Abstract
We previously reported that the secretory capacity of Pichia pastoris is limited with respect to the secretion of a 96.5-kDa bivalent anti-CD3 immunotoxin; double-copy expression generated more translation products than single-copy expression but did not increase the secretion of the immunotoxin. In Saccharomyces cerevisiae heterologous protein secretion has been reported to increase the expression of molecular chaperones, most prominently BiP/Kar2p. We therefore investigated the relationships between immunotoxin secretion and Kar2p expression in P. pastoris. We found that expression of the immunotoxin in P. pastoris increased the expression of Kar2p to levels that surpassed the retrieval capacity of the cell, leading to secretion of Kar2p into the medium. The level of Kar2p secretion was correlated with the copy number of the immunotoxin gene. Intracellular Kar2p was found to bind exclusively to the unprocessed immunotoxin containing the prosequence of α-factor in the endoplasmic reticulum. These results show that Kar2p is intimately involved in immunotoxin secretion in P. pastoris. The limited capacity of P. pastoris to retain a sufficiently high level of intracellular Kar2p may be a factor restricting the production of the immunotoxin.
The yeast Pichia pastoris has been developed into a highly successful system for the production of recombinant proteins having different origins (5). However, the efficiency of heterologous secretion can vary widely among the expressed proteins. Some eukaryotic proteins, such as human serum albumin and insulin, can be secreted at very high levels (grams per liter), whereas the secretion levels of many other proteins are significantly lower or even undetectable.
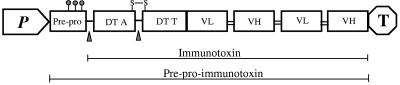
We have expressed a bivalent anti-CD3 immunotoxin in P. pastoris with a moderate secretion level (about 5 mg/liter in shake flask cultures and 37 mg/liter in fermentation cultures) (46, 47). Attempts to increase the production of this protein by using protease inhibitors and protease-deficient strains and by increasing the copy number of the gene were not successful (21). Interestingly, strains with two copies of the immunotoxin gene generated more translation product, but there was not an increase in the secretion of the intact protein, indicating that the expressing host has a limited capacity to fold and/or secret the immunotoxin. Structurally, this multidomain immunotoxin has several interesting features (Fig. 1). The N terminus consists of the diphtheria toxin (DT) catalytic domain (DT A), followed by the toxin translocation domain ending at DT residue 390, followed by two tandem scFv domains separated by a (G4S)3 linker (40). The junction between the catalytic and translocation domains contains an accessible Kex2 cleavage site spanned by a disulfide loop. To facilitate secretion, the toxin gene is preceded by the Saccharomyces cerevisiae α-mating factor preprosequence. Based on our understanding of the processing and secretion of α-factor in S. cerevisiae (37), we expect that after entering the endoplasmic reticulum (ER), the preproimmunotoxin undergoes initial processing to remove the presequence, followed by folding and N-linked glycosylation at the three potential sites in the prosequence region. Potential glycosylation sites of the immunotoxin have been removed (22). The proimmunotoxin could then be transported from the ER to the Golgi complex, where it might undergo further modification of oligosaccharide chains before it is finally processed to yield the immunotoxin by proteolytic cleavage at the Kex2 site introduced between the prosequence and the immunotoxin. Therefore, the molecular size of intracellular immunotoxin in both nonreducing and reducing gels should provide information on the transition of the immunotoxin from the early ER to the Golgi compartments.
FIG. 1.
Schematic representation of the immunotoxin expression cassette in P. pastoris strains. The immunotoxin expression cassette contains a promoter, the preprosequence of α-mating factor, the immunotoxin gene, and the transcription termination sequence for AOX1. The immunotoxin is a multidomain protein containing the DT catalytic domain (DT A), followed by the toxin translocation domain ending at DT residue 390, followed by four Fv domains (VL and VH) of an anti-CD3 antibody, UCHT1. The Fv domains are joined by a 15-amino-acid (G4S)3 linker. N-glycosylation and Kex2 cleavage sites are indicated by lollipop symbols and triangles, respectively. S-S, disulfide bridge.
In order to obtain a better understanding of the limitation of secretory capacity for the immunotoxin in P. pastoris, we decided to investigate the relationships between the major ER protein folding chaperone BiP/Kar2p and the immunotoxin during the immunotoxin transit through the ER and Golgi compartments. BiP was originally described as the immunoglobulin heavy chain binding protein (12) and the glucose-regulated protein, Grp78 (30). In the yeast S. cerevisiae, BiP is the product of a karyogamy gene, KAR2 (26, 31). BiP/Kar2p interacts transiently with stretches of folding protein units to prevent intra- and intermolecular interactions that could lead to permanent misfolding or aggregation. As an integral part of the ER quality control system, BiP/Kar2p binds more persistently to misfolded or unassembled proteins and prevents them from exiting the ER (10). BiP/Kar2p also plays an active role in the translation of precursor polypeptides across the ER membrane (23, 48) and in the ER-associated degradation of unfolded protein (15, 43).
Under normal growth conditions, BiP/Kar2p is constitutively expressed. Its synthesis can be further induced by various stress conditions, such as heat shock, inhibition of glycosylation by tunicamycin (26, 31), and expression of foreign or mutant proteins (16, 17, 41, 45). Under these stress conditions, unfolded proteins accumulate in the ER, and a complex signal pathway, called the unfolded protein response (UPR), is activated to reduce the accumulation of these proteins (24, 28, 35).
BiP/Kar2p and other ER resident soluble proteins, such as protein disulfide isomerase, have a COOH-terminal tetrapeptide retention signal that is usually KDEL in mammalian cells and HDEL in S. cerevisiae (29). ER retention of these proteins is achieved by receptor (Edr2p)-mediated retrieval (8, 19, 20, 34, 39). It has been shown that in S. cerevisiae the Erd2p-mediated retention can be saturated by overexpression of an HDEL-tagged pro-α-factor (6, 34) or Kar2p and by other conditions that activate the UPR (e.g., treatment with β-mercaptoethanol or tunicamycin [1], leading to extracellular secretion of Kar2p).
