Abstract
In several types of bacteria, the Kdp ATPase (comprising of the KdpABC complex) is an inducible, high-affinity potassium transporter that scavenges K+ from the environment. The cyanobacterium Anabaena sp. strain L-31 showed the presence of not one but two distinct kdp operons in its genome. The kdp1 consisted of kdpA1B1G1C1D genes, whereas the kdp2 contained the kdpA2B2G2C2 genes. Among the regulatory genes, the kdpD open reading frame of Anabaena sp. strain L-31 was truncated compared to the kdpD of other bacteria, whereas a kdpE-like gene was absent in the vicinity of the two kdp operons. In response to K+ limitation (<0.05 mM external K+), only kdp2 (and not kdp1) expression could be detected as a 5.3-kb transcript on Northern blots, indicating that kdpA2B2G2C2 genes constitute a polycystronic operon. Unlike E. coli, addition of osmolytes like NaCl, or a change in pH of the medium did not enhance the kdp expression in Anabaena sp. strain L-31. Interestingly, the Anabaena sp. strain L-31 kdp2 operon was strongly induced in response to desiccation stress. The addition of K+ to K+-starved cultures resulted in repression and degradation of kdp2 transcripts. Our results clearly show that kdp2 is the major kdp operon expressed in Anabaena sp. strain L-31 and may play an important role in adaptation to K+ limitation and desiccation stress.
Potassium, the major intracellular cation in bacteria, is involved in various physiological processes such as turgor adaptation (15), activation of cellular enzymes (29), and pH homeostasis (12). In nitrogen-fixing cyanobacteria such as Anabaena spp., K+ also regulates gene expression and vital metabolic processes such as photosynthesis and nitrogen fixation (1, 2). Despite very low levels of K+ available in most environments (0.1 to 10 mM), the bacteria maintain a very high concentration of K+ (0.2 to 0.6 M) within their cells and have evolved several distinct K+ uptake and efflux systems to regulate their internal K+ concentration (15). In Escherichia coli the constitutively expressed TrkG, TrkH, and Kup uptake systems have a low affinity for K+ but are competent to maintain the required levels K+ under normal physiological conditions (20). Under conditions of severe K+ limitation or osmotic upshift or when the low-affinity transporters are unable to meet the cell's demand for K+, the high-affinity KdpATPase (K+-dependent ATPase) is expressed (4). The Km of E. coli KdpATPase for K+ is 2 μM and the cells expressing Kdp can reduce the K+ concentration in the medium to as low as 50 nM (28).
Earlier studies had shown the kdp homologs to be widely distributed among the gram-negative bacteria (32). Genome sequencing has now shown kdp homologs to be present also in cyanobacteria, gram-positive bacteria, and the Archaea. We have previously demonstrated the presence of KdpB-like polypeptide in three different strains of the nitrogen-fixing cyanobacteria Anabaena spp. (6). The KdpB was induced under conditions of K+ limitation, and the KdpB protein was shown to be located in Anabaena membranes (3). In order to understand the organization and the regulation of expression of kdp genes, we undertook the cloning and sequencing of the kdp operon from Anabaena sp. strain L-31. Degenerate primers based on evolutionarily conserved amino acid stretches within different Kdp proteins from several bacteria were used to amplify Anabaena sp. strain L-31 chromosomal DNA. Interestingly, sequence analysis of kdp-like PCR fragments showed presence of not one but two distinct kdp operons (kdp1 and kdp2) in Anabaena sp. strain L-31. We examined here the expression of the two kdp operons under a variety of environmental stress conditions. Our data show that Anabaena sp. strain L-31 kdp2 (and not kdp1) is the major kdp operon transcriptionally activated in response to K+ limitation and desiccation stress. Unlike E. coli and other bacteria, osmotic upshift (both ionic and nonionic) did not induce kdp expression in Anabaena sp. strain L-31.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A filamentous, heterocystous, nitrogen-fixing freshwater cyanobacterium Anabaena sp. strain L-31 isolated in our laboratory (31) was used under axenic conditions. The Anabaena sp. strain L-31 cultures were maintained in the BG-11 medium (13) without combined nitrogen under continuous illumination (30 μE/m2/s) and aeration (2.0 liters/min). The K2HPO4 from BG-11 was replaced with equimolar Na2HPO4 to obtain BG-11/K0 medium. For K+ limitation experiments, Anabaena sp. strain L-31 cells were inoculated into BG-11/K5 (BG-11/K0 plus 5 mM KCl) and allowed to grow (with aeration) to a chlorophyll a concentration of 4 to 6 μg/ml. The cells were harvested by centrifugation, washed five times with 5 volumes of BG-11/K0 containing appropriate concentrations of KCl, and inoculated in identical medium at a concentration of ca. 4 to 6 μg of chlorophyll a/ml. The BG-11N+ medium was prepared by the addition of NaNO3 (17 mM) to the BG-11/K5 or BG-11/K0 media. The initial pH of all media, unless otherwise specified, was adjusted to 7.0. For RNA isolation, Anabaena sp. strain L-31 cells were harvested after 16 h or at the time points indicated in Results. For osmotic upshift, the BG-11/K0 or BG-11/K0.05 (BG-11/K0 plus 0.05 mM KCl) medium was supplemented with 65 mM NaCl or 110 mM sucrose or 12% polyethylene glycol (PEG). For desiccation experiments, Anabaena sp. strain L-31 cells grown in BG-11/K5 were thoroughly washed with and resuspended in BG-11/K0.1 medium. After 16 h the cells were filtered (0.45-μm pore size, mixed cellulose ester; Millipore) using a vacuum assembly. The filter paper disks containing the Anabaena sp. strain L-31 cells were allowed to desiccate in air at room temperature for different time intervals under illumination (30 μE/m2/s).
