Abstract
The HRX gene (also called MLL, ALL-1, and Htrx) at chromosome band 11q23 is associated with specific subsets of acute leukemias through translocations that result in its fusion with a variety of heterologous partners. Two of these partners, ENL and AF9, code for proteins that are highly similar to each other and as fusions with HRX induce myeloid leukemias in mice as demonstrated by retroviral gene transfer and knock-in experiments, respectively. In the present study, a structure-function analysis was performed to determine the molecular requirements for in vitro immortalization of murine myeloid cells by HRX-ENL. Deletions of either the AT hook motifs or the methyltransferase homology domain of HRX substantially impaired the transforming effects of HRX-ENL. The methyltransferase homology domain was shown to bind non-sequence specifically to DNA in vitro, providing evidence that the full transforming activity of HRX-ENL requires multiple DNA binding structures in HRX. The carboxy-terminal 84 amino acids of ENL, which encode two predicted helical structures highly conserved in AF9, were necessary and sufficient for transformation when they were fused to HRX. Similarly, mutations that deleted one or both of these conserved helices completely abrogated the transcriptional activation properties of ENL. This finding correlates, for the first time, a biological function of an HRX fusion partner with the transforming activity of the chimeric proteins. Our studies support a model in which HRX-ENL induces myeloid transformation by deregulating subordinate genes through a gain of function contributed by the transcriptional effector properties of ENL.
Chromosomal aberrations affecting band 11q23 are the most commonly observed cytogenetic alterations in infant acute lymphoblastic leukemias and in subtypes of acute myeloid leukemias (9, 29). Similar genetic lesions are prevalent in secondary leukemias arising after treatment of neoplastic diseases with topoisomerase II inhibitors (14, 15). Chromosomal translocations of 11q23 to 30 different cytogenetic loci have been found in leukemic cells (for reviews, see references 3 and 43). A molecular analysis revealed the presence of a large reading frame coding for 3,968 amino acids spanning the common chromosomal breakpoint at 11q23. Because of a regionally limited but significant homology to the Drosophila trithorax gene (TRX), the gene was designated HRX (also named ALL-1, HTRX, or MLL) (13, 17, 42, 47). TRX is responsible for the maintenance, but not initiation, of HOX gene expression in flies (5, 23). Knockout studies of mice suggest a similar role for HRX in mammals (46).
To date, 12 different fusion partners involved in chromosomal translocations affecting HRX have been cloned and sequenced (reviewed in references 3 and 43). Additionally, internal duplications within the amino-terminal part of HRX and specific deletions of exon 8 have been identified in leukemic blast cells (25, 34, 35). Despite this wealth of information, the genetic mechanisms by which mutations in HRX cause transformation are still elusive. All identified fusion transcripts join HRX in frame with a respective partner and support the potential expression of a full-length fusion protein. This is indicative of a functional contribution of the fusion partner and suggests a gain-of-function mechanism, although alternative mechanisms have been proposed (22, 27, 34, 35, 46). Indeed, an emerging theme is that many of the known fusion partners seem to be capable of interacting with the RNA polymerase II transcription machinery. Several fusion partners (AF4, AF9, and ENL) appear to be bona fide transcription factors and have been shown to transactivate certain promoters in vivo (28, 32). Two fusion partners are directly involved in transcriptional regulation. These are the RNA polymerase II elongation factor ELL and the transcriptional coactivator CBP (CREB binding protein) (37, 38, 40).
Further support for a decisive role of the fusion partners comes from knock-in studies, showing that mice expressing an HRX-AF9 fusion under the natural HRX promoter develop acute myeloid leukemia whereas mice engineered to express an HRX-Myc tag fusion show no sign of disease (10). Recently, we demonstrated that retrovirus-mediated gene transfer of HRX-ENL into murine hematopoietic progenitors led to their immortalization and that this activity required the presence of the ENL moiety of the fusion protein (24). To further study the contribution of HRX and ENL, we took advantage of this myeloid methylcellulose assay that identifies the transforming properties of HRX-ENL. We tested a series of HRX-ENL deletion mutants to investigate the participation of several conserved sequence motifs to the oncogenic activity of the fusion product. We demonstrate here that a biological function of an HRX fusion partner is critical for the oncogenic activity of the resulting fusion product. These results support a model in which 11q23 translocations create HRX fusion proteins with a new combination of functional domains that transform hematopoietic cells by a gain-of-function mechanism.
MATERIALS AND METHODS
Plasmid and mutant construction.
The 6-kb HRX-ENL cDNA (42) was inserted into the EcoRI site of the retroviral vector pMSCV-neo (pMSCV/HRX-ENL [24]) and used to construct a series of nested deletions by the method of Henikoff (19). Double-stranded oligonucleotides containing unique NsiI and NruI sites were inserted either at the amino terminus, at the carboxy terminus, or into the PflMI site 153 nucleotides upstream of the fusion point between HRX and ENL (numbering according to sequences deposited under GenBank accession no. L04284 and L04285). An additional FLAG tag sequence (ATGGACTACAAGGACGACGATGACAAG) was encoded in the amino-terminally inserted oligonucleotide. TAA codons in all three reading frames were incorporated into the carboxy-terminal oligonucleotide. After digestion with NsiI and NruI and exonuclease III treatment, the resulting deletion clones were sequenced and suitable constructs were selected. The following clones were used in this study (numbers in parentheses indicate deleted nucleotides): pΔHN1 (1 to 854), pΔHN2 (1 to 1841), pΔHN3 (1 to 2494), pΔHC1 (3800 to 4184), pΔHC2 (3112 to 4184), pΔHC3 (2118 to 4184), pΔEC1 (5952 to 5999), and pΔEC2 (5818 to 5999) (Fig. 1).