MATERIALS AND METHODS
P. pastoris strains.
We used DT-resistant P. pastoris strains that had a mutation in elongation factor 2 (21) for expression of the bivalent anti-CD3 immunotoxin (40). The immunotoxin expression cassette used in this study has been described previously (46) and is shown in Fig. 1. Briefly, it contains a promoter, the preprosequence of α-mating factor, the immunotoxin gene (40) that encodes the DT A chain and the DT translocation domain followed by two tandem scFv chains, and the transcription termination sequence for AOX1 (alcohol oxidase 1). Strains PAOX-1c and PAOX-2c were originally called mutEF2JC307-8(2) and mutEF2JC303-5(2c-1), respectively (21). These strains are elongation factor 2 mutants that express the immunotoxin under control of the AOX1 promoter (PAOX1) and contain one and two immunotoxin expression cassettes, respectively. In this study, we made two new strains that express the immunotoxin constitutively under control of the promoter of the glyceraldehyde-3-phosphate dehydrogenase gene (PGAP) characterized by Waterham et al. (44). We replaced the PAOX1 in the immunotoxin expression cassette with PGAP and introduced one PGAP-immunotoxin cassette with HIS4 as a selection marker into ef-2 his4 (21) to generate strain PGAP-1c. Two PGAP-immunotoxin cassettes, one with HIS4 and the other with ARG4, were introduced into mutEF2JC303-5 (ef-2 his4 arg4) (21), generating strain PGAP-2c. Three other P. pastoris strains were obtained from Invitrogen Corporation (Carlsbad, Calif.): wild-type strain X-33; GS115 Albumin, which secrets serum albumin; and GS115 β-gal, which expresses β-galactosidase intracellularly. Both proteins are expressed under the control of PAOX1.
Cell culturing, protein expression, and media.
P. pastoris strains were first grown for 3 days on plates (purchased from K · D Medical, Columbia, Md.) containing synthetic defined medium (SD) without the amino acid of the selection marker (e.g., SD without histidine for the strains expressing one copy of the immunotoxin gene, serum albumin, or β-galactosidase or X-33; SD without histidine and arginine for the strains expressing two copies of the immunotoxin gene). For immunotoxin expression under the control of PGAP, cells from single colonies were transferred into growth medium YPD (1% yeast extract, 2% peptone, 2% dextrose) and grown overnight. Cells were then pelleted and resuspended in fresh YPD at the original volume. The cell suspensions were inoculated to an optical density at 600 nm (OD600) of 0.3 into buffered expression medium with dextrose (B-EMD) (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 7.0, 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, 2% dextrose, 2% Casamino Acids) and incubated at 28°C with shaking. For protein expression under the control of PAOX1, cells were grown for 4 h in growth medium YPG (1% yeast extract, 2% peptone, 1% glycerol) after initial growth for 24 h in YPD. Cells were then pelleted and resuspended at an OD600 of 10 or 20 in buffered expression medium with methanol (B-EMM) (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 7.0, 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, 0.5% methanol, 2% Casamino Acids).
Protein samples, SDS-PAGE, and Western blotting.
The NuPAGE electrophoresis and blotting system (Invitrogen) was used for protein analysis. Culture supernatant containing secreted proteins was mixed with 4× NuPAGE LDS sample buffer before electrophoresis or storage at −70°C. For preparation of cell extracts, cells were pelleted and washed with phosphate-buffered saline (PBS) and then resuspended at a density of 5% (wt/vol) in PBS containing Complete Mini protease inhibitors from Roche, Basel, Switzerland (1/50 of a stock solution that was prepared by dissolving one tablet in 1 ml of water). The cell suspensions were then mixed with an equal volume of 2× NuPAGE LDS sample buffer and subjected to three cycles of boiling (5 min) and freezing on dry ice. Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with NuPAGE 4 to 12% bis-Tris gels under nonreducing conditions and transferred to nitrocellulose membranes by electroblotting. The membranes were blocked with 4% nonfat dry milk in Tris-buffered saline-Tween before incubation with primary antibodies (1:1,000) and horseradish peroxidase-labeled secondary antibodies (1:5,000). The antigen-antibody complex was detected with the TMB membrane peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
Three polyclonal antibodies were used as the primary antibodies. Goat anti-DT and rabbit anti-G4S antibodies were used in our previous studies (47). An antibody directed against Kar2p of P. pastoris was prepared (by Bio-Synthesis Inc., Lewisville, Tex.) using a 20-residue peptide (PGGQGFDDDDGDFDYDYDYD) derived from the sequence immediately before the C-terminal HDEL of Kar2p. The peptide was conjugated to keyhole limpet hemocyanin before it was injected into rabbits.
Metabolic labeling and immunoprecipitation.
P. pastoris cells expressing the immunotoxin under the control of PAOX1 were first grown overnight in YPD and then grown for 3 h in buffered synthetic complete medium without methionine and cysteine (BSC−MC) containing 1.34% yeast nitrogen base, 0.18% synthetic complete supplement mixture without methionine and cysteine (Qbiogene, Irvine, Calif.), 100 mM potassium phosphate, pH 7.0, 4 × 10−5% biotin, and 1% glycerol. Then 10 OD600 units of the cells was pelleted and resuspended in 10 ml BSC−MC containing 0.5% methanol instead of glycerol. After incubation at 28°C for 1 h, 100 μl of Redivue Pro-mix (14.3 mCi/ml; 35S-labeled methionine-cysteine mixture from Amersham Biosciences, Piscataway, N.J.) was added, and the culture was then incubated for 7 min. After labeling, 1.5 ml of the culture was transferred to ice before it was mixed with NaN3 (10 mM) and protease inhibitors (1/50 of a stock solution). This sample was the zero-chase-time (zero-time) sample. The rest of the culture was mixed with 20 ml B-EMM plus methionine and cysteine (10 mM) before centrifugation at room temperature. The cells were then resuspended in 8.5 ml B-EMM and incubated at 28°C with vigorous shaking. Then at various chase times, aliquots (1.5 ml, divided equally into two tubes) were transferred to ice and mixed with NaN3 and protease inhibitors immediately. The samples were then centrifuged, and the supernatants were collected and stored at 4°C. The cells were resuspended in 350 μl of cell extraction buffer (50 mM Tris-HCl, pH 8.0, 5% glycerol, 1% SDS, 20 mM N-ethylmaleimide, protease inhibitors) and subjected to five cycles of agitation for 30 s (Bead Beater; BioSpec, Bartlesville, Okla.) and incubation for 5 min on ice in the presence of glass beads (about 100 μl). The suspensions were then centrifuged at 10,000 × g for 4 min, and the supernatants were transferred to fresh tubes for immunoprecipitation or stored at −70°C.