E. coli DH5α [Δ(argF-lac)U169 supE44 φ80d lacZΔM15 recA1 endA1 gyrA96 thi-1 relA1 hsdR17] was used as host to maintain all the plasmids used in the present study.
Cloning of kdp operons from Anabaena sp. strain L-31.
Degenerate primers based on evolutionary conserved amino acid stretches within different Kdp proteins from several bacteria such as E. coli, Synechocystis sp. strain PCC 6803, Clostridium acetobutylicum, and Alicyclobacillus acidocaldarius were used to amplify Anabaena sp. strain L-31 kdp genes from chromosomal DNA. The PCR products were cloned sequenced, and the deduced amino acid sequence was subjected to BLAST search (5). PCR with kdp-specific primers generated two different kdpAB-like DNA products (AB1 and AB2) that showed only 70% nucleotide identity to each other and also hybridized to different restriction enzyme-digested Anabaena sp. strain L-31 DNA fragments on Southern blots (Fig. 1 and 2). This indicated the presence of two kdp operons in Anabaena sp. strain L-31. The kdp-like DNA fragments obtained from PCR were used as probes to clone the Anabaena sp. strain L-31 kdp operons by chromosomal walking (data not shown). The entire kdp1 and kdp2 operons cloned as described above were completely sequenced (GenBank accession numbers AF213466 and AY753299 for kdp1 and kdp2, respectively).
FIG. 1.
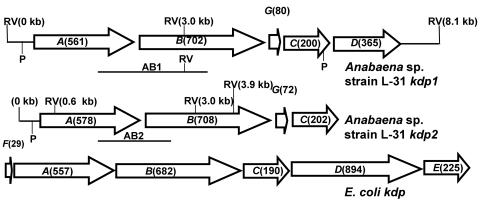
Two kdp operons from Anabaena sp. strain L-31. The arrangement of the kdp genes within the Anabaena sp. strain L-31 kdp operons is schematically depicted. The E. coli kdp operon is also shown for comparison. ORFs are shown as arrowheads indicating the direction of transcription. The expected number of amino acids for each ORF is shown in parentheses. The positions of putative promoters (P) are indicated. The chromosomal locations of the two DNA probes (AB1 and AB2) used for Southern hybridization experiments (see Fig. 2) are shown below their respective operons. The location of the EcoRV restriction enzyme site within the kdp operons is indicated (RV).
FIG. 2.
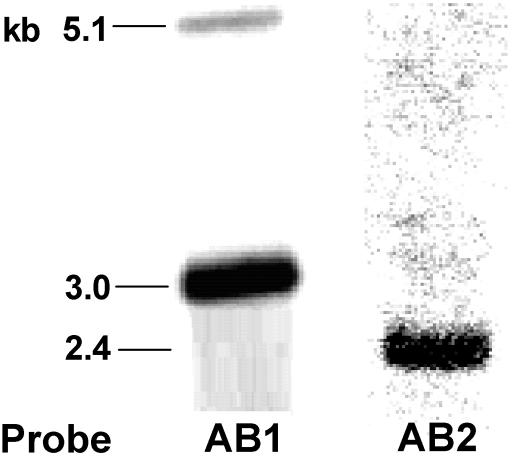
Southern hybridization analysis. The DNA probes AB1 (2.1 kb) and AB2 (1.5 kb) (see Fig. 1) were generated by PCR, labeled with DIG, and hybridized separately to the EcoRV-digested Anabaena sp. strain L-31 chromosomal DNA (3 μg) on Southern blots. The hybridization signals obtained with the two probes and their respective sizes are shown.
