plated on yeast extract-peptone-dextrose (YPD) plates and incubated at 25°C. Nonsectoring red colonies were picked and streaked on His− plates to examine their histidine auxotrophy. His+ cells were further streaked on YPD plates three times to ensure their nonsectoring phenotype. The selected nonsectoring His+ mutants were transformed with plasmid pRS414, pPRP19-pRS414, or pprp19-pRS414 (prp19-1 or prp19-4, depending on the parent each mutant was isolated from), and transformants were streaked on YPD plates to examine their sectoring phenotype for grouping.
Cloning of the SNT309 gene.
The SNT309 gene was isolated by complementation of the synthetic lethal phenotype of the snt309 mutant with a YCp50-based Sau3A genomic library obtained from M. Rose and P. Novick. The pPRP19-ADE3-URA3 plasmid in the original snt309 mutant (snt309 URA3) was replaced with plasmid pPRP19-ADE3-TRP1 by plasmid shuffling (48), and the resulting snt309 TRP1 strain was used for cloning of the gene. After transformation of the library plasmids, colonies sectored on YPD plates were selected and restreaked on YPD plates. White colonies were selected and examined for their temperature sensitivity to distinguish between the PRP19 and SNT309 genes. Plasmids were isolated from temperature-sensitive white colonies and transformed into E. coli. Plasmids isolated from E. coli transformants were analyzed by PCR with oligonucleotides N3 and N39 and restriction mapping to distinguish between plasmid pprp19-HIS3 and the library plasmid. Subsequent subcloning was carried out with plasmid pRS416. DNA sequencing was performed by the deletion method (19) with the Sequenase kit (U.S. Biochemical Corp.).
Gene replacement.
The snt309::LEU2 allele was created by insertion of a 2-kb DNA fragment containing the LEU2 gene into the AccI site of plasmid pHR9651 within the SNT309 ORF. The resulting plasmid, pHR9652, was digested with PvuII prior to transformation into diploid strain SEY6210.5. Correct integration was confirmed by Southern blot analysis. Haploid strains harboring disrupted SNT309 genes were isolated by sporulation of the heterozygous diploid strain followed by dissection of tetrads.
Northern blot analysis.
Isolation of total yeast RNA and Northern blot analysis were done as described by Vijayraghavan et al. (55). A 210-bp XhoI-ClaI DNA fragment was used to prepare the actin intron probe by random primer labeling. A 250-bp EcoRI-HindIII DNA fragment from plasmid pActΔIVS was used to prepare the message probe.
In vitro transcription, translation, and far-Western blot analysis.
Plasmid SNT309-pGEM1 was linearized at the EcoRI site and used for in vitro transcription with SP6 RNA polymerase. In vitro transcription and translation reactions were carried out as described by Tarn et al. (51). Far-Western blot analysis was performed as described by Tarn et al. (51) except that 2 × 105 to 4 × 105 cpm of 35S-labeled Prp19p or Snt309p per ml was used in the incubation with blots.
Construction of the epitope-tagged strains.
Single-stranded DNA was isolated from plasmid pHR95 or pFL.PRP4 (3) for oligonucleotide-directed insertion. Oligonucleotides N117 and N74 were used to insert 27 nucleotides immediately behind the 3′ end of the SNT309 and PRP4 coding regions, respectively. The genes with the insertion were recloned into yeast integrative plasmid pRS406. The recombinant plasmids were used to displace the wild-type genes of S. cerevisiae BJ2168 with the epitope-tagged counterpart as described by Winston et al. (58).
Splicing reactions, immunoprecipitation, and complementation assays.
Splicing assays were performed as described by Lin et al. (34) with uncapped actin pre-mRNA used as the substrate. Immunoprecipitation was carried out as described by Cheng et al. (13) for anti-Prp19p antibody and by Tarn et al. for antihemagglutinin (anti-HA) antibody (52). Complementation of the Prp19p-immunodepleted extract was performed as described by Tarn et al. (52).
Expression and purification of Snt309p from E. coli.
Snt309p and His-tagged Snt309p (His-Snt309p) were expressed in E. coli under the control of the T7 promoter. For production of antibodies, total lysates prepared from induced cells were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the Snt309p protein was eluted from the gel for immunization of rabbits. For functional assays, the His-Snt309p protein was purified through a His-Bind column (Novagen) under denaturing conditions according to the manufacturer’s manual.
Nucleotide sequence accession number.
The nucleotide sequence of the YPR101w ORF encoding 175 amino acids has been submitted to GenBank under accession no. U32445.
RESULTS
A synthetic lethal approach to isolate components interacting with Prp19p.
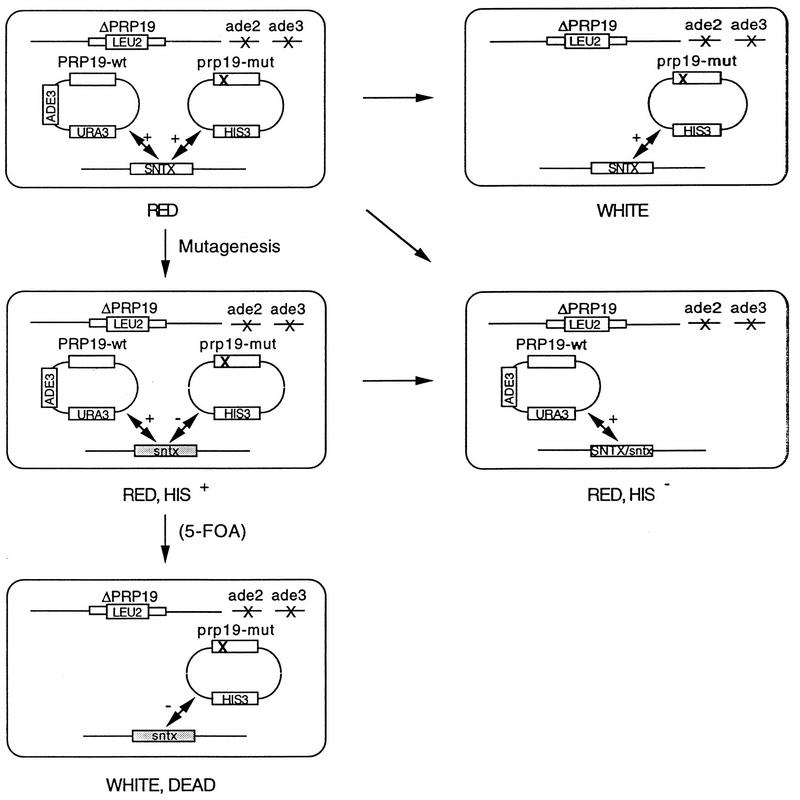
We used the ade2-ade3 red-white sectoring system (7, 22) to screen for mutants synthetic lethal to prp19 mutants in order to identify components in the Prp19p-associated complex. In the screening scheme as shown in Fig. 1, a large part of the chromosomal PRP19 gene was deleted and replaced with a DNA fragment containing the LEU2 gene. The essential PRP19 function was provided by two plasmids, one containing the wild-type PRP19 and ADE3 genes and the URA3 marker (plasmid pPRP19-ADE3-URA3) and the other containing the mutant prp19 gene and the HIS3 marker (plasmid pprp19-pRS413). Two prp19 temperature-sensitive alleles were used for screening. The prp19-1 mutant contains a point mutation changing valine to isoleucine at the 14th amino acid residue, whereas the prp19-4 mutant protein has a deletion of 20 amino acid residues from the C terminus (unpublished results).