Immunoprecipitation of the immunotoxin was carried out using the anti-DT antibody and protein G coupled to magnetic beads (Dynabeads protein G; Dynal Biotech, Oslo, Norway). For precipitation from the cell extracts, 100 μl of cell extract was first mixed with 500 μl of IP buffer (1× PBS, pH 7.4, 5 mM EDTA, 1% Triton X-100) plus protease inhibitors and then with 10 μl of antibody. For the culture supernatants, 500-μl samples were mixed with 20 μl 25% Triton X-100, 50 μl of 10× PBS, pH 7.4, before the antibody was added. The mixtures were incubated for 2 h at room temperature. Then the antigen-antibody complex was precipitated by mixing the preparation for 1 h at room temperature with 60 μl of a Dynabeads-protein G suspension that had been washed three times in IP buffer. The Dynabeads carrying the antigen-antibody complex were sedimented by magnetic force, washed three times (5 min each), and then resuspended in 60 μl of 1× NuPAGE LDS sample buffer. The bead suspensions were boiled for 5 min, and the protein solutions separated from the beads were then transferred to fresh tubes for analysis.
Coimmunoprecipitation of the immunotoxin with Kar2p was performed using the procedures described above, but with the buffers used by Simons et al. (36). Cells were disrupted in HBS buffer (200 mM NaCl, 50 mM HEPES, pH 7.6) containing 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) (Pierce, Rockford, Ill.), protease inhibitors, and 30 U/ml apyrase (Sigma-Aldrich, St. Louis, Mo.). Cell extracts were diluted 1:4 in HBS buffer before they were mixed with the anti-Kar2p antibody. The Dynabeads suspension was washed with HBS buffer containing 0.5% CHAPS.
Protein imaging and quantitation.
Western blots and stained protein gels were photographed with a digital camera (Coolpix 5000; Nikon), and protein bands of interest were quantified with the NIH Image J software. The images of radioactive proteins in SDS-PAGE gels were collected and analyzed using the Storm System and ImageQuant software (Amersham Biosciences, Piscataway, N.J.).
RESULTS
P. pastoris Kar2p sequence and specificity of the anti-Kar2p antibody.
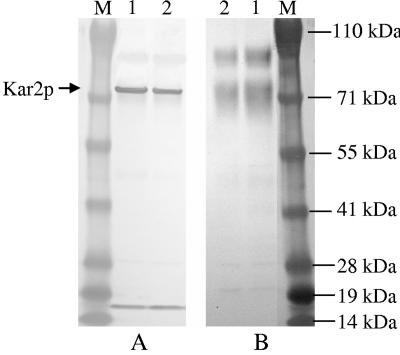
Using the sequence of S. cerevisiae Kar2p, we obtained the DNA sequence of P. pastoris Kar2p from the P. pastoris genome assemblies present in the ergo database (the accession number of the DNA sequence of KAR2 in the GenBank database is AY965684). The amino acid sequence of P. pastoris Kar2p exhibits 76% identity with the sequence of the S. cerevisiae homolog, and P. pastoris Kar2p has a calculated molecular mass of 74 kDa. The polyclonal antibody against the peptide derived from Kar2p recognized two major protein species from a P. pastoris cell lysate on a Western blot: a 74-kDa protein (Kar2p) and a protein with a molecular mass between 14 and 19 kDa (Fig. 2A). Binding of the antibody to these proteins was blocked by the peptide (Fig. 2B). This antibody did not recognize S. cerevisiae Kar2p (data not shown).
FIG. 2.
Specificity of the anti-Kar2p antibody. Western blots of cell extracts from X-33 grown overnight in YPD (lane 1) and YPG (lane 2) were incubated with the rabbit anti-Kar2p antibody (1:1,000 dilution) as the primary antibody in the absence of the Kar2p peptide (A) or in the presence of the Kar2p peptide at a concentration of 10 mg/ml (B). Lane M contained molecular weight standards.
Constitutive expression of the immunotoxin under the control of PGAP.
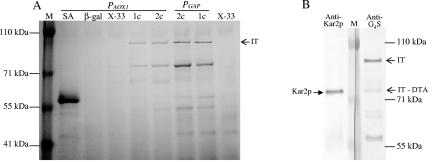
Expression of the immunotoxin in P. pastoris under the control of the promoter for alcohol oxidase (PAOX1) has been reported previously (46). However, in shake flask cultures it is difficult to control the methanol inducer concentration due to the volatility of methanol, and it is difficult to obtain sufficient aeration. Therefore, we chose to also use a constitutive promoter for our studies on the relationships between immunotoxin production and Kar2p expression in P. pastoris. We placed the immunotoxin gene, including the preprosequence of α-factor (46), under control of the promoter of the glyceraldehyde-3-phosphate dehydrogenase gene (PGAP). Under conditions described in Materials and Methods, the average level of immunotoxin expression under the control of PGAP was about 5 to 10 mg/ml, and it was roughly 1.5- to 2-fold higher than the expression level under the control of PAOX1 (Fig. 3A). As we previously reported (21), there was no increase in the amount of immunotoxin in the culture medium of strains carrying two copies of the immunotoxin gene (PGAP-2c and PAOX-2c). Interestingly, a second prominent Coomassie blue-stained band at about 75 kDa was also found in the immunotoxin-secreting strains but not in the X-33 wild-type strain and strains that express cytosolic β-galactosidase or secreted serum albumin.
FIG. 3.