Southern blotting and hybridizations.
Anabaena sp. strain L-31 chromosomal DNA was prepared as described earlier (7). Spooled Anabaena sp. strain L-31 chromosomal DNA digested with desired restriction enzymes was resolved by electrophoresis on 0.8% agarose gels, transferred to a positively charged nylon membrane (Roche) by capillary blotting, and processed as instructed (DIG System User's Guide [Roche]). The kdp genes were labeled with digoxigenin (DIG) by random priming and used as probes for Southern blots. The hybridization was performed in DIG Easy Hyb buffer (Roche) at 42°C overnight. Posthybridization, the membranes were washed twice with solution A (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) at room temperature and twice with solution B (0.1× SSC, 0.1% SDS) at 68°C. The chemiluminescent detection was subsequently carried out according to the instructions provided by the manufacturer for filter hybridizations (Roche).
Isolation of Anabaena sp. strain L-31 total RNA.
A total of 50 ml of Anabaena sp. strain L-31 cells (4 to 6 μg of chlorophyll a/ml) was harvested by centrifugation at the indicated time points and immediately shock frozen in liquid nitrogen. Then, 1 ml of RNA isolation reagent (RNAwiz; Ambion) was added to the cells, and the RNA was isolated according to the recommended protocol. The RNA pellet was resuspended in DNase I reaction buffer (0.1 M sodium acetate [pH 5.0], 5 mM MgSO4) and subjected to DNase I (RNase free, 5 U) treatment. At the end of 45 min, 2 M LiCl (final concentration) was added to the tubes, and the tubes were kept at −20°C for at least 1 h. The RNA was collected by centrifugation (15,000 × g, 20 min) at 4°C. The RNA pellet was washed with 75% ethanol, air dried, and dissolved in RNase-free water.
Northern blotting and dot blot hybridization.
For Northern blotting experiments, 15 μg total RNA was electrophoretically resolved on denaturing formaldehyde agarose gels using MOPS buffer (0.02 M 3-[N-morpholino]propanesulfonic acid, 5 mM sodium acetate [pH 7.0], 0.01 M disodium EDTA). After electrophoresis the gels were washed with diethyl pyrocarbonate-treated water, and the RNA was transferred to positively charged nylon membrane by capillary blotting with 10× SSC. For dot blots, 4 μg of RNA was spotted directly onto a nylon membrane with a micropipette. Prehybridization was carried out in DIG Easy Hyb buffer (Roche) for at least 1 h at 50°C. The individual kdp genes were amplified by PCR, labeled with DIG, and hybridized to the RNA in DIG Easy Hyb buffer (overnight at 50°C). After hybridization the membranes were washed twice with solution A (2× SSC, 0.1% SDS) for 15 min at room temperature and twice with solution B (0.1× SSC, 0.1% SDS) for 15 min at 66°C. The chemiluminescent detection was subsequently carried out according to the instructions provided by the manufacturer for filter hybridizations (Roche).
RT-PCR.
Reverse primers specific for kdpA1, kdpC1, and kdpA2 were used to reverse transcribe 0.5 μg of total RNA using the enhanced avian reverse transcriptase (RT; Sigma). The following forward and reverse primers were used: GCCACCATGTGCGGCGCAGTC (RTkdpA1fwd) and TAAAACTATGCCGGCGGTGAT (RtkdpA1rev) for kdpA1, ATGTTACAAGGCTGGATACAA (RtkdpA2fwd) and CCCTGCTAATGTTTCCGGCAC (RtkdpA2rev) for kdpA2, and ATCTCCTTGATTCGAGAACTT (RtkdpC1fwd) and CCCATTCCCTAATCTTATCGAT (RtkdpC1rev) for kdpC2. The RT products were directly used as templates for amplification with the specific forward and reverse primers. Additional PCRs were performed with RNA without the RT reaction (negative control) and chromosomal DNA (positive control) using the same master mix containing all of the necessary components. The amplification products were resolved by electrophoresis on agarose gels and detected by staining with ethidium bromide.