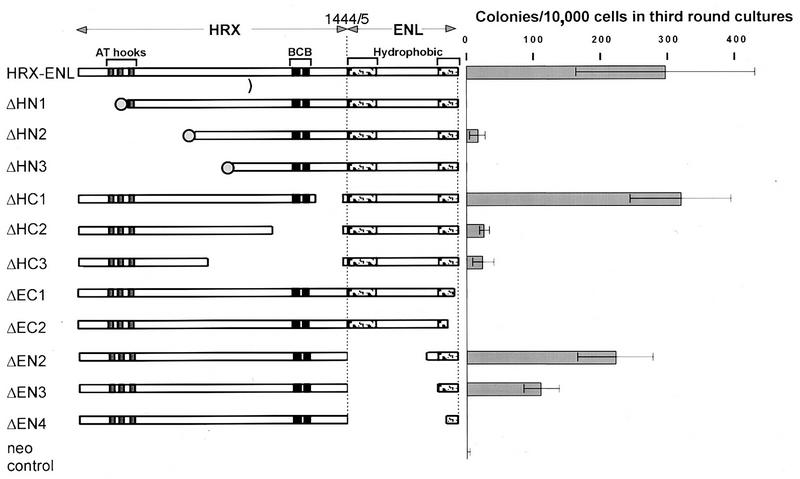
FIG. 1.
In vitro proliferative effects of HRX-ENL mutants. HRX-ENL chimeric proteins are shown schematically, with filled boxes denoting the three AT hook motifs homologous to HMG-I(Y) proteins and the BCB domain that shows similarity with mammalian DNMT. Stippled boxes indicate amino- and carboxy-terminal regions of ENL that show homology with AF9 and more distantly with yeast ANC1. An amino-terminal FLAG epitope tag is depicted by a filled circle. Gaps indicate deletions. Numbers at the top indicate amino acids at fusion sites in HRX and ENL, respectively. The proliferative capacity of hematopoietic progenitors transduced with different constructs was measured in a myeloid methylcellulose assay. The bar graph represents the number of colonies per 104 input cells in a third round of serial methylcellulose plating (mean ± standard error of the mean, n ≥ 3).
Additional mutants of pMSCV/HRX-ENL were generated by fusing various carboxy-terminal portions of ENL directly to HRX. ENL fragments were amplified by PCR and inserted into the ApaLI site at the junction of HRX and ENL. Clones obtained this way are labeled pΔEN2 (5613), pΔEN3 (5749), and pΔEN4 (5887), with the numbers in parentheses indicating the first nucleotide of ENL sequence in the clone. All PCR products were completely sequenced to exclude errors introduced by Taq polymerase.
Comparable C-terminal portions of ENL were fused to the GAL4 DNA binding domain (amino acids 1 to 147) in the vector pSG424 (33). For DNA binding studies of the BCB (basis-cysteine-basic) region, HRX sequences encoding amino acids 1147 to 1244 were amplified by PCR and inserted into the correct reading frame of pGEX-3X (pGST-MT; Pharmacia, Uppsala, Sweden). The sequences of constructs and oligonucleotides used for these studies are available upon request.
Western and Southwestern blotting.
Nuclear extracts of transiently transfected Bosc23 cells were prepared by using high-salt buffer (500 mM NaCl, 20 mM HEPES [pH 7.5], 0.5 mM EDTA, 0.1% Triton X-100, 0.5 mM sodium vanadate, 2 mM NaF, 2 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, aprotinin [40 μg/ml], leupeptin [20 μg/ml], and pepstatin A [40 μg/ml]) and centrifugation. Protein samples (40 to 60 μg per lane) were electrophoresed in a sodium dodecyl sulfate (SDS)–5% polyacrylamide gel and blotted onto polyvinylidene fluoride membranes (Immobilon-P; Millipore, Bedford, Mass.) by tank blotting using an alkaline transfer buffer {10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; pH 11.0), 0.1% SDS, 1% methanol}. Membranes were blocked in 5% nonfat dry milk in phosphate-buffered saline with 0.2% Tween 20 and incubated with either the monoclonal antibody HRX107, an anti-FLAG antibody (Kodak, Rochester, N.Y.), or a rabbit polyclonal serum raised against ENL (7). Proteins were detected with horseradish peroxidase-conjugated secondary antibodies in a standard chemiluminescence Western blotting protocol (ECL system; Amersham, Arlington Heights, Ill.). GAL4 fusion proteins were transferred in Towbin buffer (15.6 mM Tris, 120 mM glycine, 15% methanol) and detected with a monoclonal antibody against the GAL4 DNA binding domain (Clontech, Palo Alto, Calif.).
For Southwestern blotting, glutathione S-transferase (GST)–BCB or GST proteins were purified by affinity chromatography on glutathione-Sepharose (Pharmacia) as recommended by the manufacturer. Proteins were spotted onto nitrocellulose membranes, which were then blocked for 1 h at room temperature in incubation buffer (30 mM HEPES [pH 7.9], 50 mM NaCl, 5 mM dithiothreitol, 10 μM ZnSO4, 0.5% bovine serum albumin). pBluescript bacterial plasmid and sonicated salmon sperm DNA were radiolabeled by random priming; poly(dI-dC) (Sigma, St. Louis, Mo.) was end labeled by using T4 polynucleotide kinase by following standard procedures. Probes were added to the incubation buffer without further denaturation and allowed to bind to the immobilized proteins for 1.5 h at room temperature. Following two washes in incubation buffer, the membranes were exposed to X-ray film.
Infection of primitive progenitors and methylcellulose colony forming assays.