FIG. 1.
Scheme for screen of mutants synthetic lethal to prp19 temperature-sensitive mutants.
The parent strains HR112 and HR412, which carry prp19-1 and prp19-4 alleles, respectively, were mutagenized by UV and then grown at 25°C on YPD plates. Nonsectoring red colonies were selected and examined for histidine auxotrophy. Since PRP19 is an essential gene, cells which lose the prp19 mutant-containing HIS3 plasmid must keep the wild-type PRP19-containing ADE3-URA3 plasmid and give nonsectoring phenotypes. Therefore, cells of histidine auxotrophy were excluded. Those exhibiting His+ phenotypes were further streaked on YPD plates to ensure their nonsectoring phenotype. Using such a scheme, we isolated 81 nonsectoring His+ mutants after screening 93,000 mutagenized colonies.
To examine whether nonsectoring phenotypes depend on the presence of prp19 mutations, these mutants were transformed by use of a plasmid with a TRP1 marker carrying either wild-type PRP19 (pPRP19-pRS414), the mutant prp19 (pprp19-pRS414), or no PRP19 gene (pRS414), and the transformants were examined for sectoring phenotype on YPD plates. By this method, these mutants were classified into three groups. Among them, 50 mutants, class I mutants, exhibited nonsectoring phenotypes regardless of the type of the plasmid used for transformation. Class II mutants (a total of 16) sectored when transformed with plasmids carrying either the wild-type PRP19 or the mutant prp19 gene but not with the vector alone, whereas class III mutants (a total of 15) sectored only when transformed with the plasmid carrying the wild-type PRP19 gene. All 15 class III mutants came from the parent strain HR412 carrying the prp19-4 allele.
The class III mutants were further examined by being streaked on 5-fluoro-orotic acid (5-FOA) plates to ensure their synergistic lethal phenotype (Fig. 1). Of 15 mutants, 9 did not grow on 5-FOA plates (data not shown), indicating an exclusive need for the presence of the wild-type PRP19 gene for growth.
Cloning of the SNT309 gene.
The wild-type allele of the gene conferring synthetic lethality in strain snt309 was cloned by complementation of the nonsectoring phenotype. The snt309 mutant was backcrossed with strain CH1462, and the daughter was used for cloning of the gene. An ORF encoding 175 amino acid residues (YPR101w) that complemented the snt309 mutation was identified. The nucleotide and translated amino acid sequences are shown in Fig. 2. The putative Snt309p protein has no homology to sequences in the database, nor any discernible motif.
FIG. 2.

Nucleotide and encoded protein sequences of the SNT309 gene. The gene encodes a protein of 175 amino acid residues. No identifiable motif is found in the protein sequence. Neither the nucleotide nor the protein sequence has homology to an existing sequence in the database.
Interaction of Snt309p with Prp19p.
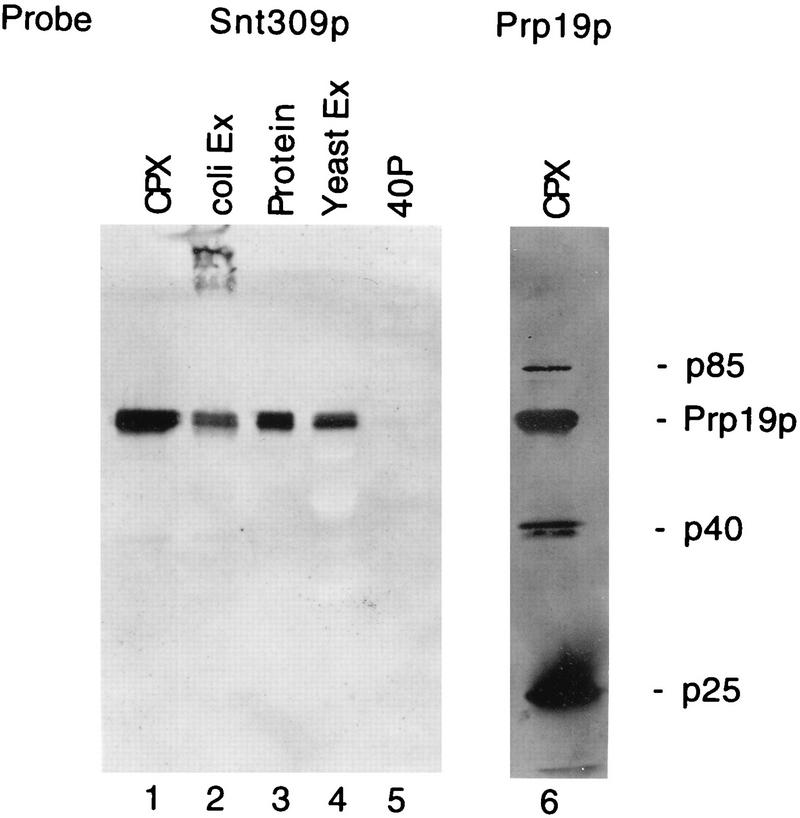
Since synthetic lethality may provide genetic evidence that two proteins physically interact with each other or have overlapping functions (7, 22), we first examined direct interaction between Snt309p and Prp19p by far-Western blot analysis. The SNT309 gene was cloned into transcription vector pGEM-1 for in vitro transcription and translation. The Prp19p proteins from various sources were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transblotted to polyvinylidene difluoride membranes for far-Western blot analysis using the in vitro-translated 35S-labeled Snt309p as a probe. As shown in Fig. 3, Snt309p indeed interacted with Prp19p isolated as a protein complex by affinity chromatography (lane 1), Prp19p expressed in E. coli (lane 2), Prp19p purified from the yeast overproducer strain (lane 3), and overproduced Prp19p in yeast whole-cell extract (lane 4). Interaction was not detected in the 40% ammonium sulfate pellet fraction of the whole-cell extract (40P), which was estimated to contain 5 ng of Prp19p (lane 5). Each of the other lanes contained approximately 250 ng of Prp19p, and about 100 μg of total proteins was in each of lanes 2, 4, and 5. No other proteins in crude extracts or 40P, when 100 μg of total proteins was loaded, seemed to significantly interact with Snt309p (Fig. 3, lanes 4 and 5). Figure 3, lane 6, shows a blot of the Prp19p-associated complex probed with the 35S-labeled Prp19p protein. Three proteins in the Prp19p-associated complex in addition to Prp19p itself could directly interact with Prp19p (51). In contrast, no other protein in the complex was found to interact with Snt309p (Fig. 3, lane 1).
