Abstract
A new scanning electron microscopic method was developed for gaining both phylogenetic and morphological information about target microbes using in situ hybridization with rRNA-targeted oligonucleotide probes (SEM-ISH). Target cells were hybridized with oligonucleotide probes after gold labeling. Gold enhancement was used for amplification of probe signals from hybridized cells. The hybridized cells released a strong backscatter electron signal due to accumulation of gold atoms inside cells. SEM-ISH was applied to analyze bacterial community composition in freshwater samples, and bacterial cell counts determined by SEM-ISH with rRNA-targeted probes for major phyla within the domain Bacteria were highly correlated to those by fluorescent in situ hybridization (FISH). The bacterial composition on surface of river sediment particles before and after cell dispersion treatment by sonication was successfully revealed by SEM-ISH. Direct enumeration of bacterial cells on the surface of sonicated sediment particles by SEM-ISH demonstrated that members of Cytophaga-Flavobacterium existed tightly on the surface of particles. SEM-ISH allows defining the number and distribution of phylogenetically defined cells adherent to material surfaces, which is difficult in FISH, and it gives new insight into electron microscopic studies of microorganisms in their natural environment.
Scanning electron microscopy (SEM) has been used widely in environmental microbiology for characterization of the surface structure of biomaterials, as well as measurement of biological responses to biomaterials, including cell attachment and changes in morphology (1, 37, 38, 42). SEM is very useful for defining the number and distribution of microorganisms adherent to surfaces. Size and some basic morphometric parameters were identified through the application of image analysis techniques to SEM (26, 39). One limitation of SEM in environmental microbiology is that it cannot provide phylogenetic or genetic information about target microbes. When target microbes are observed by SEM, they must be distinguished from nontarget cells by morphological characteristics. However, microorganisms in natural samples, as well as pure culture, are morphologically highly diversified. Immunogold labeling marks cellular components (13, 21, 36). Antibody-labeled colloidal gold particles react to surface antigen and allow observation of specific microorganisms under SEM.
In contrast, molecular studies based on rRNA analysis have led to an understanding of the microbial diversity and community composition of aquatic environments. For reliable quantitative characterization of community structure, in situ hybridization (ISH) with rRNA-targeted fluorescent oligonucleotide probes (FISH) is an increasingly popular method. FISH permits the identification and quantification of individual cells and has demonstrated great power in the analysis of bacterial community composition in several environments (2, 4, 15).
In order to detect, identify, and enumerate target microbes by electron microscopy using genetic information, we developed an ISH technique using SEM (SEM-ISH). Target cells were hybridized with biotin-labeled oligonucleotide probes and recognized by streptavidin attached to an ultrasmall gold immunoprobe. Gold enhancement was used for amplification of probe signals from hybridized cells. rRNA-targeted probes for identification of microbial cells in freshwater environments were used for specific detection based on phylogenetic information at the single-cell level.
MATERIALS AND METHODS
Bacterial strains.
The strains used in the present study were Aeromonas hydrophila ATCC 7966, Aeromonas sobria ATCC 43979, Brevundimonas diminuta IFO3140, Comamonas testosteroni R5, Escherichia coli W3110, and E. coli JM109. The strains were cultured at 37°C in aerobic Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl [pH 7.0]). Cells were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.2]), and suspended in freshly filtered 4% paraformaldehyde in PBS for 16 h. After fixation, cells were washed twice in PBS and suspended in 50% ethanol with PBS. Fixed cells were stored at −20°C until use.
Environmental samples.
Surface natural river water samples were taken from Kitahashi in the Neyagawa River, Juhachijo in the Kanzakigawa River, and Kuwazu in the Inagawa River in the northern part of Osaka, Japan. Kitahashi is located in a commercial area, Osaka Business Park. Juhachijo and Kuwazu are located in an industrial area. These sites were considered to be polluted by organic carbon (15, 18). Total organic carbon is 5 to 7 mg/liter in these sites. Pond water was taken from the garden of the Graduate School of Pharmaceutical Sciences, Osaka University. The pond is 10 by 15 m and ca. 1.2 m deep. The water samples were collected in sterile 500-ml glass bottles and carried to the laboratory on ice. The densities of heterotrophic bacterioplankton in the river (Kitahashi, Juhachij, Kuwazu) and pond (Osaka University) samples determined with DAPI (4′,6′-diamidino-2-phenylindole) at a final concentration of 1 μg ml−1 were 1.4 × 107, 2.0 × 107, 5.3 × 105, and 1.0 × 106 cells ml−1, respectively. Portions (40 ml) of the samples were fixed with 4% paraformaldehyde at 4°C for 16 h. After fixation, 0.5- to 10 ml-portions were filtered through gelatin [0.1% gelatin, 0.01% CrK(SO4)2]-coated polycarbonate white filter (0.2-μm pore size, 25-mm diameter; ADVANTEC) and rinsed twice with filtered deionized water. Sediment was taken from sand oxidized surface (upper 2 cm) at Kuwazu. The sand particles were fixed with 4% paraformaldehyde at 4°C for 16 h. The fixed samples were washed twice with filtered deionized water and dried by air. Then samples were stored at −20°C.