RESULTS
Organization of the kdp operons in Anabaena sp. strain L-31.
By using PCR with kdp-specific degenerate primers, we detected the presence of not one but two kdp operons in Anabaena sp. strain L-31. Both of the kdp operons have been cloned and sequenced by standard molecular biology techniques (GenBank accession numbers AF213466 and AY753299 for kdp1 and kdp2, respectively). The arrangement of individual kdp genes within the two Anabaena sp. strain L-31 kdp operons is shown in Fig. 1 and closely resembles the arrangement of kdp operons observed in Anabaena sp. strain 7120 (21). Sequence analysis showed the presence of five open reading frames (ORFs; kdpA1, kdpB1, kdpG1, kdpC1, and kdpD) in kdp1 (9) and four ORFs (kdpA2, kdpB2, kdpG2, and kdpC2) in kdp2. In all cyanobacterial kdp operons sequenced thus far, no genes with overlapping ORF have been observed. The position of putative promoters (based on homology search) identified upstream of kdpA1, kdpA2, and kdpD is depicted in Fig. 1. The BLAST search with deduced KdpABC amino acid sequences from both Anabaena sp. strain L-31 kdp operons showed very good homology to other bacterial KdpABC proteins (Table 1). Many kdp operons from other bacteria show the presence of small ORFs encoding for hydrophobic peptides, e.g., kdpF in E. coli (17) and M. tuberculosis (14) and kdpZ and kdpY in C. acetobutylicum (10). Although a kdpF-like gene was absent from the two Anabaena sp. strain L-31 kdp operons, an additional ORF, kdpG, was found located between kdpB and kdpC. Anabaena sp. strain L-31 kdpG appeared to encode a hydrophobic protein with two transmembrane segments. The KdpG, which is unique to cyanobacterial genomes, may play a role in stabilization or activity of the KdpATPase complex, as has been shown for KdpF and KdpY-Z in other organisms (10, 17). A naturally short kdpD ORF was observed downstream of kdpC1, while no such ORF was observed in the kdp2 operon of Anabaena sp. strain L-31. The kdpD ORF encoded a protein of only 365 amino acids corresponding to the KdpD N-terminal domain of E. coli, whereas the C-terminal histidine kinase domain was missing from Anabaena sp. strain L-31 KdpD (9). We have evidence that Anabaena sp. strain L-31 KdpD is synthesized constitutively irrespective of the K+ concentration in the medium (8a), which implies that kdpD gene is transcribed from its own promoter independent of the kdpA1 promoter. No kdpE like gene was found in the vicinity of the two kdp operons.
TABLE 1.
Predicted homology (% identity) of Anabaena sp. strain L-31 Kdp proteins with corresponding KdpA, KdpB, or KdpC proteins from other bacteria
| Anabaena sp. strain L-31 Kdp | % Identity with:
|
||||
|---|---|---|---|---|---|
| Anabaena sp. strain PCC 7120 Kdp1 | Anabaena sp. strain PCC 7120 Kdp2 | Synechocystis sp. strain PCC 6803 Kdp | Escherichia coli Kdp | Mycobacterium tuberculosis Kdp | |
| KdpA1 | 94 | 64 | 66 | 44 | 38 |
| KdpA2 | 65 | 91 | 65 | 43 | 40 |
| KdpB1 | 92 | 72 | 75 | 58 | 54 |
| KdpB2 | 72 | 91 | 73 | 59 | 50 |
| KdpC1 | 85 | 51 | 59 | 46 | 45 |
| KdpC2 | 54 | 91 | 61 | 48 | 41 |
kdp2 is the major kdp operon expressed under K+ limitation.
Anabaena sp. strain L-31 kdpA1B1C1 showed 60 to 75% nucleotide homology with the corresponding genes from the kdp2 operon. Southern blotting and hybridization experiments were performed with probes AB1 and AB2 (see Fig. 1 for details) to ascertain the ability of the Anabaena sp. strain L-31 kdp1 gene (DNA) probe to cross-hybridize with the corresponding kdp2 genes and vice versa. The probe AB1 showed hybridization signals of 5.1 and 3.0 kb on Southern blots (Fig. 2) of EcoRV-digested Anabaena sp. strain L-31 chromosomal DNA. When probe AB2 was used, neither the 5.1- nor the 3.0-kb signal was observed, although a 2.4-kb signal was detected (Fig. 2). This demonstrated the inability of the probes to cross-hybridize with the corresponding DNA from the other kdp operon under the stringency conditions used (see Materials and Methods). Hence, Northern blotting (of the Anabaena sp. strain L-31 total RNA), followed by hybridization to kdp1 or kdp2 DIG-labeled DNA probes, was used to monitor the expression of one kdp operon vis a vis the other.