Relative secretion levels for immunotoxin and Kar2p. Protein expression was performed as described in Materials and Methods for 22 h. (A) Coomassie blue-stained SDS gel of culture supernatants of P. pastoris strains expressing genes for cytosolic β-galactosidase (lane β-gal) or secreted serum albumin (lane SA) and for immunotoxin (lanes 1c, one copy; lanes 2c, two copies) under the control of PAOX1 or PGAP. Lanes X-33 contained wild-type strain X-33 culture supernatants. IT, immunotoxin. (B) Western blots of culture supernatant of the strain expressing one copy of the immunotoxin gene (PGAP-1c). The blots were probed with the anti-Kar2p antibody (left blot) or anti-G4S antibody (right blot). Unlike anti-DT antibody, anti-G4S reacts equally well with intact immunotoxin and immunotoxin devoid of the diphtheria toxin A chain (IT-DTA).
Kar2p was secreted from P. pastoris expressing the immunotoxin.
In order to determine the origin of the ∼75-kDa protein present in the medium of P. pastoris cultures expressing the immunotoxin, we probed Western blots of P. pastoris PGAP-1c culture supernatants with two polyclonal antibodies, an antibody directed at the immunotoxin G4S linker in the scFv heavy and light chains of the immunotoxin (47) and the antibody described above directed at P. pastoris Kar2p. We reasoned that the ∼75-kDa protein could represent an immunotoxin breakdown product, such as immunotoxin devoid of the diphtheria toxin A chain, which could occur by reduction of the disulfide attaching the A chain to the rest of the immunotoxin after cleavage of the immunotoxin Kex2 site. Alternatively, the band could represent secreted Kar2p, a situation that has been observed under certain conditions in S. cerevisiae (1, 34). The ∼75-kDa protein probed with anti-G4S stained at only one-fifth the intensity of the 96.3-kDa intact immunotoxin band (Fig. 3B), indicating that the lower band contained mostly another protein. The lower band stained strongly when it was probed with anti-Kar2p antibody, indicating that the other protein was Kar2p.
Secretion of the immunotoxin increased Kar2p secretion.
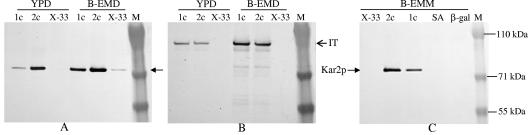
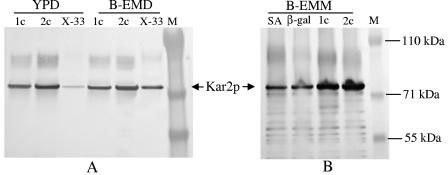
In normal growth medium (YPD or YPG), P. pastoris did not secret a detectable amount of Kar2p into the medium. When the cells were cultured in the richer buffered expression medium (B-EMD), a trace of Kar2p could be detected by Western blotting in the culture medium of wild-type strain X-33 (Fig. 4A). However, under the same culture conditions, P. pastoris strains expressing the immunotoxin under the control of PGAP secreted at least 10 times more Kar2p than wild-type strain X-33 secreted, as judged by densitometry. The amount of Kar2p secreted into the medium was correlated with the copy number of the immunotoxin gene and was influenced by the culture medium (Fig. 4A). In B-EMD, PGAP-2c secreted about 1.7-fold more than PGAP-1c secreted, which in turn secreted 10-fold more than the wild-type strain secreted. In YPD, PGAP-2c secreted about six times more than PGAP-1c secreted. Secretion of the immunotoxin was also affected by the culture medium; about threefold more immunotoxin was produced in B-EMD than in YPD (Fig. 4B).
FIG. 4.
Secretion of the immunotoxin increases Kar2p secretion. Protein expression was performed for 4 h under conditions described in Materials and Methods. P. pastoris strains expressing immunotoxin (lanes 1c, one copy; lanes 2c, two copies) under PGAP control (A and B) were incubated in YPD and buffered (pH 7.0) richer expression medium with dextrose (B-EMD), whereas strains expressing immunotoxin, serum albumin (lane SA), or β-galactosidase (lane β-gal) under PAOX1 control (C) were induced in expression medium containing methanol (B-EMM). Lanes X-33 contained wild-type strain X-33. Samples of culture supernatant were analyzed by SDS-PAGE and Western blotting using anti-Kar2p (A and C) or anti-DT (B) as the primary antibody.
Expression of the immunotoxin under the control of PAOX1 had the same effect on Kar2p secretion as expression under the control of PGAP had. In the buffered expression medium with methanol instead of glucose as the carbon source (B-EMM), PAOX-2c secreted 2.5-fold more Kar2p than PAOX-1c secreted, whereas wild-type strain X-33 did not secrete a detectable amount (Fig. 4C). Interestingly, the P. pastoris strain that expresses and secretes serum albumin at a very high level (>1 g/liter according to Invitrogen's data) and the strain expressing cytosolic β-galactosidase did not secrete Kar2p at a detectable level (Fig. 4C).
Expression of the immunotoxin increased the intracellular Kar2p level.
The intracellular level of Kar2p was relatively low in the wild-type strain grown in YPD and was ∼fivefold greater when the organism was grown in B-EMD (Fig. 5A). Intracellular Kar2p levels were further elevated in single-copy and double-copy immunotoxin-secreting strains in both media. In YPD the intracellular Kar2p level increased sixfold with single-copy immunotoxin expression and ninefold with double-copy expression under the control of PGAP. In B-EMD the increases in Kar2p expression compared with the wild-type strain were ∼1.5-fold for PGAP-1c and ∼2.5-fold for PGAP-2c.
FIG. 5.
Expression of the immunotoxin increases the intracellular Kar2p level. The P. pastoris strains and conditions for protein expression and SDS-PAGE used were the same as those described in the legend to Fig. 4. Cell extracts of 4-h protein-expressing cultures were analyzed by Western blotting using anti-Kar2p as the primary antibody. (A) Samples of strains expressing immunotoxin under PGAP control. (B) Samples of strains expressing immunotoxin and other proteins under PAOX1 control.