Oligonucleotide probes.
The following oligonucleotide probes were used: (i) EUB338 complementary to a region of the 16S rRNA conserved in the domain bacteria (3); (ii) NON338, negative control (41); (iii) ALF1b, complementary to a region of the 16S rRNA specific the alpha subclass of Proteobacteria (22); (iv) BET42a, complementary to a region of the 23S rRNA specific for the beta subclass of Proteobacteria (22); (v) GAM42a, complementary to a region of the 23S rRNA specific for the gamma subclass of Proteobacteria (22); (vi) CF319, complementary to a region of the 16S rRNA specific Cytophaga-Flavobacterium phylum (23); and (vii) ES445, complementary to a region of the 16S rRNA specific for Escherichia-Shigella (16). Probes were labeled with biotin or CY3 at the 5′ end.
SEM-ISH. (i) SEM-ISH on membrane filter for water samples.
The polycarbonate filters with bacterial cells from water samples were cut into 12 sections. Each filter section was transferred to a microtube (volume, 1.8 ml) and dehydrated by vacuum. For EUB338, NON338, ALF1b, BET42a, and GAM42a probes, hybridization and washing were performed as described by Alfreider et al. (2). For ES445 probe, hybridization was performed in a moisture chamber at 41°C for 2 h with hybridization buffer (0.45 M NaCl, 20 mM Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate) containing 2 ng of probe/μl. The washing step was done at 41°C for 30 min with washing buffer (0.08 M NaCl, 20 mM Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate) (16).
Samples were incubated with 100 μl of alkali solution (0.5 M NaOH, 1.5 M NaCl) for 10 min at room temperature (ca. 25°C) in order to allow regents for gold labeling to enter the cell and neutralized with 100 μl of acidic solution (0.5 M HCl, 1.5 M NaCl). The filter sections were rinsed with deionized water. After blocking with 100 μl of PBS containing 0.1% gelatin and 0.1% Tween 20, Nanogold-streptavidin (Nanoprobes, Yaphank, N.Y.; 1:1,000 in 1% bovine serum albumin in PBS) was reacted for 30 min at room temperature. The filter sections were soaked in PBS containing 0.1% gelatin and 0.1% Tween20 at room temperature for 10 min. Afterward, the filter sections were soaked in filtered deionized water. For gold autometallography, the filter sections were soaked in 100 μl of the solution mixture of GOLDENHANCE-EM (Nanoprobes) and incubated for 10 min at room temperature in the dark. The filter sections were soaked in PBS containing 0.1% gelatin and 0.1% Tween 20 for 10 min and in filtered deionized water for 10 min.
(ii) SEM-ISH for sediment particles.
Sediment particles of 0.5 to 1.5 mm were transferred to a microtube (volume, 1.8 ml) and resuspended in filtered deionized water. Samples were sonicated for 5 min at 400 kHz and 125 W using a JUS-S01 bath sonicator (JEOL Datum, Ltd., Tokyo, Japan). Samples were then washed three times with filtered deionized water. Both sonicated sediment particles and untreated particles were subjected to SEM-ISH. The sediment particles were transferred to a new microtube (volume, 1.8 ml), and SEM-ISH in microtube was performed as described for SEM-ISH on membrane filters for water samples.
SEM.
Slides, polycarbonate filters, or sediment particles were put on an SEM pore (JEOL Datum) connected to an LV cooling holder (JEOL Datum) and incubated with 50 μl of t-butyl alcohol at room temperature for 10 min. The holder was covered with a cooling cap (JEOL Datum) and chilled with liquid nitrogen for 30 s. After the cooling cap was removed, the holder was put immediately in the specimen chamber of the scanning electron microscope (JSM 5610L; JEOL Datum). Freeze-drying was carried out in the chamber. After 20 min, samples were observed in reduced vacuum (30 Pa), i.e., low vacuum mode. For observation under high vacuum, samples were further sputter coated with evaporated gold for 2 min using the Quick Coater (Sanyu Denshi Co, Ltd., Tokyo, Japan). The accelerating voltage was 5 to 20 kV for both low and high vacuum mode imaging. Specimens were placed 14 mm from the base detector. Electron micrographs were obtained at magnifications of ×60 to ×20,000 using both secondary electron (SE) and backscatter electron (BSE) detectors for high vacuum imaging and BSE detectors for low vacuum imaging. Composite images of SE and BSE images were constructed with Adobe Photoshop 4.0.1J (Adobe Systems, Inc.). For quantitative evaluation, at least 200 cells in different fields were counted in triplicate. Counting results were always corrected by substracting signals observed with the NON338 probe.