FIG. 3.
Snt309p can interact with Prp19p in far-Western blots. Prp19p isolated as a protein complex (lane 1), expressed in E. coli (lane 2), as a purified protein (lane 3), in yeast extract from an overproducing strain (lane 4), and in a 40P fraction from a regular strain (lane 5) probed with in vitro-translated, 35S-labeled Snt309p protein in far-Western blots and the Prp19p-associated complex probed with labeled Prp19p (lane 6) are shown. Lanes 2, 4, and 5 each contain approximately 100 μg of total proteins. CPX, complex; Ex, extract.
Disruption of the SNT309 gene caused a temperature-sensitive phenotype.
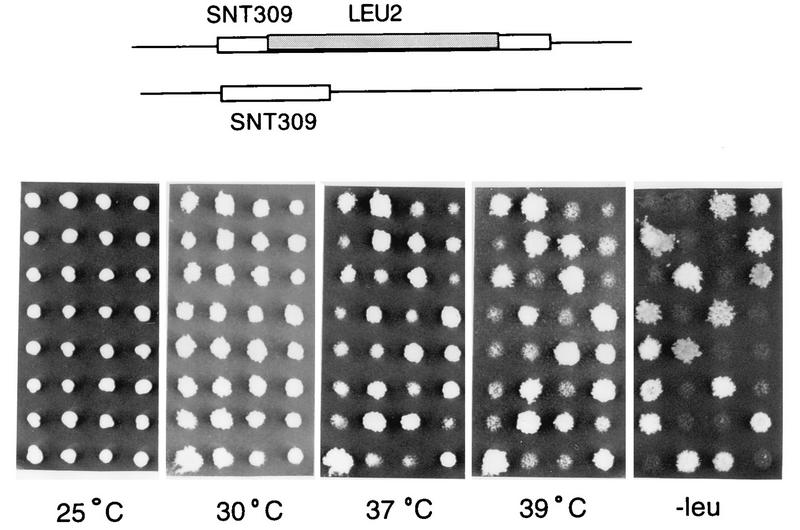
To see whether SNT309 is essential for growth, a null allele of SNT309 was constructed by inserting a fragment of the LEU2 gene near the 5′ end of the SNT309 gene. Dissection of diploid strains containing one copy of wild-type SNT309 and one copy of the null allele of SNT309 yielded four viable tetrads with 2:2 segregation for leucine auxotrophy as shown in Fig. 4, indicating that the SNT309 gene is not essential for growth. However, in contrast to the wild type (Leu− cells in tetrads), the SNT309-disrupted cells (Leu+ cells in tetrads) grew well at 30°C but grew poorly at 37°C and did not grow at 39°C. The same temperature-sensitive phenotype was observed when the entire ORF of SNT309 was removed (data not shown).
FIG. 4.
Disruption of the SNT309 gene results in a temperature-sensitive phenotype. A diploid strain carrying one copy of the wild-type and one copy of the LEU2-disrupted SNT309 gene was sporulated for tetrad analysis. Shown are nine tetrads after transfer to a fresh YPD plate and growth at 25°C. They were then replica plated to test for leucine auxotrophy and growth phenotype at various temperatures.
Snt309p is a novel splicing factor.
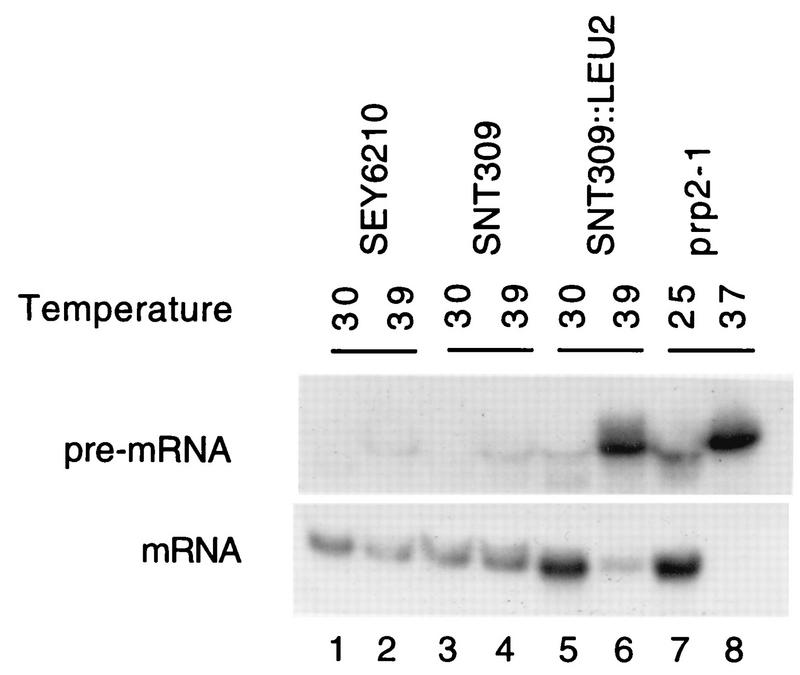
To see whether Snt309p is involved in pre-mRNA splicing, we examined whether the SNT309-disrupted yeast strain is defective in splicing at the nonpermissive temperature. The SNT309-disrupted yeast cells were grown at 30°C until mid-log phase and then shifted to 39°C for 2 h. RNA was isolated for Northern blot analysis using the actin intron or the mature message as a probe. As a control, RNA was isolated from the prp2-1 mutant strain grown at 25°C prior to being shifted to 37°C for 2 h. Figure 5 shows that precursor actin RNA accumulated in large amounts in the SNT309-disrupted strain after being shifted to 39°C (Fig. 5, lane 6), as in the case of the prp2-1 mutant after being shifted to 37°C (lane 8) (55), whereas only small amounts of precursor accumulated when cells were grown at the permissive temperatures (lanes 5 and 7). Wild-type strains and the SEY6210 parent (Fig. 5, lanes 1 and 2) and Leu− spores isolated from tetrad dissection (lanes 3 and 4) also accumulated only small amounts of precursor RNA at 39°C (lanes 2 and 4). Consistently, very small amounts of the mature message were detected in the SNT309-disrupted strain after the shift to 39°C. This indicates that pre-mRNA splicing is blocked at higher temperatures in the absence of Snt309p, although it is not clear whether splicing is blocked at the first or the second step of the reaction since the actin precursor and the splicing intermediate, lariat intron-exon 2, could not be resolved in this gel system. Thus, although Snt309p is not essential for yeast growth under normal conditions, it may play a subsidiary role in pre-mRNA splicing and may represent a novel splicing factor.