Again, expression of the immunotoxin under the control of PAOX1 had an effect on the intracellular level of Kar2p similar to the effect under the control of PGAP. In B-EMM, the two immunotoxin-expressing strains, PAOX-1c and PAOX-2c, had about 2.5-fold and 3-fold more Kar2p, respectively, than the strain expressing serum albumin or β-galactosidase (Fig. 5B).
Immunotoxin processing and secretion.
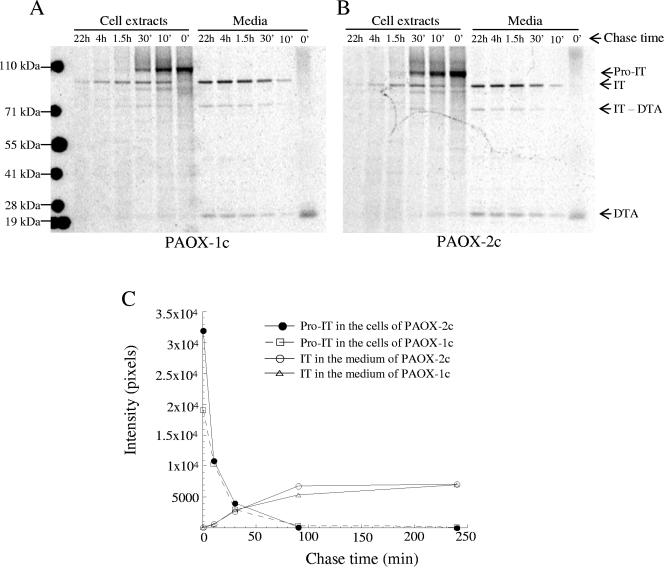
SDS gel electrophoresis of the metabolically labeled and anti-DT antibody-precipitated proteins revealed the kinetics of immunotoxin processing and secretion in the PAOX strains. During the 7-min labeling, the immunotoxin synthesized in PAOX-1c (zero-time cell extract) (Fig. 6A) migrated predominantly at the position corresponding to a protein with a molecular mass of ∼105 kDa, which is the same size as the size of the unprocessed immunotoxin that contains the prosequence of α-factor (21). A band corresponding to a small polypeptide with a molecular mass of about 21 kDa also appeared in the medium of the zero-time sample. Although this molecular mass is about the same as that of the DT A chain that can appear in the culture medium of P. pastoris expressing the immunotoxin (21), the band was not DT A because it was also present in the labeling medium of wild type strain X-33 and was not present when the sample was analyzed under reducing conditions. At the 10-min chase time, the intensity of the proimmunotoxin band was about 54% of the intensity at zero time, indicating that about one-half of the proimmunotoxin seen at zero time disappeared during the first 10 min of the chase (Fig. 6A and C). Some of the proimmunotoxin was converted to mature immunotoxin as mature immunotoxin appeared both in the medium and in the cell extract. The amount of immunotoxin secreted into the medium was approximately 3% of the amount of proimmunotoxin at zero time, whereas the amount remaining in the cell was about 10%. The rest of the proimmunotoxin was broken down, as shown by the presence both in the medium and in the cell of protein species with molecular weights lower than that of the mature immunotoxin. As the chase progressed, less and less proimmunotoxin remained in the cell, and concurrently more and more immunotoxin was secreted into the medium. At the 4-h chase time, almost all proimmunotoxin was processed, and the immunotoxin accumulation in the medium was the highest (the amount was about 37% of the amount of proimmunotoxin at the zero chase time for the single-copy strain). The amount of mature immunotoxin in the cell reached the maximum value after 30 min of chasing (∼11% of the proimmunotoxin at the zero chase time) and then gradually fell, and surprisingly some immunotoxin even remained in the cell for 22 h. Apart from the immunotoxin, there were two other prominent immunotoxin-related species in the medium; one was the ∼21-kDa DT A chain, and the molecular weight of the other was the molecular weight of immunotoxin devoid of DT A.
FIG. 6.
Immunotoxin processing and secretion. Immunotoxin was expressed under PAOX1 control in P. pastoris strains containing one copy (strain PAOX-1c) (A) and two copies (strain PAOX-2c) (B) of the gene. Cells were labeled for 7 min, chased for the times indicated, and then lysed. The cell lysates and culture media were immunoprecipitated with anti-DT antibody before SDS-PAGE and phosphorimaging. The primary translation product of the immunotoxin gene contains α-factor pre- and prosequences. The presequence is removed when the polypeptide enters the ER to generate proimmunotoxin (Pro-IT), which is then further processed to yield mature immunotoxin (IT). Two major immunotoxin breakdown products were identified: immunotoxin without the diphtheria toxin A chain (IT-DTA) and the DT A chain (DTA). The intensities of the Pro-IT and IT bands in panels A and B were measured by using ImageQuant software, and the results are shown in panel C.
The profile of immunotoxin processing and secretion in the PAOX-2c strain was similar to that in the PAOX-1c strain (Fig. 6B and C), except that at zero chase time, the PAOX-2c strain had about 1.7 times more proimmunotoxin than the PAOX-1c strain at the same time. After the first 10 min of chasing, the amount of proimmunotoxin in PAOX-2c dropped about 76%, to almost the same amount seen in PAOX-1c at this time, yet there was no increase in the amount of immunotoxin converted. At the subsequent chase times, there was virtually no difference in the rate of the proimmunotoxin processing and immunotoxin conversion between PAOX-1c and PAOX-2c. However, the amount of breakdown products, mostly DT A and the aggregated material, was significantly greater in PAOX-2c, which is consistent with the results that we reported previously (21).
Binding of Kar2p to the immunotoxin.