FISH.
The polycarbonate filters with bacterial cells from water samples was cut into 12 sections, the filter section was transferred to a microtube (volume, 1.8 ml) and dehydrated by vacuum. For sediment samples, particles of 0.5 to 1.5 mm were transferred to microtube (volume, 1.8 ml) and resuspended in filtered deionized water. Samples were sonicated for 1.5 min at 400 kHz and 125 W using a JUS-S01 bath sonicator. Then supernatant was filtered through gelatin [0.1% gelatin, 0.01% CrK(SO4)2]-coated polycarbonate white filter (0.2-μm pore size, 25-mm diameter; ADVANTEC) and rinsed twice with filtered deionized water. Hybridization and washing were carried out in microtube under the same conditions as described above. Samples were counterstained with DAPI. Filters were mounted in immersion oil for observation by epifluorescence microscopy (E-400; Nikon, Tokyo, Japan) with the Nikon filter sets UV-2A and HQ:CY3 for UV and green excitation, respectively. Image were acquired by a cooled charge-coupled device camera (Cool Snap; Roper Photometrics) and stored as digital files. More than 1,000 DAPI-stained cells were counted per sample. Counting results were always corrected by substracting signals observed with the NON338 probe.
RESULTS AND DISCUSSION
rRNA-targeted SEM-ISH.
In order to observe microbes with a targeted sequence by electron microscopy, cells must be labeled with metal. For phylogenetic identification, bacterial cells were hybridized with rRNA-targeted probes and labeled with gold in the present study. After hybridization with biotin-labeled oligonucleotide probes, target microbes were recognized by streptavidin attached to Nanogold (12). In order to amplify the signal by enlarging the gold particle, gold enhancement was carried out. In this reaction, gold ions in solution are catalytically deposited onto Nanogold particles as metallic gold (Au0). Particles grow in size as development time elapses. Consequently, hybridized cells contain gold deposited inside them.
Conventionally, SEM is used in a high-vacuum mode and specimens are first coated with an electron conductive layer of gold or carbon in order to dissipate surface charges which can accumulate on the specimen. SEM images have been constructed by SE signals that are derived from metal on the cell surface. The energy of the SE is relatively weak, and it is difficult to detect SE signals released from gold inside the cell. The energy of BSE is higher than the SE by 1 order (13); therefore, BSE was used for the detection of gold inside cells in the present study. Bacterial cells were first observed by low-vacuum SEM in order to obtain BSE signal from hybridized cells effectively. The low-vacuum SEM operates under conditions of reduced vacuum in the specimen chamber (32).
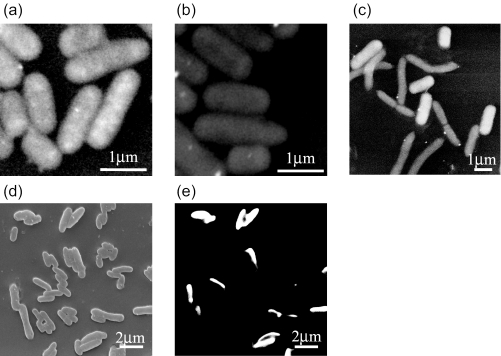
Representative low-vacuum SEM images of cells hybridized with 16S rRNA targeted probe are shown in Fig. 1. E. coli cells hybridized with EUB338 probe released a strong BSE signal due to accumulation of gold atoms inside cells (Fig. 1a). Cells which did not hybridize with probes showed a weaker BSE signal (Fig. 1b). Figure 1c shows the results of SEM-ISH with the GAM42a probe for bacterial mixtures of Brevundimonas diminuta and E. coli W3110. E. coli, which belongs to the γ-Proteobacteria, demonstrated a strong signal due to the high density of gold inside cells. However, B. diminuta, which belongs to α-Proteobacteria, did not show this signal, thus allowing easy distinction between the cells giving positive results and those giving negative results.
FIG. 1.
Low-vacuum (a, b, and c) and high-vacuum (d and e) SEM images of bacterial cells after ISH: E. coli hybridized with EUB338 probe (a) and NON338 probe (b), mixture of B. diminuta and E. coli hybridized with GAM42a probe (c), mixture of E. coli and A. sobria cells hybridized with ES445 probe (d and e). The same microscopic fields are shown with SE (d) and BSE images (e).