FIG. 5.
The SNT309-disrupted yeast strain accumulates pre-mRNA in vivo. The SEY6210, SNT309-nondisrupted, and SNT309-disrupted strains isolated from the same tetrad were grown at 30°C until mid-log phase and then shifted to 39°C and grown for 2 h. RNA was extracted from cells grown at both 30 and 39°C and hybridized with an actin intron probe to reveal the pre-mRNA or with an actin message probe. The prp2-1 mutant was grown at 25°C and then shifted to 37°C. Each lane contained 10 μg of total RNA.
Snt309p is a spliceosomal component.
Although the SNT309-disrupted cells were defective in splicing in vivo at higher temperatures, it is possible that this defect is not a deficiency in splicing efficiency per se, but rather is an indirect effect along with other biological functions. Thus, it is necessary to examine the function of Snt309p in splicing in vitro. Since Prp19p is tightly associated with the spliceosome during the splicing reaction, we examined whether Snt309p is also a spliceosomal component.
The Snt309p protein was first tagged with the HA epitope at its carboxy terminus so that the protein could be monitored with the anti-HA antibody. Splicing reactions were carried out with extracts prepared from the epitope-tagged strain, and the reaction mixtures were subjected to immunoprecipitation with the anti-HA antibody. As shown in Fig. 6, precursor RNA, splicing intermediates, and IVS, but only a small amount of the mature message, were precipitated from the reaction carried out in the Snt309p HA-tagged extract (lane 7). In the absence of the antibody, no RNA was precipitated (Fig. 6, lane 6). Preincubation of the antibody with excessive amounts of the HA peptide prevented precipitation of the RNA by the antibody (Fig. 6, lane 8), indicating that precipitation was specific to the HA epitope. Furthermore, no RNA was precipitated by the antibody when extracts were prepared from a nontagged strain (Fig. 6, lane 3). These results indicate that Snt309p is a spliceosomal component. We therefore conclude that Snt309p is directly involved in the splicing reaction and is a novel spliceosomal component.
FIG. 6.
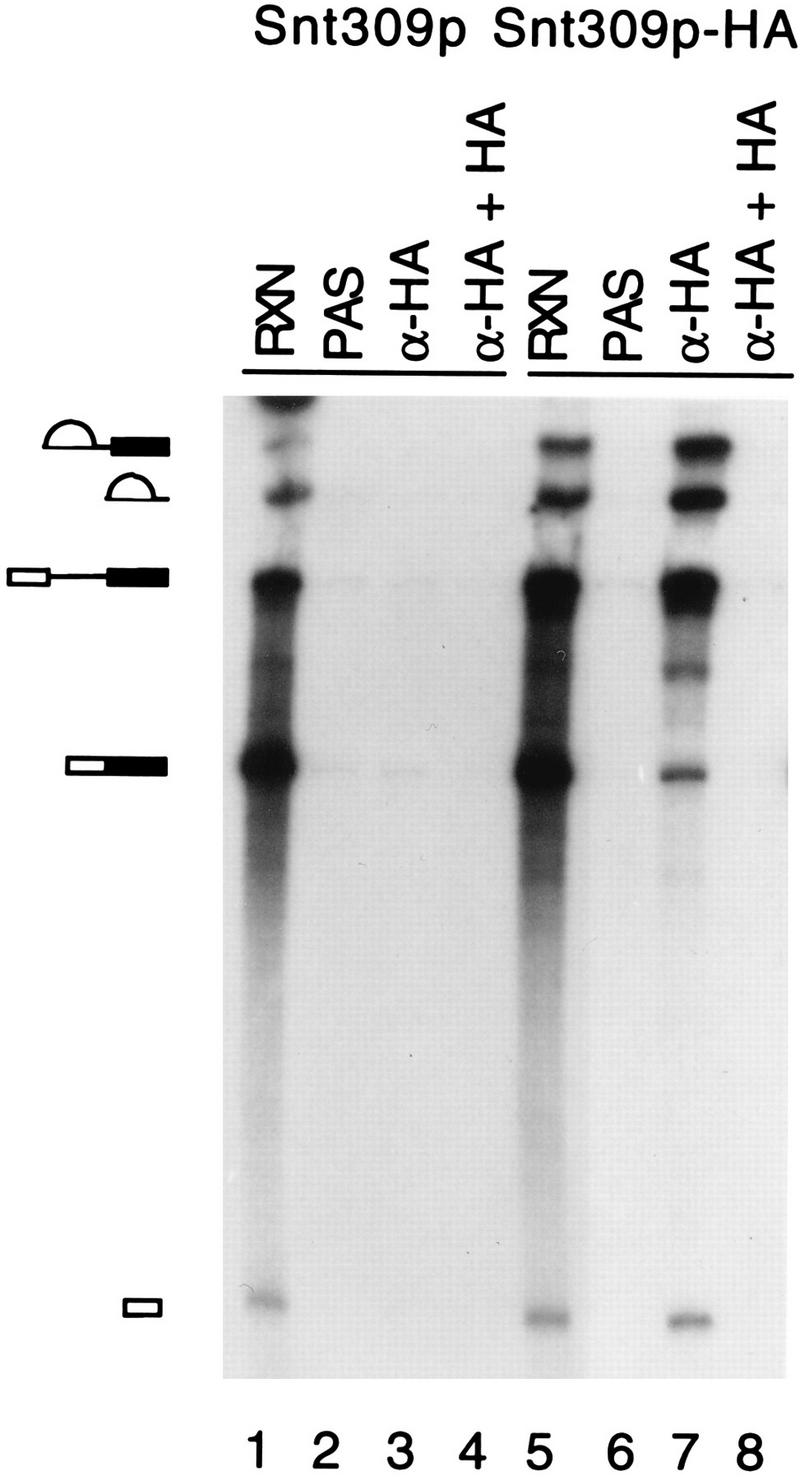
The Snt309p protein is associated with the spliceosome during the splicing reaction. Mixtures of the splicing reactions (20 μl) carried out in the extract prepared from the wild-type and Snt309p HA-tagged strains (lanes 1 and 5) were precipitated with the anti-HA antibody (lanes 3 and 7) or without the antibody (lanes 2 and 6). The antibody was also preincubated with excessive amounts of the HA peptide before immunoprecipitation (lanes 4 and 8). The pre-mRNA, E1, IVS, and IVS-E2, but only very small amounts of the mature message, were precipitated in the HA-tagged extract. Lanes 1 and 5 contain 2 μl of the splicing reaction mixtures. Diagrams on the left correspond to the structures in Fig. 7A. α-HA, anti-HA antibody; RXN, 1 to 2 μl of the splicing reaction mixture; PAS, protein A-Sepharose.