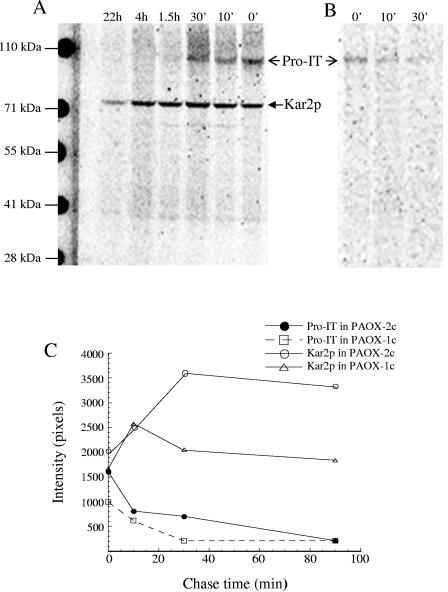
To determine whether Kar2p interacted with the immunotoxin in the cell, coimmunoprecipitation experiments were performed under mild detergent conditions and in the presence of apyrase to preserve Kar2p binding. Figure 7A shows that using the anti-Kar2p antibody, two major proteins were precipitated from the PAOX-2c cell extract; one was the 74-kDa Kar2p protein, and the other was the proimmunotoxin because it had a molecular mass of ∼105 kDa and reacted with anti-DT antibody (Fig. 7B). The proimmunotoxin band was mostly seen at the first three times and was strongest at zero time. Although processed intact immunotoxin was present in the cell, it was not precipitated by the Kar2p antibody. This result, together with the fact that under the same coimmunoprecipitation conditions the immunotoxin-Kar2p complex was not found in the culture medium in which both proteins were present, demonstrates that Kar2p does not bind to the folded protein. When coimmunoprecipitation was performed in the presence of the peptide used to raise the Kar2p antibody, neither Kar2p nor proimmunotoxin was detected, indicating that the precipitated proimmunotoxin had been associated with Kar2p (data not shown).
FIG. 7.
Association of Kar2p with the immunotoxin precursor. Cells of P. pastoris strain PAOX-2c were pulse-chase-labeled as described in the legend to Fig. 6 and subsequently lysed in 2% CHAPS in the presence of 30 U/ml apyrase to preserve Kar2p binding. The cell lysates were immunoprecipitated with the anti-Kar2p antibody. The precipitates were then subjected to SDS-PAGE and phosphorimaging (A) or to immunoprecipitation by anti-DT antibody before SDS-PAGE analysis (B). Anti-DT immunoprecipitation was performed after the anti-Kar2p precipitates were boiled for 5 min in the presence of 1% SDS and then diluted 1:6 in IP buffer (see Materials and Methods). The relative amounts of proimmunotoxin (Pro-IT) coprecipitated by Kar2p from PAOX-1c and PAOX-2c are shown in panel C.
The Kar2p bands (Fig. 7A and C) were stronger that the bands representing the proimmunotoxin, indicating that not all Kar2p was associated with proimmunotoxin. At zero time, the intensity of the Kar2p band was about 1.3-fold greater than that of the proimmunotoxin band. Considering the fact that the number of Met and Cys residues in Kar2p is 2.5-fold less than the number in proimmunotoxin, the actual number of Kar2p molecules precipitated might have been more than three times greater than the number of proimmunotoxin molecules precipitated. Interestingly, while the amount of proimmunotoxin precipitated gradually decreased as the chase progressed, the amount of Kar2p precipitated increased about 1.5-fold from 0 to 30 min before it started to decrease slowly. This suggests that in the double-copy strain a substantial fraction of the newly synthesized Kar2p was not available for antibody precipitation until after a 10- to 30-min lag period. This phenomenon was observed but was less pronounced in the single-copy strain (Fig. 7C).
DISCUSSION
We investigated the efficiency of the immunotoxin secretion from P. pastoris cells by monitoring with time the conversion of the proimmunotoxin to the immunotoxin in strains expressing one and two copies of the immunotoxin gene (Fig. 6). We found that initially the two-copy strain had about 1.7-fold more proimmunotoxin in a cell than the one-copy strain had but did not generate more immunotoxin. Most of the proimmunotoxin disappeared during the first 10 min, before it reached the Golgi complex. This strongly indicates that the ER has a limited capacity for processing the immunotoxin.
The yeast secretory pathway is capable of rapid folding and processing of native proteins; for instance, the half-time for transporting α-factor mating pheromone and invertase from the ER to the Golgi complex after synthesis is less than 2 min (14, 32). Such rapid processing made the detection of glycosylated pheromone precursors possible only in mutants defective at various stages in protein transport or in protein processing events (9, 14). Although there have been numerous reports of α-factor preprosequence-mediated secretion of heterologously expressed proteins in yeast, including P. pastoris (5), examples of pulse-chase kinetic studies on formation, conversion, and translocation of precursors are limited. In a study of α-factor-leader-directed secretion of recombinant human-insulin-like growth factor I (IGFI) from S. cerevisiae, Steube et al. (38) found that conversion of the primary translation products to the differently glycosylated IGFI precursors and to the mature protein occurs very rapidly, with 2 min of pulse time, and almost all precursors were processed after about 5 min of chase time. However, in this case secretion of the mature IGFI from the cell was very slow; only after 30 to 40 min did IGFI appear in the medium. By contrast, conversion of the proimmunotoxin to mature protein was much slower; it took more than 1.5 h for the precursor form to be completely processed. On the other hand, secretion of mature immunotoxin was relatively quick since immunotoxin appeared outside the cells within 10 min of chasing (Fig. 6).
Slow maturation of the immunotoxin was accompanied by prolonged association of Kar2p with the proimmunotoxin (Fig. 7). The immunotoxin translocation domain contains several very hydrophobic stretches (3), and therefore it is not surprising that folding of the immunotoxin requires chaperones like Kar2p. The Kar2p/proimmunotoxin complex remained detectable for more than 1.5 h after the proimmunotoxin was synthesized. Although some immunotoxin started to appear in the medium after 7 min, most (about 70% of the total secreted immunotoxin) exited from the cell between 10 min and 1.5 h, and some even took about 4 h to be exported. The profile of immunotoxin secretion and its association with Kar2p suggests that the folding process for the proimmunotoxin is slow and that the ER can maintain the immunotoxin in a folding-competent or secreting status for 1.5 h. The lack of association between Kar2p and processed immunotoxin, as opposed proimmunotoxin (Fig. 7), and the fact that Kar2p was secreted from wild-type strain X-33 when the cells were cultured in buffered rich expression medium (Fig. 4) indicated that Kar2p secretion is not physically coupled to immunotoxin secretion.