The low-vacuum SEM imaging system permits both gold particles and background to be viewed at the same time, although surface detail of the cells is not visible. In order to improve the resolution of cell surface and obtain topographic information effectively, hybridized cells were observed under high-vacuum SEM. In high-vacuum SEM, an SE image is obtained of the cell surface and the BSE image is used to detect the probe signals, which appear as bright objects against a black background (10, 33). High-vacuum SEM images of a mixture of E. coli W3110 and A. sobria after ISH with the ES445 probe and labeling with gold are shown in Fig. 1d and e. SE images demonstrated the surface topography of all cells (Fig. 1d). In BSE images, E. coli cells hybridized with the ES445 probe are identified as bright spots due to the enhanced signal from the gold labeling (Fig. 1e). In hybridized cells, gold inside cells was larger in quantity than on the cell surface, and thus it resulted in higher BSE signal. In the same microscopic field, both images could be viewed side by side.
Generally, the sensitivity in detecting BSE signal depends on the penetration depth of the primary beam, which is a function of the accelerating voltage used. In our study, at least 15 kV of accelerating voltage was necessary in order to obtain a sufficient BSE signal from hybridized cells. Although the time for high-vacuum SEM required 1 h after samples were prepared for low-vacuum SEM, high-vacuum SEM gave better resolution at high magnification.
The specificity of each probe in SEM-ISH was examined for six bacterial strains (Table 1). Each probe was hybridized with only targeted species. No strong hybridization signal was observed with other nontargeted species used in the present study with the specified conditions. rRNA-targeted SEM-ISH enabled to identify specific bacteria under the scanning electron microscope.
TABLE 1.
Specificity of each probe as determined by SEM-ISH
| Strain (subclass of Proteobacteria) | Presence (+) or absence (−) of signal
|
|||||
|---|---|---|---|---|---|---|
| EUB338 | NON338 | ALF1b | BET42a | GAM42a | ES445 | |
| Brevundimonas diminuta (alpha) | + | − | + | − | − | − |
| Comamonas testosteroni (beta) | + | − | − | + | − | − |
| Aeromonas hydrophila (gamma) | + | − | − | − | + | − |
| Aeromonas sobria (gamma) | + | − | − | − | + | − |
| Escherichia coli JM109 (gamma) | + | − | − | − | + | + |
| Escherichia coli W3110 (gamma) | + | − | − | − | + | + |
Enumeration of bacterial cells in environmental samples using rRNA-targeted probes under an electron microscope.
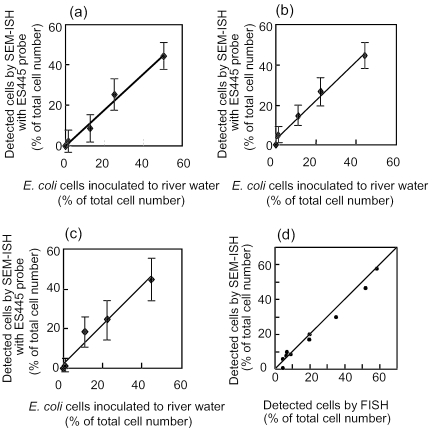
For further quantitative evaluation, E. coli JM109 cells were introduced into natural river water samples. Before quantitative evaluation of SEM-ISH for river water samples, the abundance of E. coli in the river water samples was confirmed by FISH and PCR to be <0.1% of total bacteria. Then, serial dilutions of fixed E. coli cells were added to 40 ml of fixed river water samples and subjected to SEM-ISH. Using the ES445 probe, E. coli cells were detected in each dilution tube. A good correlation between the number of E. coli cells in the sample and the number of cells revealed by SEM-ISH was demonstrated (Fig. 2a, b, and c) (a, Juhachijo, y = 0.89x + 0.27, R2 = 0.988, n = 12; b, Kuwazu, y = 0.97x + 2.8, R2 = 0.988, n = 12; c, Kitahashi, y = 0.99x + 2.1, R2 = 0.973, n = 12).
FIG. 2.
Recovery of E. coli cells through SEM-ISH and relationship between bacteria in river and pond water detected by SEM-ISH and FISH. (a to c) E. coli JM109 cells were added to river water samples (a, Juhachijo; b, Kuwazu; c, Kitahashi) at different concentrations (1.25, 12.5, 25, and 50% of total cells) and enumerated after ISH with the ES445 probe. (d) Indigenous bacteria in river and pond water were subjected to both SEM-ISH and FISH with EUB338, CF319, ALF1a, BET42a, and GAM42b probes, and the cell numbers were compared.