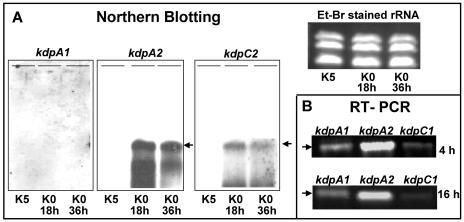
In E. coli and several other bacteria the kdp operon is induced by K+ limitation and repressed by high levels of K+ (20). Earlier studies in our laboratory showed that high K+ (5 mM) repressed KdpB expression in Anabaena spp. (3, 6). Based on this observation the Anabaena sp. strain L-31 cells were grown in BG-11/K5 (to completely suppress kdp expression) and transferred to BG-11/K0 medium after thorough washing in the same medium. When hybridized with the kdpA1 probe, no signal was detected in the BG-11/K5 or BG-11/K0 total RNA (Fig. 3A). However, when the same RNA was probed with kdpA2 or kdpC2, a strong signal of 5.3 kb was observed in the K+ starved samples while none was observed in the BG-11/K5 samples (Fig. 3A). This indicated that kdp2 and not kdp1 was the kdp operon induced under K+ limiting conditions and that the kdpA2B2G2C2 did form a polycistronic operon.
FIG. 3.
Differential expression of the kdp operons in Anabaena sp. strain L-31. (A) Northern blotting. Total RNA was isolated from Anabaena sp. strain L-31 cells grown in BG-11/K5 or BG-11/K0 for 18 and 36 h and electrophoretically resolved (15 μg per lane) on formaldehyde agarose gels. First, 600 bp of kdpA1 ORF, 1 to 400 bp of kdpA2 ORF, and 200 to 600 bp of kdpC2 ORF were amplified by PCR, labeled with DIG, and used as kdpA1, kdpA2, or kdpC2 probes, respectively. The RNA was transferred to nylon membrane and subsequently hybridized to the individual probes. The 5.3-kb transcript is indicated by an arrow. Equal loading of RNA was confirmed by ethidium bromide staining of rRNA from various samples. (B) RT-PCR. Total RNA was isolated from cells grown in BG-11/K0 for 4 or 16 h after inoculation and subjected to RT reaction with primers specific for Anabaena sp. strain L-31 kdpA1, kdpA2, or kdpC1 genes. The RT reaction was subsequently subjected to PCR, and the amplification products were resolved by electrophoresis and stained with ethidium bromide.
A more sensitive RT-PCR technique was used to verify whether kdp1 was expressed at all. Reverse primers (see Materials and Methods) specific for kdpA1, kdpC1, and kdpA2 were utilized to reverse transcribe RNA isolated from Anabaena sp. strain L-31 cells grown in BG-11/K5 or BG-11/K0. On PCR amplification with the specific forward primers, kdpA1 and kdpC1 amplification products were detected in BG-11/K0 RNA. However, the amount of kdpA1/C1 PCR products was severalfold lower than that obtained with kdpA2-specific primers (Fig. 3B), clearly showing that kdp2 is the major kdp operon expressed under conditions of K+ starvation in Anabaena sp. strain L-31. No PCR product was observed when RNA isolated from Anabaena sp. strain L-31 cells grown in BG-11/K5 was subjected to RT-PCR with the above-mentioned primers (data not included). PCRs without the RT reaction (negative control) yielded no amplification products (data not shown).
Response of Anabaena sp. strain L-31 kdp2 operon to K+ starvation.
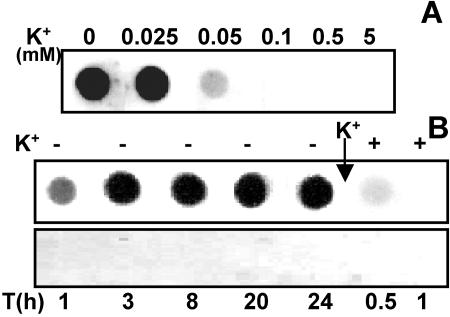
To determine the threshold of external K+ concentration that allowed kdp2 expression, Anabaena sp. strain L-31 cells that had been grown in BG-11/K5 were inoculated into BG-11 medium with different concentrations of K+ as indicated (Fig. 4A). The total RNA was isolated from all the cultures after 16 h and hybridized to the kdpA2 probe. The highest external K+ concentration that allowed kdpA2 expression was 0.05 mM K+, whereas maximal expression was observed at <0.025 mM K+ (Fig. 4A).