Upregulation of BiP/Kar2p is the first and most-characterized event of the UPR. It has been shown that overexpression of secreted mutant or foreign proteins, particularly those with slower folding characteristics, elevates intracellular BiP/Kar2p levels (7, 13, 16, 17, 41, 45). In line with these reports, we found that secretion of overexpressed immunotoxin increased Kar2p expression in P. pastoris (Fig. 5). However, under the same expression conditions, secretion of serum albumin did not result in an increase even though the level of secretion was much higher than that of the immunotoxin. It has been demonstrated that BiP/Kar2p also plays a key role in regulation of the UPR mediated by Ire1p, an ER resident transmembrane kinase/nuclease (4, 25) whose activation leads to production of a transcription factor, Hac1p, which in turn drives the UPR (24). Under normal conditions, Kar2p associates with the luminal domain of Ire1p, keeping Ire1p in an inactive form. When the unfolded proteins accumulate in the ER, BiP/Kar2p dissociates from Ire1p and binds to unfolded proteins, resulting in activation of Ire1p and consequently the UPR (2, 27). We postulated that slow folding and prolonged association of the immunotoxin with Kar2p would reduce the amount of free Kar2p available for binding Ire1p, consequently triggering the UPR. This postulate is supported by the fact that in the P. pastoris strains expressing two copies of the immunotoxin gene, more primary products were produced, more precursors were found to be associated with Kar2p, and more Kar2p was expressed than in strains expressing one copy of the gene (Fig. 5 and 6).
We found that expression of the immunotoxin not only elevated the intracellular level but also increased the extracellular secretion of Kar2p, and the higher level of Kar2p secretion coincided with greater activation of the UPR as strains expressing two copies of the immunotoxin gene secreted more Kar2p than strains expressing one copy secreted (Fig. 4). Previous studies have demonstrated that in S. cerevisiae Erd2p-dependent retrieval of HDEL proteins is saturable when HDEL-tagged proteins are overexpressed (1, 6, 34). Therefore, we suggest that in the case of immunotoxin secretion in P. pastoris, UPR upregulation of HDEL proteins, prominently Kar2p, surpasses the capacity of ER retention, at least Erd2p-dependent retrieval. Although the Erd2 gene was upregulated by the Ire1p-mediated UPR in S. cerevisiae, at least five genes coding for ER resident HDEL proteins were upregulated (42). Among these genes, the Kar2, Pdi1, and Eug1 genes exhibited about twofold-greater increases in transcription than the Erd2 gene exhibited. The Kar2 gene also showed significant upregulation in Δire1 and ΔHac1 mutant strains (4, 42), indicating that it can also be regulated by Δire1 and ΔHac1 independent mechanisms (18, 33). Furthermore, it has been shown in mammalian cells that BiP is regulated at the posttranscription level, which allows the cells to produce more protein in response to the accumulation of unfolded protein in the ER (11).
Since BiP/Kar2p plays a key role in the UPR, not only as an ER chaperone but also as an ER stress sensor, limitations in retention of BiP/Kar2p and possibly other HDEL proteins, such as Pdi1p, may be a factor that restricts heterologous secretion of certain overexpressed proteins. This would be a fruitful area for further studies considering the wide use of P. pastoris in heterologous protein secretion.
REFERENCES
- 1.Belden, W. J., and C. Barlowe. 2001. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell 12:957-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 3.Choe, S., M. J. Bennett, G. Fujii, P. M. Curmi, K. A. Kantardjieff, R. J. Collier, and D. Eisenberg. 1992. The crystal structure of diphtheria toxin. Nature 357:216-222. [DOI] [PubMed] [Google Scholar]
- 4.Cox, J. S., C. E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197-1206. [DOI] [PubMed] [Google Scholar]
- 5.Cregg, J. M., J. L. Cereghino, J. Shi, and D. R. Higgins. 2000. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 16:23-52. [DOI] [PubMed] [Google Scholar]
- 6.Dean, N., and H. R. Pelham. 1990. Recycling of proteins from the Golgi compartment to the ER in yeast. J. Cell Biol. 111:369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorner, A. J., L. C. Wasley, and R. J. Kaufman. 1989. Increased synthesis of secreted proteins induces expression of glucose-regulated proteins in butyrate-treated Chinese hamster ovary cells. J. Biol. Chem. 264:20602-20607. [PubMed] [Google Scholar]
- 8.Elmendorf, H. G., and K. Haldar. 1993. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. EMBO J. 12:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emter, O., B. Mechler, T. Achstetter, H. Muller, and D. H. Wolf. 1983. Yeast pheromone alpha-factor is synthesized as a high molecular weight precursor. Biochem. Biophys. Res. Commun. 116:822-829. [DOI] [PubMed] [Google Scholar]
- 10.Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 355:33-45. [DOI] [PubMed] [Google Scholar]
- 11.Gülow, K., D. Bienert, and I. G. Haas. 2002. BiP is feed-back regulated by control of protein translation efficiency. J. Cell Sci. 115:2443-2452. [DOI] [PubMed] [Google Scholar]
- 12.Haas, I. G., and M. Wabl. 1983. Immunoglobulin heavy chain binding protein. Nature 306:387-389. [DOI] [PubMed] [Google Scholar]
- 13.Hohenblum, H., B. Gasser, M. Maurer, N. Borth, and D. Mattanovich. 2004. Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol. Bioeng. 85:367-375. [DOI] [PubMed] [Google Scholar]
- 14.Julius, D., R. Schekman, and J. Thorner. 1984. Glycosylation and processing of prepro-α-factor through the yeast secretory pathway. Cell 36:309-318. [DOI] [PubMed] [Google Scholar]
- 15.Kabani, M., S. S. Kelley, M. W. Morrow, D. L. Montgomery, R. Sivendran, M. D. Rose, L. M. Gierasch, and J. L. Brodsky. 2003. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell 14:3437-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauffman, K. J., E. M. Pridgen, F. J. Doyle, 3rd, P. S. Dhurjati, and A. S. Robinson. 2002. Decreased protein expression and intermittent recoveries in BiP levels result from cellular stress during heterologous protein expression in Saccharomyces cerevisiae. Biotechnol. Prog. 18:942-950. [DOI] [PubMed] [Google Scholar]