The accuracy of enumeration of indigenous bacterial cells in pond and river water by SEM-ISH was examined with EUB338, CF319, ALF1b, BET42a, and GAM42a probes. The detection rate for bacterial cells determined by SEM-ISH with the EUB338 probe ranged from 47 to 57%. Probes for major phyla within the domain Bacteria permitted affiliation of more than 90% of the EUB338 counts with known bacterial groups. The results were compared to those obtained by FISH with CY3-labeled probes (Fig. 2d). A good correlation between the number of cells detected by FISH in the aquatic samples and the number of cells revealed by SEM-ISH was demonstrated (y = 0.42 + 0.93x, R2 = 0.983, n = 10). That is, SEM-ISH could be applied to indigenous cells in aquatic samples, as well as standard FISH with CY3-labeled probe. In addition, SEM-ISH on a polycarbonate filter allowed effective concentration of target cells from aquatic samples and enhanced the quantitative analysis.
However, 40 to 50% of the total cells in the samples still could not be detected by SEM-ISH with the EUB338 probe or by FISH. Our preliminary experiments with FISH showed that an abundance of archaea, planctomyces that were reported to be present in other aquatic environments (6, 8, 11, 34), were ca. 1% of the total cells in the samples used in the present study. Therefore, large portion of cells undetectable by SEM-ISH with EUB338 may have been impermeable to biotin-labeled probes or Nanogold-streptavidin or perhaps did not have a ribosomal content high enough for detection by this technique. In order to detect cells with low metabolic activity, SEM-ISH in combination with direct viable counting (16), use of rRNA targeted polynucleotide transcript probes (28), enzymatic signal amplification (14, 19, 29), and in situ DNA amplification methods (25, 40) may be effective as previously demonstrated for these FISH methods.
Microbial communities in the natural environment are complex, and their permeability of cell wall structure is not uniform. The application of enzymatic or other chemical treatments may be required for in situ identification of cells that are impermeable to probes or streptavidin (30).
Bacterial cells distributed on the surface of river sediment particles.
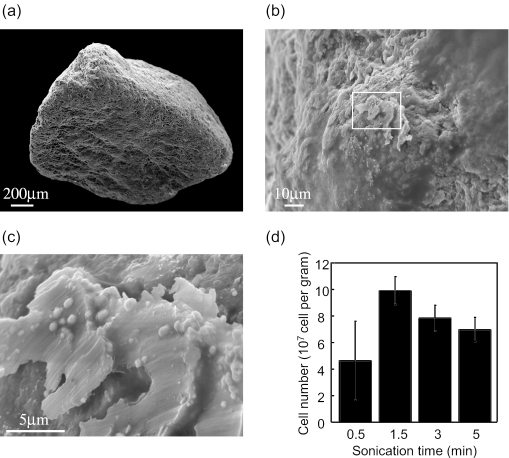
River microbial communities play important role in global nutrient cycles, and bacteria in river sediments may be major contributors. FISH analysis of sediment samples usually requires cell dispersion from sediment particles by sonication (20, 31). One limitation of community analysis of sediment samples by FISH is that the composition of the bacterial community which cannot be recovered by sonication remains undefined. In the present study, bacterial community that attached on the surface of river sediment samples before and after sonication was investigated by using SEM-ISH. Some research groups use 20 to 30 s of mild sonication for marine sediment samples (20). Pretreatment seems to depend on both samples and equipment availability for sonication. We examined the effect of sonication time on cell recovery from river sediment particles (Fig. 3). Representative SE images of sediment sand particles and cells on the particle surface are shown in Fig. 3a, b, and c. The recovered cell number was maximized after 1.5 min of treatment in the bath sonicator (Fig. 3d). Longer treatment times caused a decrease in cell number probably due to cell destruction caused by excess treatment. Thus, 1.5 min of sonication was used for bacterial community analysis. For SEM-ISH analysis, both untreated sediment particles and sonicated particles were used.
FIG. 3.
SE images of river sediment particles at magnifications of ×60 (a), ×1,000 (b), and ×6,000 (c) and effect of sonication time on cell number recovered from sediment particles (d).
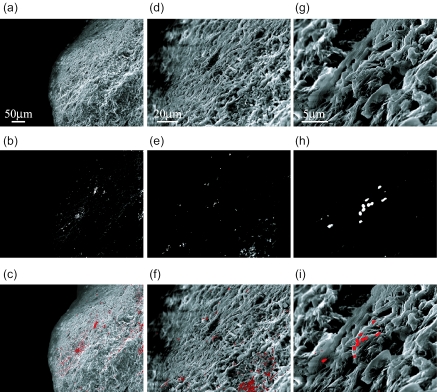
Sediment particles without sonication were subjected to SEM-ISH, and the cells on the particles surface that hybridized with EUB338 were visualized under SEM. SE images of sediment particles were obtained at magnifications of ×250 (Fig. 4a), ×1,000 (Fig. 4d), and ×5,000 (Fig. 4g) and confirmed the presence of bacterial cells by using SE signals that correspond to topographic information. By comparing the BSE images from the same microscopic fields, the probe signals from the hybridized cells were detected (Fig. 4b, e, and h). The composite images are represented in Fig. 4c, f, and i. SEM-ISH with the EUB338 probe proved that bacterial cells were distributed all over the surface of particles.