The production of retroviral supernatants, infection of primitive hematopoietic progenitors, and their culture in methylcellulose were conducted as previously described (24), with the following modifications. Bone marrow cells from 5-fluorouracil-treated B.A1 mice (C57BL/Ka.AKR/J) depleted in mature myeloid and lymphoid cells were cultured overnight with interleukin-3 (IL-3), IL-6 (10 ng/ml), and stem cell factor (100 ng/ml). The cells were then infected by spinoculation, which was repeated on the next day. On the day following the second spinoculation, an equivalent of 2 × 103 to 5 × 103 cells of the starting lineage-depleted population was seeded per 1.1 ml of Methocult M3230 methylcellulose medium (Stem Cell Technologies Inc., Vancouver, British Columbia, Canada) supplemented with 10 ng each of murine recombinant IL-3, IL-6, and granulocyte-macrophage colony-stimulating factor (R&D Systems, Minneapolis, Minn.) per ml and 100 ng of murine recombinant stem cell factor (gift from Sandoz) per ml with or without 1.3 mg of G418 (Gibco BRL, Gaithersburg, Md.) per ml. Comparable percentages of G418-resistant colonies ensured that the retroviral titers obtained with the different constructs were similar. This was confirmed by performing a parallel titration on NIH 3T3 cells. Growth of the cells transduced with the different constructs was assessed during the course of four rounds of serial methylcellulose cultures by scoring the number of colonies and studying their morphology. Results presented in Fig. 1 correspond to the cloning efficiency at the third passage measured as the mean of results with duplicate dishes seeded with 10,000 cells pooled from secondary colonies. Averages and error margins were determined from at least three independent transduction experiments (except for construct ΔHC1, studied in only two experiments).
Transcriptional transactivation assays.
REH cells (3 × 106) were electroporated with 1 μg each of GAL4 fusion construct and pGAL4-TKcat reporter plasmid DNAs in RPMI–10% fetal calf serum containing 5 μg of DEAE-dextran per ml at 250 V and 960 μF in a 0.4-mm cuvette (Bio-Rad electroporator). At 48 h after electroporation, expression of the chloramphenicol acetyltransferase (CAT) reporter gene was assayed by a standard chloramphenicol conversion assay. Results and error margins were determined from four independent experiments.
RESULTS
Retroviral transduction of primary myeloid colony-forming cells with mutated forms of HRX-ENL.
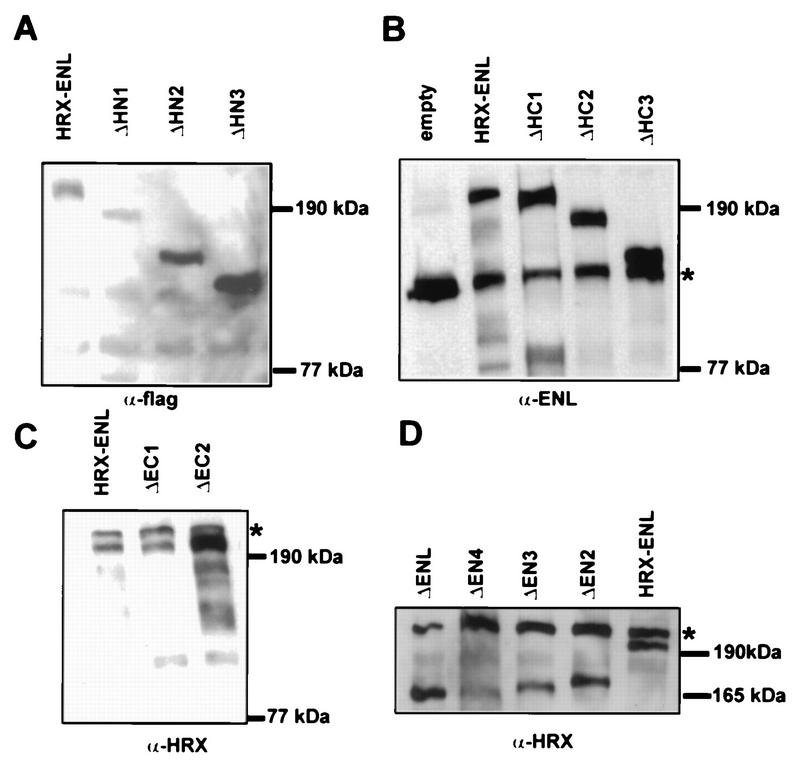
To investigate the molecular requirements for HRX-ENL-mediated transformation, a structure-function analysis was performed to determine which portions of HRX and ENL were necessary for in vitro immortalization of murine hematopoietic cells. Various mutated forms of HRX-ENL (Fig. 1) were constructed in the retroviral vector pMSCV/neo for expression under the control of the retroviral long terminal repeat (18). High-titer retroviral stocks were generated by transient transfections of the plasmid constructs into the ecotropic packaging cell line Bosc23 and then used for infection of primary bone marrow cells that were enriched for progenitor and stem cells (24). In previous studies, we showed that expression of HRX-ENL under these conditions resulted in the immortalization and transformation of murine myelomonocytic precursors (24). In the present study, the numbers and morphologic appearances of colonies at the third round of serial plating in methylcellulose cultures were used as criteria for assessing the in vitro transformation abilities of HRX-ENL proteins. Appropriate expression of each protein was confirmed by Western blot analysis of nuclear extracts prepared from transfected Bosc23 cells. All proteins expressed by the engineered constructs displayed the expected apparent molecular weights in SDS-polyacrylamide gel electrophoresis (Fig. 2) and were correctly imported into the nucleus. Expression levels of mutant proteins were comparable to that obtained for HRX-ENL. Several constructs contained additional amino-terminal sequences encoding the FLAG epitope tag, which did not impair the transformation ability of full-length HRX-ENL (data not shown).
FIG. 2.
Analysis of mutant HRX-ENL protein expression levels in transfected Bosc23 cells. Nuclear proteins (40 to 60 μg per lane) isolated from cells transiently transfected with the indicated constructs were subjected to Western blot analysis using either a monoclonal anti-FLAG (specificity M2) antibody (A), a polyclonal antiserum against ENL (B), or a monoclonal anti-HRX antibody (HRX107) (C and D). Asterisks indicate endogenous ENL (B) or breakdown products (7) from endogenous HRX (C and D). The positions of molecular mass markers are indicated.
HRX-ENL requires its AT hook and methyltransferase homology domains to exert its full transforming effects on primary myeloid progenitors.