- 17.Kozutsumi, Y., M. Segal, K. Normington, M. J. Gething, and J. Sambrook. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462-464. [DOI] [PubMed] [Google Scholar]
- 18.Leber, J. H., S. Bernales, and P. Walter. 2004. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2:E235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, H. I., S. Gal, T. C. Newman, and N. V. Raikhel. 1993. The Arabidopsis endoplasmic reticulum retention receptor functions in yeast. Proc. Natl. Acad. Sci. USA 90:11433-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis, M. J., and H. R. Pelham. 1990. A human homologue of the yeast HDEL receptor. Nature 348:162-163. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y. Y., J. H. Woo, and D. M. Neville, Jr. 2003. Targeted introduction of a diphtheria toxin resistant mutation into the chromosomal EF-2 locus of Pichia pastoris and expression of immunotoxin in the EF-2 mutants. Protein Expr. Purif. 30:262-274. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y. Y., I. Gordienko, A. Mathias, S. Ma, J. Thompson, J. H. Woo, and D. M. Neville, Jr. 2000. Expression of an anti-CD3 single-chain immunotoxin with a truncated diphtheria toxin in a mutant CHO cell line. Protein Expr. Purif. 19:304-311. [DOI] [PubMed] [Google Scholar]
- 23.Lyman, S. K., and R. Schekman. 1997. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88:85-96. [DOI] [PubMed] [Google Scholar]
- 24.Ma, Y., and L. M. Hendershot. 2001. The unfolding tale of the unfolded protein response. Cell 107:827-830. [DOI] [PubMed] [Google Scholar]
- 25.Mori, K., W. Ma, M. J. Gething, and J. Sambrook. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74:743-756. [DOI] [PubMed] [Google Scholar]
- 26.Normington, K., K. Kohno, Y. Kozutsumi, M. J. Gething, and J. Sambrook. 1989. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 57:1223-1236. [DOI] [PubMed] [Google Scholar]
- 27.Okamura, K., Y. Kimata, H. Higashio, A. Tsuru, and K. Kohno. 2000. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 279:445-450. [DOI] [PubMed] [Google Scholar]
- 28.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 29.Pelham, H. R. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15:483-486. [DOI] [PubMed] [Google Scholar]
- 30.Pouyssegur, J., R. P. Shiu, and I. Pastan. 1977. Induction of two transformation-sensitive membrane polypeptides in normal fibroblasts by a block in glycoprotein synthesis or glucose deprivation. Cell 11:941-947. [DOI] [PubMed] [Google Scholar]
- 31.Rose, M. D., L. M. Misra, and J. P. Vogel. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211-1221. [DOI] [PubMed] [Google Scholar]
- 32.Schauer, I., S. Emr, C. Gross, and R. Schekman. 1985. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J. Cell Biol. 100:1664-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schröder, M., R. Clark, and R. J. Kaufman. 2003. IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol. Microbiol. 49:591-606. [DOI] [PubMed] [Google Scholar]
- 34.Semenza, J. C., K. G. Hardwick, N. Dean, and H. R. Pelham. 1990. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61:1349-1357. [DOI] [PubMed] [Google Scholar]
- 35.Shen, X., K. Zhang, and R. J. Kaufman. 2004. The unfolded protein response—a stress signaling pathway of the endoplasmic reticulum. J. Chem. Neuroanat. 28:79-92. [DOI] [PubMed] [Google Scholar]
- 36.Simons, J. F., S. Ferro-Novick, M. D. Rose, and A. Helenius. 1995. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sprague, G. F., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 675-777. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 38.Steube, K., B. Chaudhuri, W. Marki, J. P. Merryweather, and J. Heim. 1991. α-Factor-leader-directed secretion of recombinant human-insulin-like growth factor I from Saccharomyces cerevisiae. Precursor formation and processing in the yeast secretory pathway. Eur. J. Biochem. 198:651-657. [DOI] [PubMed] [Google Scholar]
- 39.Tang, B. L., S. H. Wong, X. L. Qi, S. H. Low, and W. Hong. 1993. Molecular cloning, characterization, subcellular localization and dynamics of p23, the mammalian KDEL receptor. J. Cell Biol. 120:325-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J., S. Stavrou, M. Weetall, J. M. Hexham, M. E. Digan, Z. Wang, J. H. Woo, Y. Yu, A. Mathias, Y. Y. Liu, S. Ma, I. Gordienko, P. Lake, and D. M. Neville, Jr. 2001. Improved binding of a bivalent single-chain immunotoxin results in increased efficacy for in vivo T-cell depletion. Protein Eng. 14:1035-1041. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga, M., A. Kawamura, and K. Kohno. 1992. Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J. Biol. Chem. 267:17553-17559. [PubMed] [Google Scholar]
- 42.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, B., Y. Ye, and T. A. Rapoport. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 44.Waterham, H. R., M. E. Digan, P. J. Koutz, S. V. Lair, and J. M. Cregg. 1997. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37-44. [DOI] [PubMed] [Google Scholar]
- 45.Watowich, S. S., R. I. Morimoto, and R. A. Lamb. 1991. Flux of the paramyxovirus hemagglutinin-neuraminidase glycoprotein through the endoplasmic reticulum activates transcription of the GRP78-BiP gene. J. Virol. 65:3590-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo, J. H., Y. Y. Liu, A. Mathias, S. Stavrou, Z. Wang, J. Thompson, and D. M. Neville, Jr. 2002. Gene optimization is necessary to express a bivalent anti-human anti-T cell immunotoxin in Pichia pastoris. Protein Expr. Purif. 25:270-282. [DOI] [PubMed] [Google Scholar]
- 47.Woo, J. H., Y. Y. Liu, S. Stavrou, and D. M. Neville, Jr. 2004. Increasing secretion of a bivalent anti-T-cell immunotoxin by Pichia pastoris. Appl. Environ. Microbiol. 70:3370-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young, B. P., R. A. Craven, P. J. Reid, M. Willer, and C. J. Stirling. 2001. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 20:262-271. [DOI] [PMC free article] [PubMed] [Google Scholar]