FIG. 4.
High-vacuum SEM images of bacteria attached on surface of river sediment particles detected by ISH with the EUB338 probe. The same microscopic fields are shown with SE image (a, d, and g), BSE image (b, e, and h), and their composite image (c, f, and i). The topographic information was obtained with SE images (a, d, and g), and cells hybridized with the EUB338 probe were detected with the BSE image (b, e, and h). The middle areas of the micrographs a, b, and c were magnified in micrographs d, e, and f, whose middle area was further magnified in micrographs g, h, and i. Magnifications: a, b, and c, ×250; d, e, and f, ×1,000; g, h, and i, ×5,000.
Fluorescence microscopy and confocal laser scanning microscopy have been key tools in examining complex microbial communities attached on various materials in their natural habitat effectively. Potential problems with these fluorescent techniques include autofluorescence (4, 24), which results from natural substances within plant tissue, organic debris, soil particles, etc. This may hamper the accurate enumeration of target microbes in complex microbial communities. SEM-ISH allowed visualization of cells attached on sediment particles without interference by autofluorescence or requiring ultrathin sectioning of sediment particles.
Bacterial community composition of river sediment.
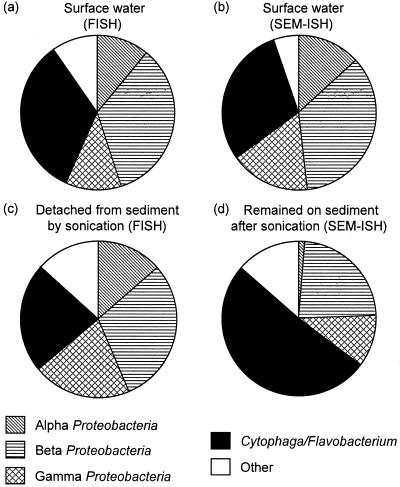
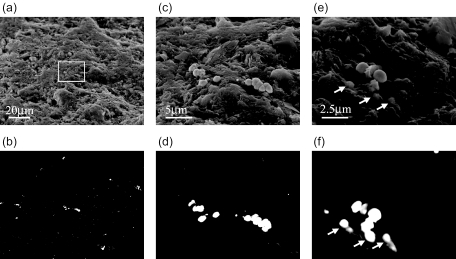
Community composition in sediment samples in which cells were detached by sonication, were investigated and compared to those in surface river water. FISH demonstrated that ca. 60% of DAPI-stained cells hybridized with the EUB338 probe in the samples, and the Cytophaga-Flavobacterium cluster and beta subclass Proteobacteria were abundant in both surface water (Fig. 5a) and sediment (Fig. 5c) samples. Similar results were obtained by SEM-ISH (Fig. 5b). SEM-ISH for untreated sediment samples also revealed significant abundance of the Cytophaga-Flavobacterium cluster on the surface of sediment particles and confirmed a wide distribution over the particle surface (Fig. 6a and b). A microcolony of members of Cytophaga-Flavobacterium on the particle surface was also detected by SEM-ISH with a CF319 probe (Fig. 6c and d). When observed at high magnification, certain bacterial cells were found to be buried in the particles, as indicated by arrows in Fig. 6e and f. Cell boundaries were unclear; thus, it was difficult to distinguish individual cells within the particle structure using only SE signals. BSE images provided the probe signals from the hybridized cells in the same microscopic fields and clarified the existence of buried cells. SEM-ISH with rRNA-targeted probes identified the buried cells based on rRNA sequence.
FIG. 5.
Overview of the bacterial community structure of surface river water (a and b) and river sediments (c and d) as revealed by FISH (a and c) and SEM-ISH (b and d). The fractions shown indicated the percentage of total cells (a and b) and the relative percentages of EUB338 counts (c and d), respectively. Bacterial community detached from sediment particles by sonication (c) and bacterial community remained on surface of sediment particles after sonication (d) were classified by FISH and SEM-ISH with rRNA-targeted probes, respectively.
FIG. 6.
High-vacuum SEM images of bacteria attached on surface of river sediment particles detected by ISH with the CF319 probe. The same microscopic fields are shown with SE image (a, c, and e), BSE image (b, d, and f). The topographic information was obtained with SE images (a, c, and e), and cells hybridized with the CF319 probe were detected with the BSE image (b, d, and f). Microcolony in micrographs a and b was magnified in micrographs c and d. Certain bacterial cells were buried in the particles as indicated by arrows (e and f). Magnifications: a and b, ×1,000; c and d, ×5,000; e and f, ×10,000.