A series of deletion mutants was used to establish the portions of HRX required for in vitro transformation by HRX-ENL. Three of these constructs (ΔHN1, ΔHN2, and ΔHN3) contained increasing deletions of the amino terminus of HRX-ENL. They removed part (ΔHN1) or all (ΔHN2 and ΔHN3) of the AT hook motifs, which have homology with HMG-I(Y) proteins, but did not extend into the region of similarity with DNA methyltransferases (DNMT) (Fig. 1). Progenitors transduced with two of the mutants (ΔHN1 and ΔHN3) did not demonstrate any proliferative advantage compared to cells infected with the empty vector, as they had practically completely exhausted their clonogenic potential after two rounds of methylcellulose plating. Cells transduced with the ΔHN2 mutant generated a slightly higher number of tertiary colonies than the control (16 ± 12 versus 2.8 ± 1.8). These colonies were comprised of tightly aggregated cells reminiscent of the compact primitive colonies generated by full-length HRX-ENL. They were, however, smaller than those induced by HRX-ENL and generally did not proliferate upon a fourth round of plating, suggesting that this construct had lost most of its proliferative effect. Taken together, these observations indicated that amino-terminal HRX sequences containing the AT hook motifs make an essential contribution to the in vitro oncogenic properties of HRX-ENL.
We conducted a similar analysis of mutants with deletions initiating at different internal sites in the coding region of HRX and terminating at the HRX-ENL fusion site (ΔHC1, ΔHC2, and ΔHC3). Deletion of 128 HRX amino acids near the fusion region between HRX and ENL (mutant ΔHC1) but sparing the methyltransferase homology domain showed no adverse effects on transforming activity, as the colonies generated were similar in appearance and numbers to those obtained with full-length HRX-ENL (Fig. 1). In contrast, larger deletions that encompassed the region of homology with methyltransferases (ΔHC2 and ΔHC3) substantially diminished the transformation potency of HRX-ENL. However, they did not appear to completely eliminate it, since the numbers of third-generation colonies were slightly, but reproducibly, above that observed in vector control cultures (26 ± 8 and 24 ± 16 for ΔHC2 and ΔHC3, respectively). The few colonies that were observed displayed a somewhat diffuse morphology unlike that of the compact colonies induced by HRX-ENL. Cells harvested from these colonies displayed a limited proliferation capability and did not generally yield daughter colonies in subsequent platings. Thus, the methyltransferase homology domain as well as the AT hook region of HRX appeared to be essential for the in vitro oncogenic effects of HRX-ENL.
The methyltransferase homology domain of HRX binds DNA.
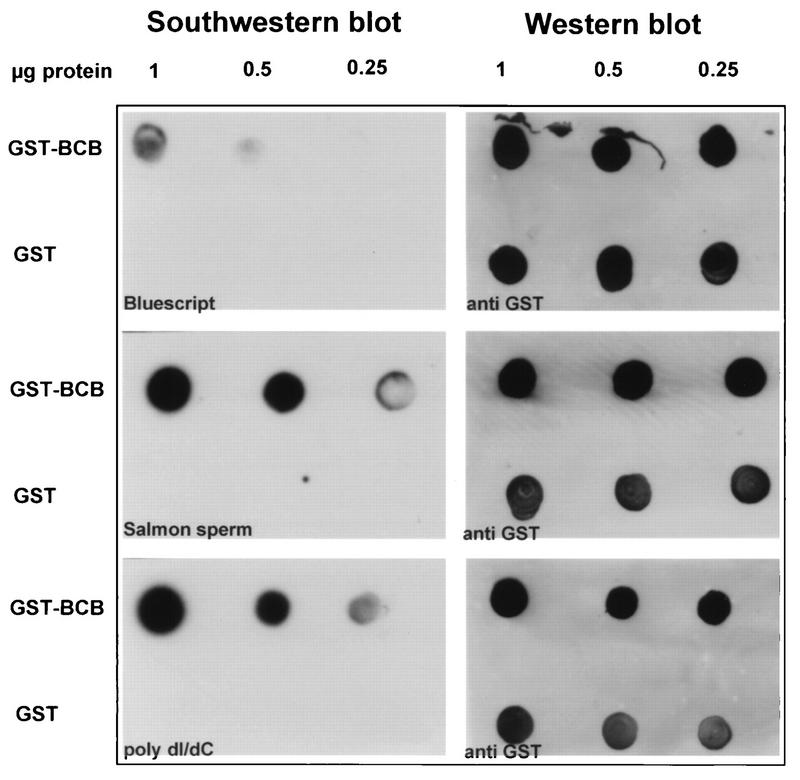
Of the two HRX-derived domains of HRX-ENL established above to be fundamental for in vitro proliferation, the one containing the AT hook motifs has previously been shown to possess DNA binding properties (6). The other HRX domain is conserved in DNMT (Fig. 3A) within a region necessary for specific binding to methylated DNA (4), but its actual contributions to DNA recognition have not been established. The sequence of this homology region includes short clusters of cysteines flanked by basic amino acids (hence the term BCB, for basic-cysteine-basic motif). To test for the potential of this region to bind DNA, it was fused to GST and the purified GST-BCB protein was immobilized on membranes for Southwestern blot analyses. GST protein alone served as a negative control. Salmon sperm DNA and poly(dI-dC) were used as probes; the latter possesses conformational features that are recognized and bound by DNMT like hemimethylated DNA despite the absence of methylation. GST-BCB bound radiolabeled salmon sperm DNA and the synthetic substrate poly(dI-dC) under conditions where no binding was observed with GST alone. Less binding was obtained when the blot was probed with bacterial plasmid DNA (Fig. 4). These observations indicated that the methyltransferase homology domain can bind DNA in vitro without a pronounced sequence specificity, features that are also characteristic of the AT hook motifs (6, 30).