Snt309p is associated with the spliceosome concomitantly with or immediately after dissociation of U4.
It has been shown previously that Prp19p is associated with the spliceosome immediately after or concurrently with dissociation of U4 (53). The scheme of the spliceosome assembly pathway is shown in Fig. 7A. Since Snt309p interacted strongly with Prp19p, we questioned whether Snt309p also binds to the spliceosome at the same step as Prp19p during spliceosome assembly. It has previously been demonstrated that dissociation of U4 is highly sensitive to ATP concentration (53). At low concentrations of ATP, U4 dissociation is blocked and the U4-containing splicing complex A2-1 accumulates in larger amounts. When the ATP concentration increases, transition from complex A2-1 to A1 is rapid and only a small amount of complex A2-1 accumulates. This can be revealed by immunoprecipitation of the spliceosome with the anti-Prp4p antibody (53) or the anti-HA antibody when Prp4p is tagged with the HA epitope (Fig. 7B), since Prp4p is tightly associated with U4 and therefore is present only in complex A2-1. In contrast, Prp19p was detected in splicing complexes at higher ATP concentrations and, to a much lesser extent, at lower ATP concentrations, as revealed by precipitation of the same reaction mixtures with the anti-Prp19p antibody. To examine the step at which Snt309p binds to the spliceosome during assembly, the same ATP titration experiments were performed with extracts prepared from the Snt309p HA-tagged strain. Immunoprecipitation of splicing reaction mixtures with anti-HA and anti-Prp19p antibodies revealed that precipitation by the anti-HA antibody has a pattern similar to that of the anti-Prp19p antibody in the course of ATP titration (Fig. 7C), indicating that, like Prp19p, Snt309p binds to the spliceosome concomitantly with or immediately after dissociation of U4.
FIG. 7.
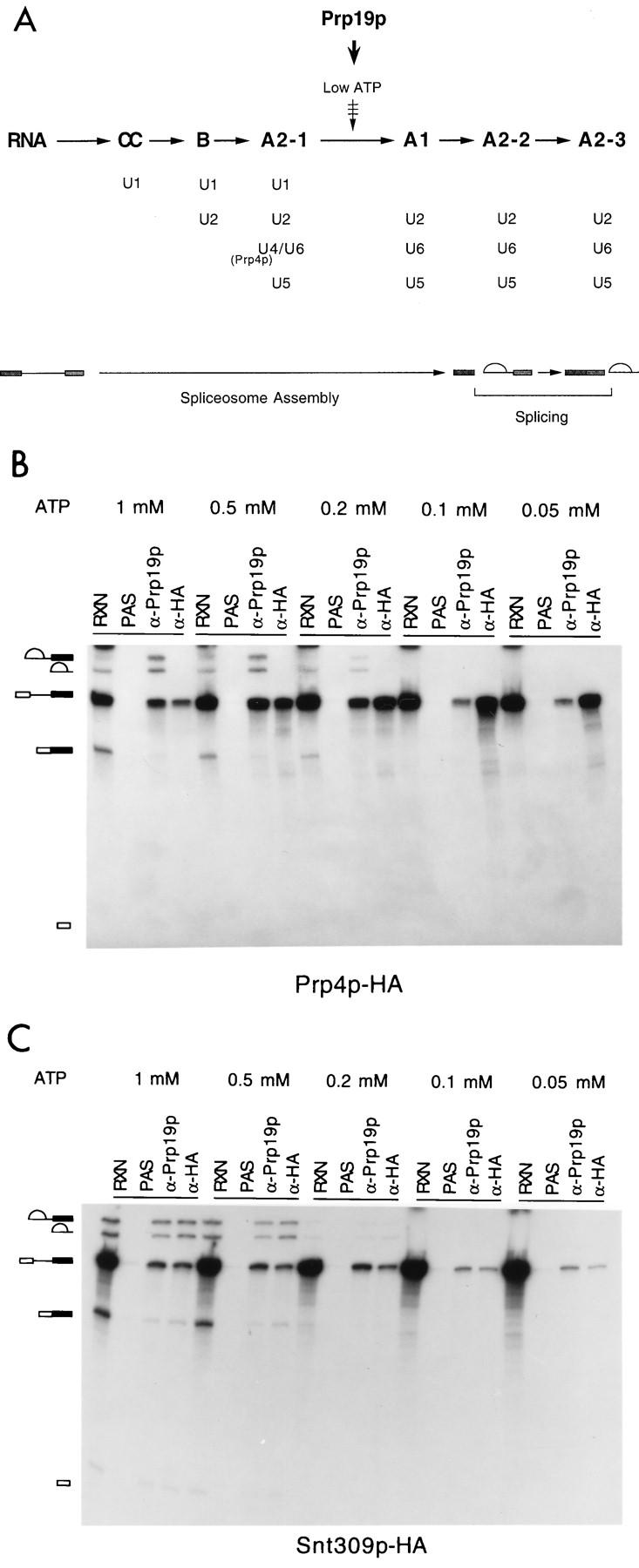
Snt309p is associated with the spliceosome in the same manner as Prp19p. (A) A scheme for spliceosome assembly showing that Prp19p becomes associated with the spliceosome during transition from complex A2-1 to A1, which can be blocked at lower ATP concentrations. Also indicated is Prp4p, which is tightly associated with the U4 snRNP and is present only in complex A2-1 during spliceosome assembly. (B and C) Splicing reactions (20-μl mixtures) carried out at various ATP concentrations in Prp4p HA-tagged or Snt309p HA-tagged extracts, respectively, were precipitated with anti-Prp19p antibody and anti-HA antibody. RXN, 1 to 2 μl of the splicing reaction mixture; PAS, protein A-Sepharose; α-Prp19p, anti-Prp19p antibody; α-HA, anti-HA antibody. Symbols: □, 5′-exon; ▪, 3′-exon; , lariot intron.