In order to investigate the composition of the bacterial community which could not be recovered by sonication, sediment particles were subjected to SEM-ISH with rRNA-targeted probes after 5 min of sonication. SEM-ISH revealed that cells still remained on particle surfaces even when excess sonication (5 min) was used. The cells hybridized with each probe were enumerated in an electron microscopic field at a magnification of ×3,000, and the community composition was represented as the percentage of the EUB338 count (Fig. 5d). Compared to results on the bacterial community detached by sonication (Fig. 5c), the Cytophaga-Flavobacterium cluster remaining on the particles constituted a higher percentage (51%) than for the population detached from the particles by sonication (23%). That is, members of Cytophaga-Flavobacterium cluster existed together tightly on the particle surface and were not easily dislodged by sonication.
High numbers of Cytophaga-Flavobacterium were also found in sea sediments, where they constituted between 15 and 25% of the EUB338 counts (20), and in water columns associated with marine aggregates (9). Cytophaga-Flavobacterium and β-Proteobacteria have been reported to dominate river biofilms, surface river water, and particle-attached communities in river (5, 8, 15, 23, 27). The members of Cytophaga-Flavobacterium may be common in freshwater system and sometimes constitute a considerable fraction. They may play an important role in the turnover of organic matter (7).
Conclusions and future prospects.
Morphological characteristics do not provide phylogenetic or genetic information about target microbes. Few methods have been available for identifying and enumerating specific bacteria or bacterial groups in the natural environment by electron microscopy except via antibody labeling or ISH technique based on transmission electron microscopy (35). In the present study, SEM-ISH was developed for scanning electron microscopic studies of complex microbial communities in natural samples based on rRNA sequences. The composition of the bacterial community on river sediment particles was revealed by SEM-ISH with rRNA-targeted probes.
SEM-ISH also offers the potential to refine microbial study with epifluorescence microscopy. The presence of enormous numbers of bacterium-sized detritus and dissolved organic carbon that aggregates abiotically to particles 0.4 to 0.8 μm in diameter may be a source of error in enumerating aquatic microbes (17). Fluorochrome-stainable particles may be retained tightly on glass slides or polycarbonate filters and detectable by image intensity enhancement. In contrast, SEM enables cell images to be magnified, and thus the shape, size, surface morphology, cell location, and adhesion property can be examined in more detail than by fluorescence microscopy. These advantages of SEM promote easier discrimination between bacterial cells and others. In addition, the ISH technique described here allows the detection of target cells on the basis of phylogenic information. The SEM-ISH technique may give us valuable information about accurate enumeration of target cells in the natural environment.
SEM has been widely used for quantitative measurement of biofilm formation under nutrient limitation or in a continuous water flow system (1, 38), morphological characterization of waterborne bacteria (37), and estimation of biomass in aquatic environments (39). Combining morphological study with SEM and ISH techniques leads to new understanding about the spatial distribution of target cells, as well as the extent of cell heterogeneity on plant, metal, alloy, bioreactor, or the three-dimensional structure of the attachment matrix.
Acknowledgments
This study was supported by the Development of Monitoring Methods for Microorganisms in the Environment Project supported by the New Energy and Industrial Technology Development Organization, Tokyo, Japan.
REFERENCES
- 1.Allan, V. J., M. E. Callow, L. E. Macaskie, and M. Paterson-Beedle. 2002. Effect of nutrient limitation on biofilm formation and phosphatase activity of a Citrobacter sp. Microbiology 148:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Alfreider, A., J. Pernthaler, R. Amann, B. Sattler, F. O. Glöckner, A. Wille, and R. Psenner. 1996. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl. Environ. Microbiol. 62:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brümmer, I. H. M., A. D. M. Felske, and I. Wagner-Döbler. 2004. Diversity and seasonal changes of uncultured Planctomycetales in river biofilms. Appl. Environ. Microbiol. 70:5094-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 10.Ferguson, D. J., J. Burns, D. Harrison, J. A. Jonasson, and J. O. McGee. 1986. Chromosomal localization of genes by scanning electron microscopy using in situ hybridization with biotinylated probes: Y chromosome repetitive sequences. Histochem J. 18:266-270. [DOI] [PubMed] [Google Scholar]
- 11.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacker, G. W. 1998. High performance Nanogold-silver in situ hybridisation. Eur. J. Histochem. 42:111-120. [PubMed] [Google Scholar]