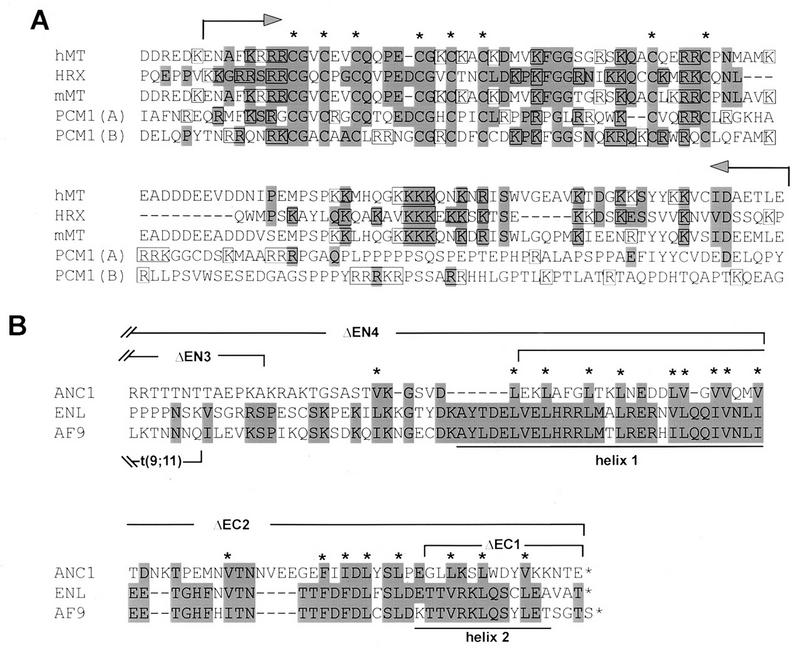
FIG. 3.
Comparative alignments of conserved regions of HRX and ENL required for the oncogenic activity of HRX-ENL. (A) Comparison of HRX amino acid sequence (residues 1141 to 1244) with mouse and human DNMT (mMT and hMT) and the PCM1 component of the MeCP1 repressor complex [PCM1(A) amino acids 162 to 280 and PCM1(B) amino acids 275 to 395]. Amino acids conserved or conservatively replaced in HRX are highlighted. Conserved cysteine residues are marked by asterisks, and basic amino acids are boxed. The portion of HRX fused to GST for the in vitro DNA binding assays is indicated by arrows. (B) Comparison of the carboxy terminus of ENL (residues 463 to 559) with those of AF9 and ANC1 (21, 27, 42, 44). The highlighted amino acids are conserved or conservatively replaced in ENL. Asterisks denote hydrophobic residues consistently found in all three proteins. Brackets denote the extent of deletions in the indicated mutant proteins. Predicted helices 1 and 2 are underlined.
FIG. 4.
DNA binding activity of the methyltransferase homology (BCB) region demonstrated by Southwestern blot analysis. The GST-BCB fusion protein (1, 0.5, and 0.25 μg) was spotted onto nitrocellulose membranes in parallel with similar amounts of GST as a control. The membranes were incubated with the indicated radiolabeled DNA probes, and bound DNAs were detected by autoradiography (left). To ensure that comparable amounts of test and control proteins were applied, GST-immunoreactive proteins were detected on the same membranes by Western blotting with an anti-GST monoclonal antibody (right).
The conserved, carboxy-terminal, transcriptional activation domain of ENL is essential for in vitro transformation of myeloid progenitors by HRX-ENL.
In previous studies, we demonstrated that fusion of HRX with ENL confers a gain of function that is required for alterations of the in vitro growth and self-renewal potentials of primary myelomonocytic precursors (24). To determine which portions of ENL were required for induction of this phenotype, we constructed a series of HRX-ENL deletion mutants that progressively removed amino- or carboxy-terminal ENL amino acids. Assessment of the colony formation of hematopoietic progenitors transduced with the resultant constructs indicated that most of the amino-terminal portion of ENL was dispensable for in vitro transformation. Two HRX-ENL constructs lacking 430 and 475 N-terminal ENL amino acids (constructs ΔEN2 and ΔEN3, respectively) retained the ability to stimulate the proliferation and replatability of progenitor cells (Fig. 1) which displayed only modest decreases in cloning efficiencies in the third round of methylcellulose cultures (ΔEN2 and ΔEN3 generated 219 ± 56 and 109 ± 26 colonies, respectively, compared to 293 ± 132 colonies obtained with HRX-ENL). The deletion in construct ΔEN3 preserved the last 84 carboxy-terminal amino acids of ENL containing two predicted helical structures that are highly conserved with HRX fusion partner AF9 (Fig. 3B). However, a deletion extending further C terminal that removed ENL amino acids comprising helix 1 but preserved helix 2 (construct ΔEN4) completely abrogated the proliferative effect of HRX-ENL. These analyses demonstrated that the 84 carboxy-terminal amino acids of ENL were sufficient to confer growth-altering properties when they were fused to HRX. This was further addressed by analyzing the effects of HRX-ENL carboxy-terminal deletions that removed one (ΔEC1) or both (ΔEC2) of the predicted helical structures conserved with AF9 (Fig. 3B). Both mutations completely abolished the ability of HRX-ENL to sustain the proliferation of primary hematopoietic cells in the third round of methylcellulose plating (Fig. 1). Therefore, the carboxy-terminal 84 amino acids of ENL are necessary and sufficient when fused to HRX to alter the in vitro growth and self-renewal properties of primary myeloid cells.