Snt309p is a component of the Prp19p-associated complex.
Our results that the Snt309p protein could directly interact with Prp19p and associated with the spliceosome at the same time as Prp19p suggest that Snt309p might be a component of the Prp19p-associated complex. The Prp19p-associated complex contains a protein of 25 kDa which interacts with Prp19p strongly, as analyzed by far-Western blotting (51). Since the in vitro-translated Snt309p migrated as a 25-kDa protein (data not shown), Snt309p is likely the Ntc25p of the complex.
To see whether Snt309p is indeed a component of the Prp19p-associated complex, we raised antibodies against the Snt309p protein for immunoblot analysis. The SNT309 gene was placed under the control of the bacterial T7 promoter, and the recombinant protein, which also has an apparent molecular weight of 25,000, was used for production of antibodies. The Prp19p-associated complex was isolated by affinity chromatography (51), fractionated by SDS-PAGE, and subjected to Western blot analysis using the anti-Snt309p antibody. Figure 8A shows that the antibody reacted with a protein of 25 kDa (lane 3), whereas the preimmune serum did not react with any protein in the complex (lane 2). Preincubation of the antibody with an excess amount of the recombinant His-tagged Snt309p protein significantly blocked the reaction of the antibody with the 25-kDa protein (Fig. 8A, lanes 4 to 6). This result shows that Snt309p is associated with Prp19p upon affinity purification of the Prp19p-associated complex.
FIG. 8.
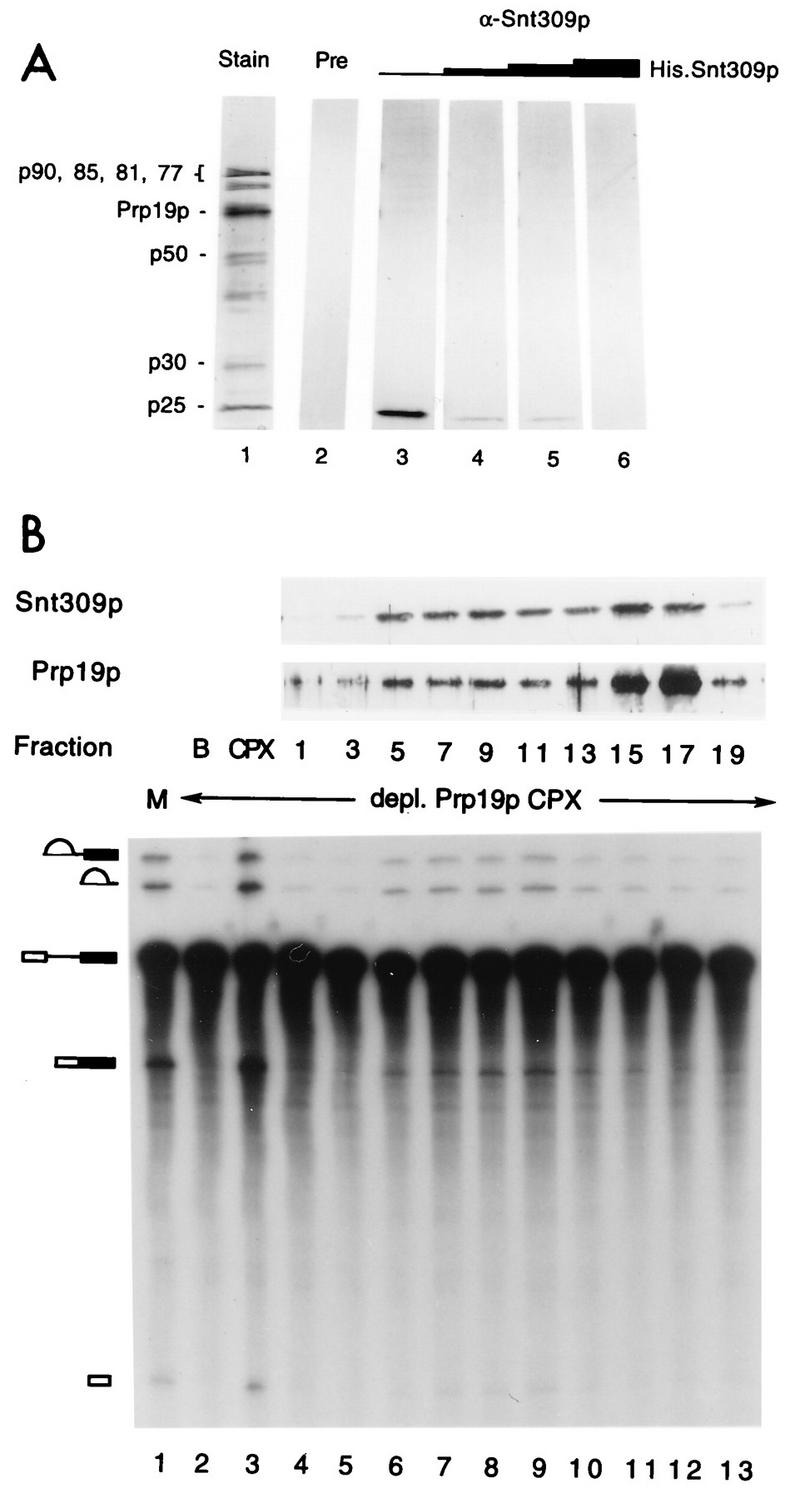
Immunoblot analysis showing that Snt309p is a component of the Prp19p-associated complex. (A) The Prp19p-associated complex was analyzed for the presence of Snt309p by immunoblot analysis using anti-Snt309p antibody (lanes 3 to 6) or preimmune serum (lane 2). The antibody was preincubated with increasing amounts of recombinant Snt309p to block the reaction (lanes 4 to 6). Lane 1, components of the Prp19p-associated complex revealed by silver staining. (B) Immunoblot analysis using anti-Snt309p and anti-Prp19p antibodies and complementation of the Prp19p-depleted extract of gradient fractions. Lane M, mock-treated extract; CPX, complementation with the Prp19p-associated complex.