- 13.Hermann, R., P. Walther, and M. Muller. 1996. Immunogold labeling in scanning electron microscopy. Histochem. Cell Biol. 106:31-39. [DOI] [PubMed] [Google Scholar]
- 14.Ishii, K., M. Muβmann, B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 15.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology 144:2085-2093. [DOI] [PubMed] [Google Scholar]
- 16.Kenzaka, T., N. Yamaguchi, B. Prapagdee, E. Mikami, and M. Nasu. 2001. Bacterial community composition and activity in urban rivers in Thailand and Malaysia. J. Health Sci. 47:353-361. [Google Scholar]
- 17.Kerner, M., H. Hohenberg, S. Ertl, M. Reckermann, and A. Spitzy. 2003. Self-organization of dissolved organic matter to micelle-like microparticles in river water. Nature 422:150-154. [DOI] [PubMed] [Google Scholar]
- 18.Kurokawa, K., K. Tani, M. Ogawa, and M. Nasu. 1999. Abundance and distribution of bacteria carrying sltII gene in natural river water. Lett. Appl. Microbiol. 28:405-410. [DOI] [PubMed] [Google Scholar]
- 19.Lebaron, P., P. Catala, C. Fajon, F. Joux, J. Baudart, and L. Bernard. 1997. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl. Environ. Microbiol. 63:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llobet-Brossa, E., R. Rossello-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, F., J. Lukasik, and S. R. Farrah. 2001. Immunological methods for the study of Zoogloea strains in natural environments. Water Res. 35:4011-4018. [DOI] [PubMed] [Google Scholar]
- 22.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 23.Manz, W., R., Amann, W., Ludwing, M., Vancanneyt, and K. H. Schleifer. 1996. Application of a suite 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 24.Manz, W., K. Wendt-Potthoff, T. R. Neu, U. Szewzyk, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridisation and confocal laser scanning microscopy. Microb. Ecol. 37:225-237. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama, F., T. Kenzaka, N., Yamaguchi, K. Tani, and M. Nasu. 2003. Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification. Appl. Environ. Microbiol. 69:5023-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa, M., K. Tani, N. Yamaguchi, and M. Nasu. 2003. Development of multicolor digital image analysis system to enumerate actively respiring bacteria in natural river water. J. Appl. Microbiol. 95:120-128. [DOI] [PubMed] [Google Scholar]
- 27.O'Sullivan, L. A., A. J. Weightman, and J. C. Fry. 2002. New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in river Taff Epilithon. Appl. Environ. Microbiol. 68:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernthaler, A., J. Pernthaler, M. Schattenhofer, and R. Amann. 2002. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sammons, R., and P. Marquis. 1997. Application of the low vacuum scanning electron microscope to the study of biomaterials and mammalian cells. Biomaterials 18:81-86. [DOI] [PubMed] [Google Scholar]
- 33.Scopsi, L., L. I. Larsson, L. Bastholm, and M. H. Nielsen. 1986. Silver-enhanced colloidal gold probes as markers for scanning electron microscopy. Histochemistry 86:35-41. [DOI] [PubMed] [Google Scholar]
- 34.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spring, S., U. Lins, R. Amann, K. H. Schleifer, L. C. S. Ferreira, D. M. S. Esquivel, and M. Farina. 1998. Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch. Microbiol. 169:136-147. [DOI] [PubMed] [Google Scholar]
- 36.Stierhof, Y. D., B. M. Humbel, and H. Schwarz. 1991. Suitability of different silver enhancement methods applied to 1 nm colloidal gold particles: an immunoelectron microscopic study. J. Electron. Microsc. Tech. 17:336-343. [DOI] [PubMed] [Google Scholar]
- 37.Sundaram, S., S. Mallick, J. Eisenhuth, G., Jr. Howard, and H. Brandwein. 2001. Retention of waterborne bacteria by membrane filters. II. Scanning electron microscopy (SEM) and fatty acid methyl ester (FAME) characterization of bacterial species recovered downstream of 0.2/0.22 micron rated filters. PDA J. Pharm. Sci. Technol. 55:87-113. [PubMed] [Google Scholar]
- 38.Sutton, N. A., N. Hughes, and P. S. Handley. 1994. A comparison of conventional SEM techniques, low temperature SEM and the electroscan wet scanning electron microscope to study the structure of a biofilm of Streptococcus crista CR3. J. Appl. Bacteriol. 76:448-454. [DOI] [PubMed] [Google Scholar]
- 39.Tani, K., J. M. Chen, N. Yamaguchi, and M. Nasu. 1996. Estimation of bacterial biovolume and biomass by scanning electron microscopic image analysis. Microb. Environ. 1:11-17. [Google Scholar]
- 40.Tani, K., K. Kurokawa, and M. Nasu. 1998. Development of a direct in situ PCR method for detection of specific bacteria in natural environments. Appl. Environ. Microbiol. 64:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallner, G., R. I. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 42.Yuda, A. A., R. M. Weiner, J. A. Johnson, C. E. De Rezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 67:4048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]