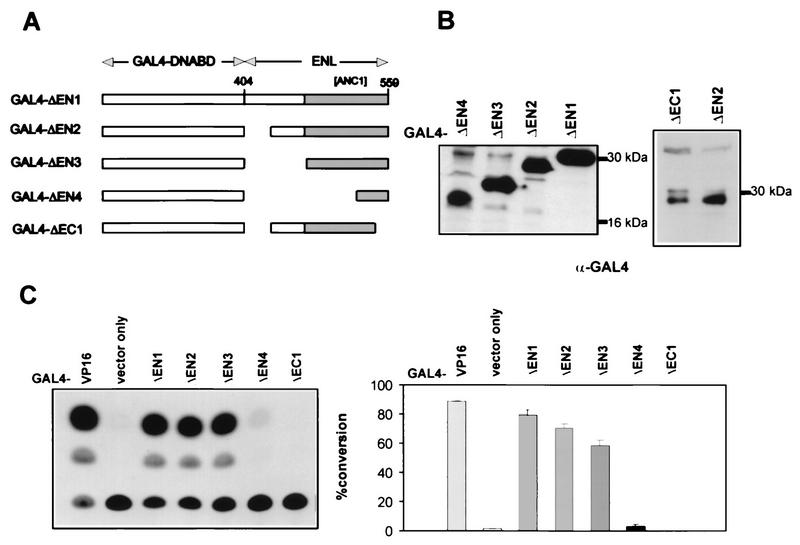
Previous studies have shown that the carboxy-terminal half of ENL displays promoter-specific transactivation properties in mammalian and yeast cells (32). We extended these analyses to determine whether the region encompassing the transactivation activity of ENL mapped with the carboxy-terminal fragment essential to the proliferative effect of HRX-ENL. Various carboxy-terminal portions of ENL were fused to the heterologous GAL4 DNA binding domain (Fig. 5A), and the integrity of the clones was confirmed by Western blot analysis of protein extracts obtained by transient transfection of Bosc23 cells (Fig. 5B). The GAL4 fusion clones were then assessed for the ability to activate transcription of a reporter gene under control of the herpes simplex virus thymidine kinase promoter after cotransfection into the REH pre-B-cell line. Under these conditions, the carboxy-terminal 156 amino acids of ENL (construct GAL4-ΔEN1) displayed transcriptional activation potential comparable to that of GAL4-VP16 (Fig. 5C). Transactivation was only modestly decreased with constructs containing smaller C-terminal fragments of ENL (GAL4-ΔEN2 and GAL4-ΔEN3), the latter containing the minimal 84 amino acids of ENL required for in vitro transformation. In contrast, a deletion extending into this conserved domain that removed helix 1 (construct GAL4-ΔEN4) reduced transactivation to less than 5%, whereas deletion of helix 2 in construct GAL4-ΔEC1 abolished it completely (Fig. 5C). These studies established a complete correlation between the transactivation properties of ENL and its contributions to the growth-altering properties of HRX-ENL.
FIG. 5.
Localization of the transcriptional transactivation region of ENL. (A) Schematic depictions of the structures of GAL4 DNA-binding-domain fusion constructs containing carboxy-terminal portions of ENL. The filled bar indicates the region of ENL similar to AF9 and ANC1. Numbers refer to amino acid positions in wild-type ENL. (B) Comparison of levels of expression by the different GAL4-ENL constructs. Total cellular proteins (60 μg) extracted from transiently transfected Bosc23 cells were subjected to Western blot analysis using a monoclonal antibody specific for the GAL4 DNA binding domain. The positions of molecular mass markers are indicated. (C) Transcriptional transactivation capability of GAL4-ENL proteins was measured by cotransfection with a plasmid carrying a CAT reporter gene under the control of the herpes simplex virus thymidine kinase minimal promoter. Shown are a representative CAT conversion assay (left) and the average result of four independent experiments (right).
DISCUSSION
The structure-function analysis reported here demonstrates for the first time a correlation between specific biochemical functions of distinguishable domains within an HRX fusion protein and the ability to transform hematopoietic cells in vitro. The DNA binding domains of HRX as well as the carboxy-terminal transactivation domain of ENL were identified as crucial for the ability of HRX-ENL to alter the growth properties of primary hematopoietic progenitor cells. These observations provide strong support for a model in which HRX-ENL induces transformation by deregulating subordinate target genes through a gain of function contributed by the transcriptional effector properties of ENL.
Among the three domains shown here to be necessary for HRX-ENL’s oncogenic activity, the AT hook region is one whose function has been best characterized due to previous studies of HMG-I(Y) proteins. HMG-I(Y) proteins are members of the high-motility group proteins and, like HRX, contain three closely spaced AT hooks. They are abundant, nonhistone components of chromatin that bind DNA in the minor groove, recognizing altered DNA secondary structures, rather than sequence, and function to bend DNA (for a review, see reference 16). In this capacity, HMG-I(Y) proteins serve as architectural transcription factors that are required for correct enhancer-dependent expression of several genes, including the T-cell receptor alpha (2) and beta interferon genes (41). The HRX AT hooks display in vitro DNA binding properties similar to those of HMG-I(Y) proteins. They bind both to cruciform and intrinsically bent, AT-rich, scaffold attachment region DNA but not to nonscaffold DNA (6). Although these similarities in DNA binding properties raise the possibility that HRX targets genomic sites similar to those targeted by HMG-I(Y) proteins, additional constraints on DNA binding are likely to result from other domains within the amino-terminal portion of HRX.
AT hook-containing proteins are also implicated in the pathogenesis of a variety of benign mesenchymal tumors, especially lipomas (39). In these tumors, chromosomal translocations result in the fusion of HMGI-C, a relative of HMG-I(Y), to a variety of fusion partners predicted to be transcriptional regulators (1, 36). HMGI-C proteins are small proteins consisting of little more than three AT hook motifs which are consistently preserved in HMGI-C chimeric proteins. These features suggest that ectopic expression of HMGI-C targets might be responsible for tumorigenesis in a subset of mesenchymal tumors. The fact that deletion of the AT hooks in HRX-ENL leads to a concomitant loss of its transforming potency underscores its potential similarity to the HMG-I fusion proteins and supports the ectopic expression of original HRX targets as one likely mechanism of leukemogenesis. However, our studies indicate that the AT hooks are not a sufficient contribution from HRX for oncogenesis since deletions of HRX-ENL that preserved the AT hook motifs but removed the BCB region led to marked reductions of growth-altering activity. Therefore, HRX contributes more than simply its AT hook motifs to oncogenic chimeras, suggesting that target gene selection may result from a combination of specificity determinants within HRX.