It has previously been demonstrated that the Prp19p-associated complex, which consists of at least eight proteins, sediments as a large complex on glycerol gradients (51). To show that Snt309p is a component of the large complex rather than in a separate complex, the isolated Prp19p-associated complex was sedimented on a 10 to 30% glycerol gradient, and the gradient fractions were examined for Snt309p by immunoblot analysis. Figure 8B shows that a fraction of the Snt309p protein cosedimented in fractions 5 to 10 with Prp19p and the complementation activity of the Prp19p-immunodepleted extract. The rest of the protein sedimented in fractions 15 to 17, which were inactive for complementation, near the top of the gradient. As fractions 5 to 10 represented the intact Prp19p-associated complex (51), Snt309p was a component of this complex. It is believed that fractions 15 to 17 represent incomplete Prp19p-associated complexes which contain only Prp19p and Snt309p and perhaps together with some other components tightly associated with Prp19p or Snt309p.
DISCUSSION
In our screen for synthetic lethal mutants to prp19 mutations, we used two types of prp19 mutants. The prp19-1 mutant protein carries a point mutation at the N-terminal region, whereas the prp19-4 mutant protein contains a deletion at the C terminus. All 15 mutants isolated in this screen were from the strain carrying the prp19-4 mutation; none were from the strain with the prp19-1 allele. Moreover, none of these mutants caused synthetic lethality to the prp19-1 mutation (data not shown). Since the prp19-1 mutation has less severe effects than the prp19-4 mutation, as judged from the growth and sectoring patterns of the mutants (data not shown), it is possible that the prp19-1 mutation is not severe enough to cause synthetic lethality in combination with mutations in other genes encoding interacting proteins. Alternatively, mutation near the N terminus of the protein, as in the case of the prp19-1 protein, may not seriously affect interaction of Prp19p with other components. The latter possibility is supported by the previous observation from far-Western blot analysis that deletion of 68 amino acid residues from the N terminus of Prp19p does not strongly affect the ability of Prp19p to interact with proteins in the Prp19p-associated complex (51).
Synthetic lethality provides genetic evidence that two proteins may physically interact with each other or have overlapping functions. Indeed, interaction between Prp19p and Snt309p can be demonstrated by far-Western blot analysis. This interaction can also be detected in vivo with the two-hybrid system (data not shown). In the Prp19p-associated complex, at least three proteins can directly interact with Prp19p, as observed in far-Western blots (Fig. 3) (51). These include a protein of 25 kDa which interacts with Prp19p most strongly among the three proteins. The facts that the anti-Snt309p antibody detected a protein of 25 kDa in the Prp19p-associated complex and that both the in vitro-translated and the E. coli-expressed Snt309p proteins had an apparent molecular weight of 25,000 (data not shown) suggest that Snt309p is the 25-kDa protein of the Prp19p-associated complex but do not exclude the possibility that Ntc25p is composed of Snt309p and some other components. Although no 25-kDa protein was detected by isolation of the Prp19p-associated complex from ΔSNT309 extracts, the amounts of other associated components were greatly diminished as well (unpublished results), suggesting that the Prp19p-associated complex might be unstable in the absence of Snt309p.
Disruption of the SNT309 gene did not affect the growth of cells under normal conditions but resulted in growth arrest at higher temperatures. At the nonpermissive temperature, these cells accumulated large amounts of unspliced precursor mRNAs, indicative of splicing deficiency. Although such in vivo analysis does not demonstrate a direct role in splicing, our immunoprecipitation results, which showed that Snt309p is associated with the spliceosome during the splicing reaction, provide a clear demonstration of its direct involvement in pre-mRNA splicing. We therefore conclude that Snt309p is a splicing factor, which is required for the splicing reaction only at higher temperatures.
During spliceosome assembly, snRNAs bind to the spliceosome in the order U1, U2, and then U4-U6 plus U5 as a pre-formed tri-snRNP complex (18, 44, 49). After all five snRNAs are associated with the pre-mRNA, U4, and perhaps also U1, is dissociated from the spliceosome. This involves a large conformational rearrangement of the spliceosome to destabilize base-pairings between U4 and U6 and between U1 and the pre-mRNA. Factors mediating this conformational change remain to be identified. Previously, we demonstrated that Prp19p becomes associated with the spliceosome immediately after or concurrently with dissociation of U4, suggesting a possible role in modulating this conformational change. During characterization of the purified Prp19p protein, no biochemical activities, including ATPase, ATP binding, and RNA annealing activities, that are possibly associated with this conformational change were detected (47a). In consideration of Prp19p being associated with a large protein complex, it is conceivable that the function of Prp19p might be dependent on its association with other components. The Snt309p protein is a component of the Prp19p-associated complex. The fact that Snt309p binds to the spliceosome in the same manner as Prp19p suggests coordinate action of these two proteins. It remains of interest to determine whether all components of the Prp19p-associated complex bind to the spliceosome simultaneously. So far, Snt309p and Prp19p are the only proteins reported to be involved in this step of the spliceosome assembly process.
The Snt309p protein, although being a spliceosomal component, is not required for yeast growth under normal conditions. Considering the prevalence of gene duplications in the yeast S. cerevisiae (24, 59), the existence of a functional homolog of Snt309p remains possible. However, a search of the yeast genome database did not identify any sequence with significant homology to Snt309p. Thus, the fact that Snt309p is dispensable for cellular growth may not be attributed to functional redundancy.
In fact, many yeast splicing factors, including Prp17p, Prp18p, Mud1p, Mud2p, Mud13p, and Snp1p, have also been shown to be encoded by nonessential genes (2, 14, 20, 21, 23, 32). It is not clear why a large number of protein splicing factors are not essential for normal cellular growth and presumably not essential for splicing in vivo. Our in vitro analysis showed that the anti-Snt309p antibody failed to inhibit the splicing reaction (data not shown), indicating that Snt309p is also not required for the splicing reaction in vitro. Unlike Prp19p, which can interact directly with several proteins in the Prp19p-associated complex, Snt309p could interact directly with only Prp19p, as observed in far-Western blots (Fig. 3). This suggests a possible role for the Snt309p protein, which might be involved in modulating interactions of Prp19p with components in the Prp19p-associated complex or in the spliceosome. In fact, in vitro analysis of the Prp19p-associated complex isolated from ΔSNT309 extracts revealed that the complex is destabilized in the absence of Snt309p and could not function properly during the splicing reaction (unpublished results). This supports the role of Snt309p in modulating interactions of Prp19p with other components in the complex (unpublished data).
ACKNOWLEDGMENTS
We thank G. Fink and C. Holm for providing strains and plasmids for the ade2-ade3 sectoring system and M. F. Tam for synthesizing oligonucleotides and peptides. We also thank M.-Y. Cheng and W.-Y. Tarn for reading the manuscript.
This work was supported by a grant from Academia Sinica and by National Science Council grant NSC85-2311-B-001-033 to S.-C.C. and by a CNRS grant to J.B.
REFERENCES
- 1.Abovich N, Legrain P, Rosbash M. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9gene encodes a protein required for U2 snRNP binding. Mol Cell Biol. 1990;10:6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 3.Banroques J, Abelson J. PRP4: a protein of the yeast U4/U6 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1989;9:3710–3719. doi: 10.1128/mcb.9.9.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beggs J D. Yeast protein splicing factors involved in nuclear pre-mRNA splicing. Mol Biol Rep. 1993;18:99–103. doi: 10.1007/BF00986763. [DOI] [PubMed] [Google Scholar]
- 5.Behrens S-E, Galisson F, Legrain P, Lührmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci USA. 1993;90:8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens S-E, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender A, Pringle J R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bindereif A, Green M R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987;6:2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjørn S P, Soltyk A, Beggs J D, Friesen J D. PRP4 (RNA4) from Saccharomyces cerevisiae: its gene product is associated with the U4/U6 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1989;9:3698–3709. doi: 10.1128/mcb.9.9.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosi R, Gröning K, Behrens S-E, Lührmann R, Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 11.Burgess S M, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1392. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 12.Cheng S-C, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 13.Cheng S-C, Tarn W-Y, Tsao T Y, Abelson J. PRP19: a novel spliceosomal component. Mol Cell Biol. 1993;13:1876–1882. doi: 10.1128/mcb.13.3.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colot H V, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 15.Cooper M, Johnston L H, Beggs J D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott D J, Bowman D S, Abovich N, Fay F S, Rosbash M. A yeast splicing factor is localized in discrete subnuclear domains. EMBO J. 1992;11:3731–3736. doi: 10.1002/j.1460-2075.1992.tb05458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank D, Patterson B, Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol Cell Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S. Unidirectional digestion with exonuclease III creates targeted break point for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 20.Hilleren P J, Kao H-Y, Siliciano P G. The amino-terminal domain of yeast U1-70K is necessary and sufficient for function. Mol Cell Biol. 1995;15:6341–6350. doi: 10.1128/mcb.15.11.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horowitz D S, Abelson J. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2959–2970. doi: 10.1128/mcb.13.5.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huffaker T C, Hoyt M A, Botstein D. Genetic analysis of the yeast cytoskeleton. Annu Rev Genet. 1987;21:259–284. doi: 10.1146/annurev.ge.21.120187.001355. [DOI] [PubMed] [Google Scholar]
- 23.Jones M H, Frank D N, Guthrie C. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc Natl Acad Sci USA. 1995;92:9687–9691. doi: 10.1073/pnas.92.21.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaback D B. Yeast genome structure. In: Wheals A E, Rose A H, Harrison J S, editors. The yeasts. London, United Kingdom: Academic Press Ltd.; 1995. pp. 179–222. [Google Scholar]
- 25.Kao H-Y, Siliciano P G. The yeast homolog of the U1 snRNP protein 70K is encoded by the SNP1 gene. Nucleic Acids Res. 1992;20:4009–4013. doi: 10.1093/nar/20.15.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao H-Y, Siliciano P G. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S-H, Smith J, Claude A, Lin R-J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992;11:2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konarska M M, Sharp P A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 29.Krämer A, Utans U. Three protein factors (SF1, SF2 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legrain P, Chapon C. Interaction between PRP11 and SPP91 yeast splicing factors and characterization of a PRP9-PRP11-SPP91 complex. Science. 1993;262:108–110. doi: 10.1126/science.8211114. [DOI] [PubMed] [Google Scholar]
- 31.Legrain P, Chapon C, Galisson F. Interactions between PRP9 and SPP91 splicing factors identify a protein complex required in prespliceosome assembly. Genes Dev. 1993;7:1390–1399. doi: 10.1101/gad.7.7b.1390. [DOI] [PubMed] [Google Scholar]
- 32.Liao X C, Tang J, Rosbash M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- 33.Lin R-J, Lustig A J, Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987;1:7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- 34.Lin R-J, Newman A J, Cheng S-C, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 35.Lockhart S R, Rymond R C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lossky M, Anderson G J, Jackson S P, Beggs J. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell. 1987;51:1019–1026. doi: 10.1016/0092-8674(87)90588-5. [DOI] [PubMed] [Google Scholar]
- 37.Maddock J R, Weidenhammer E M, Adams C C, Lunz R L, Woolford J. Extragenic suppressors of Saccharomyces cerevisiae prp4 mutations identify a negative regulator of PRPgenes. Genetics. 1994;136:833–847. doi: 10.1093/genetics/136.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniatis T, Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987;325:673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- 39.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 40.Pikielny C W, Rymond B C, Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature. 1986;324:341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- 41.Roy J, Kim K, Maddock J R, Anthony J G, Woolford J L., Jr The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995;1:375–390. [PMC free article] [PubMed] [Google Scholar]
- 42.Ruby S W, Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991;7:79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- 43.Ruby S W, Chang T-H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 44.Rymond B C, Rosbash M. Yeast pre-mRNA splicing. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 45.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 46.Shannon K, Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 47.Sharp P A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 47a.Sheu, Y.-J., and S.-C. Cheng. Unpublished results.
- 48.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steitz J A, Black D L, Gerke V, Parker K A, Krämer A, Frendewey D, Keller W. Functions of the abundant U-snRNPs. In: Birnsteil M, editor. Structure and function of major and minor SNURPS. New York, N.Y: Springer-Verlag; 1988. pp. 115–154. [Google Scholar]
- 50.Tang J, Abovich N, Rosbash M. Identification and characterization of a yeast gene encoding the U2 small nuclear ribonucleoprotein particle B" protein. Mol Cell Biol. 1996;16:2787–2795. doi: 10.1128/mcb.16.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarn W-Y, Hsu C-H, Huang K-T, Chen H-R, Kao H-Y, Lee K-R, Cheng S-C. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarn W-Y, Lee K-R, Cheng S-C. The yeast PRP19 protein is not tightly associated with small nuclear RNAs but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol. 1993;13:1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarn W-Y, Lee K-R, Cheng S-C. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci USA. 1993;90:10821–10825. doi: 10.1073/pnas.90.22.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson R C. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem Sci. 1988;13:91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- 55.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 56.Wassarman D A, Steitz J A. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 57.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 58.Winston F, Chumley F, Fink G R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- 59.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 60.Xu D, Nouraini S, Field D, Tang S-J, Friesen J D. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 61.Yean S-L, Lin R-J. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol Cell Biol. 1991;11:5571–5577. doi: 10.1128/mcb.11.11.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]