Our studies implicate at least one additional recognizable motif in HRX as vital for HRX-ENL’s oncogenic function since the inactivation of the cysteine-rich methyltransferase homology domain led to severe reductions in growth-altering effects of HRX-ENL. This motif is found in all known mammalian DNMT (26, 45), and two copies of it are present in the PCM1 component of the MeCP1 transcriptional repressor (11). It consists of a bipartite structure in which a core containing three cysteines (CGXCX2C) is repeated twice and flanked by basic residues. The homology of the HRX cysteine motif with mammalian DNMT extends further downstream into another highly basic region that is not present in PCM1 (Fig. 3A).
Although the BCB motif of human DNMT has been demonstrated to bind zinc in vitro (4), this motif does not resemble typical DNA-binding Zn finger structures. The BCB motif is part of a large amino-terminal portion of DNMT that inhibits de novo methylation of unmethylated but not hemimethylated DNA, suggesting that it contributes to recognition of DNA methylation status. Rather than direct interaction with 5-methylcytosine itself, however, DNMT is proposed to recognize distortions of DNA secondary structure resulting from methylation or other perturbations. For instance, DNMT binds comparably to the artificial substrate poly(dI-dC) as to hemimethylated DNA, presumably because poly(dI-dC) has conformational features similar to those of methylated DNA (4). This might also be a feature of the BCB domain of HRX, as it displays more binding affinity in vitro for methylated salmon sperm DNA and poly(dI-dC) than for bacterial plasmid DNA that was only sporadically dcm methylated. However, in a mobility shift assay, PCM1 does not require its BCB motif to bind and discriminate methylated from unmethylated DNA (11). Although its functional role remains to be fully elucidated, the importance of the HRX BCB motif is underscored not only by the results of our in vitro transformation analyses but also by its consistent presence in all HRX fusion proteins reported to date.
The deletion analyses performed on ENL identified its carboxy-terminal helical segments as critical for the oncogenic activity of HRX-ENL. Additional, albeit circumstantial evidence for the essential nature of this domain comes from the molecular analysis of chimeric HRX-AF9 proteins in leukemias. At least in one leukemia, only the carboxy-terminal 91 amino acids of AF9 were fused to HRX (27), resulting in a fusion protein highly analogous to our ΔEN3 mutant. An alignment of ENL and AF9 (Fig. 3B) reveals that the minimally required region that confers transformation activity when fused to HRX corresponds almost exactly to the region conserved between these proteins. Secondary structure predictions (31) indicate the presence of two highly conserved helical segments with possible amphipathic character. Mutations that deleted either of these helical regions completely abrogated the transforming activity of HRX-ENL. Furthermore, our finding that both helical segments are necessary for transactivation demonstrate a complete concordance in the structural requirements for these functions of ENL. However, ENL does not display features of a typical transcriptional activator, and we suggest that it may normally function as a coactivator, using its C-terminal helical segments to interact with unidentified components of the transcriptional machinery. Both ENL and AF9 show weak similarity in their C termini with yeast ANC1 (44), which has been identified as a component of three multicomponent transcription complexes: the SWI/SNF complex and the TFIID and TFIIF basal transcription complexes (8, 20). However, ENL has not been observed as a component of the comparable mammalian complexes (12). Furthermore, the C terminus of ANC1 does not conserve the helical secondary structures present in ENL/AF9. Thus, although our studies indicate a transcriptional role for ENL, it probably differs from the one served by ANC1.
Taken together, our studies support a gain-of-function mechanism for the oncogenic activity of HRX-ENL. We posit that the DNA binding motifs of HRX are likely to target the chimeric proteins to specific genomic sites and that the transcriptional effector properties of ENL/AF9, and other partners as well, will result in perturbations of target gene expression. Two other HRX fusion partners with known functions seem to support this hypothesis, as both are involved in transcriptional regulation. ELL functions as an RNA polymerase II elongation factor, and CBP functions as a coactivator with, among other properties, histone acetylase activity (37, 38, 40). The available genetic and colocalization studies of Drosophila suggest that TRX plays a role in cellular memory by opposing or modulating the activities of Polycomb group proteins involved in maintenance of repressed states of transcription. If this function is conserved in HRX, the acquisition of a constitutively active transcriptional motif resulting from protein fusions in 11q23 leukemias may disrupt the ability of HRX to appropriately modulate the activity of Polycomb group proteins. This could prevent Polycomb group-mediated repression of target genes whose continued expression would antagonize terminal differentiation and associated cell cycle exit of hematopoietic cells.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
R.K.S. was supported by DFG grant SL27/1-1. This work was supported in part by grant CA55029 from the National Institutes of Health.
We thank Robert Hawley for providing the MSCV vector.
REFERENCES
- 1.Ashar H R, Fejzo M S, Tkachenk A, Zhou X, Fletcher J A, Weremowicz S, Morton C C, Chada K K. Disruption of the architectural factor HMGI-C: DNA-binding AT-hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell. 1995;82:57–65. doi: 10.1016/0092-8674(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 2.Bagga R, Emerson B M. An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- 3.Bernard O A, Berger R. Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 4.Bestor T H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breen T R, Harte P J. trithorax regulates multiple homeotic genes in the bithorax and antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. doi: 10.1242/dev.117.1.119. [DOI] [PubMed] [Google Scholar]
- 6.Broeker P L, Harden A, Rowley J D, Zeleznik-Le N. The mixed lineage leukemia (MLL) protein involved in 11q23 translocations contains a domain that binds cruciform DNA and scaffold attachment region (SAR) DNA. Curr Top Microbiol Immunol. 1996;211:259–268. doi: 10.1007/978-3-642-85232-9_26. [DOI] [PubMed] [Google Scholar]
- 7.Butler L H, Slany R K, Cui X, Cleary M L, Mason D Y. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood. 1997;89:3361–3370. [PubMed] [Google Scholar]
- 8.Cairns B R, Henry N L, Kornberg R D. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary M L. Oncogenic conversion of transcription factors by chromosomal translocations. Cell. 1991;66:619–622. doi: 10.1016/0092-8674(91)90105-8. [DOI] [PubMed] [Google Scholar]
- 10.Corral J, Lavenir I, Impey L, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N J, King G, Rabbitts T H. An M11-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 11.Cross S H, Meehan R R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 12.Cui, X., R. K. Slany, and M. L. Cleary. Unpublished observations.
- 13.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukemia. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 14.Felix C A, Winick N J, Negrini M, Bowman W P, Croce C M, Lange B J. Common region of ALL-1 gene disrupted in epipodophyllotoxin-related secondary acute myeloid leukemia. Cancer Res. 1993;53:2954–2956. [PubMed] [Google Scholar]
- 15.Gill-Super H J, McCabe N R, Thirman M J, Larson R A, LeBeau M M, Pedersen-Bjergaard J, Philip P, Diaz M O, Rowley J D. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood. 1993;82:3705–3711. [PubMed] [Google Scholar]
- 16.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 17.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 18.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1995;1:136–138. [PubMed] [Google Scholar]
- 19.Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- 20.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 21.Iida S, Seto M, Yamamoto K, Komatsu H, Tojo A, Asano S, Kamada N, Ariyoshi Y, Takahashi T, Ueda R. MLLT3 gene on 9p22 involved in t(9;11) leukemia encodes a serine/proline rich protein homologous to MLLT1 on 19p13. Oncogene. 1993;8:3085–3092. [PubMed] [Google Scholar]
- 22.Joh T, Kagami Y, Yamamoto K, Segawa T, Takizawa J, Takahashi T, Ueda R, Seto M. Identification of MLL and chimeric MLL gene products involved in 11q23 translocation and possible mechanism of leukemogenesis by MLL truncation. Oncogene. 1996;13:1945–1953. [PubMed] [Google Scholar]
- 23.Kennison J A. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993;9:75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- 24.Lavau C, Szilvassy S J, Slany R, Cleary M L. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löchner K, Siegler G, Führer M, Greil J, Beck J D, Fey G H, Marschalek R. A specific deletion in the breakpoint cluster region of the ALL-1 gene is associated with acute lymphoblastic T-cell leukemias. Cancer Res. 1996;56:2171–2177. [PubMed] [Google Scholar]
- 26.Ma Q, Alder H, Nelson K K, Chatterjee D, Gu Y, Nakamura T, Canaani E, Croce C M, Siracusa L D, Buchberg A M. Analysis of the murine All-1 gene reveals conserved domains with human ALL-1 and identifies a motif shared with DNA methyltransferases. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W M, Nowell P C, Croce C M, Canaani E. Genes on chromosomes 4, 9, and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabbitts T H. Translocations, master genes, and differences between the origins of acute and chronic leukemias. Cell. 1991;67:641–644. doi: 10.1016/0092-8674(91)90057-6. [DOI] [PubMed] [Google Scholar]
- 30.Reeves R, Nissen M S. The AT-DNA-binding domain of mammalian high mobility group chromosomal proteins: a novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 31.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 32.Rubnitz J E, Morrissey J, Savage P A, Cleary M L. ENL, the gene fused with HRX in t(11;19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells. Blood. 1994;84:1747–1752. [PubMed] [Google Scholar]
- 33.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schichman S A, Caligiuri M A, Strout M P, Carter S L, Gu Y, Canaani E, Bloomfield C D, Croce C M. ALL-1 tandem duplication in acute myeloid leukemia with a normal karyotype involves homologous recombination between Alu elements. Cancer Res. 1994;54:4277–4280. [PubMed] [Google Scholar]
- 35.Schichman S A, Caligiuri M A, Gu Y, Strout M P, Canaani E, Bloomfield C D, Croce C M. ALL-1 partial duplication in acute leukemia. Proc Natl Acad Sci USA. 1994;91:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoenmakers E F P M, Wanschura S, Mols R, Bullerdiek J, Van der Berghe H, Van de Ven W J M. Recurrent rearrangements in the high mobility group protein gene, HMGI-C in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 37.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. An RNA polymeraseII elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 38.Sobulo, O. M., J. Borrow, R. Tomek, S. Reshmi, A. Harden, B. Schlegelberger, D. Housman, N. A. Doggett, J. D. Rowley, and N. J. Zeleznik-Le. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3). Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 39.Sreekantaiah C, Leong S P L, Karakousis C P, McGee D L, Pappaport W D, Villar H V, Neal D, Fleming S, Wankel A, Herrington P N, Carmona R, Sandberg A. Cytogenetic profile of 109 lipomas. Cancer Res. 1991;51:422–433. [PubMed] [Google Scholar]
- 40.Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 41.Thanos D, Maniatis T. The high motility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 42.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 43.Waring P M, Cleary M L. Disruption of a homolog of trithorax by 11q23 translocations: leukemogenic and transcriptional implications. Curr Top Microbiol Immunol. 1997;220:1–23. doi: 10.1007/978-3-642-60479-9_1. [DOI] [PubMed] [Google Scholar]
- 44.Welch M D, Drubin D G. A nuclear protein with sequence similarity to proteins implicated in human leukemias is important for cellular morphogenesis and actin cytokeletal function in Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:617–632. doi: 10.1091/mbc.5.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen R W, Vertino P M, Nelkin B D, Yu J J, el-Deiry W, Cumaraswamy A, Lennon G G, Trask B J, Celano P, Baylin S B. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Altered HOX expression and segmental identity in M11-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 47.Ziemin van der Poel S, McCabe N R, Gill H J, Espinosa R, III, Patel Y, Harden A, Rubinelli P, Smith S D, Le Beau M M, Rowley J D, Diaz M O. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]