FIG. 4.
Effect of K+ limitation on Anabaena sp. strain L-31 kdp2 expression. (A) Anabaena sp. strain L-31 cells grown in BG-11/K5 were inoculated into BG-11 with different concentrations of external K+ and grown for 16 h. Total RNA (4 μg) was spotted onto nylon membrane and hybridized to the kdpA2 probe. (B) Anabaena sp. strain L-31 cells grown in BG-11/K5 were inoculated into BG-11/K0 medium, and the total RNA isolated at the indicated time points. At the end of 24 h, KCl (5 mM) was added to the medium (shown by an arrow), and the total RNA was isolated 0.5 and 1.0 h after K+ repletion. All of the RNA samples (4 μg each) were hybridized to the kdpA2 probe (upper panel) or the kdpA1 probe (lower panel).
The kinetics of Anabaena sp. strain L-31 kdp2 expression in response to K+ starvation was also monitored (Fig. 4B). The kdp2 expression was clearly observed after 1 h of K+ limitation, and maximal expression occurred by 3 h of K+ starvation. The ability of external K+ to repress the previously induced kdp2 operon was also examined. Upon addition of 5 mM K+ to a K+-starved culture, the kdp2 expression was drastically reduced, and 30 min after K+ addition hardly any expression could be detected (Fig. 4B). The kdpA1 expression could not be detected on dot blots even after prolonged (24 h) K+ starvation.
Effect of osmotic stress on Anabaena sp. strain L-31 kdp2 expression.
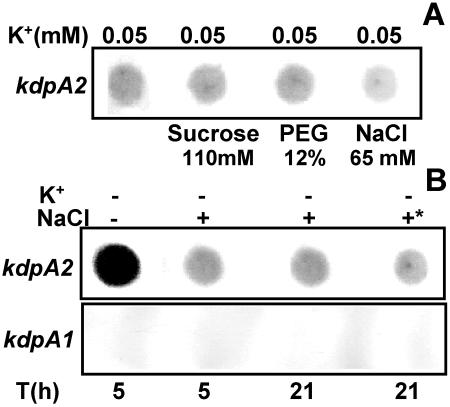
Osmotic stress, particularly ionic upshock, is known to induce kdp expression in the presence of moderate external K+ in E. coli (8, 25, 30). To check whether the same held true for Anabaena, the kdp expression in response to NaCl and other nonionic osmolytes was examined. Initially, the kdp expression was monitored in BG-11/K0.05, i.e., the highest K+ concentration at which Anabaena sp. strain L-31 kdp2 expression could be detected (Fig. 5A). Sucrose and PEG did not enhance kdp2 expression, and NaCl, in fact, decreased kdp2 expression (Fig. 5A).
FIG. 5.
Effect of osmotic stress on Anabaena sp. strain L-31 kdp expression. (A) Anabaena sp. strain L-31 cells were inoculated into BG-11/K0.05 medium containing different osmolytes as indicated. Total RNA (4 μg each) was hybridized to the kdpA2 probe. (B) Combined effect of extreme K+ limitation and NaCl stress on kdp2 expression. Anabaena sp. strain L-31 cells grown in BG-11/K5 were inoculated into BG-11/K0 with or without NaCl (65 mM) for the indicated time intervals. In another experiment, the cells grown in BG-11/K0 medium for 5 h were supplemented with NaCl (65 mM), and the culture was incubated for a further 16 h with NaCl (indicated by an asterisk). Total RNA was isolated and hybridized to the kdpA2 (upper panel) or kdpA1 (lower panel) probe.
Whether osmotic stress increased kdp2 expression over and above that seen with extreme K+ deprivation (K0) was also determined. Sucrose and PEG did not enhance kdp2 expression (data not shown), but with NaCl a significant reduction in the kdp2 expression was again observed. The addition of NaCl at the beginning of K+ starvation or at a later point during K+ starvation had the same effect of reducing kdp2 expression (Fig. 5B). These RNA samples were also hybridized to the kdpA1 DNA probe, but no detectable kdp1 expression was observed (Fig. 5B).
Expression of Anabaena sp. strain L-31 kdp2 in response to other environmental stresses.
A decrease in pH of the medium is known to induce kdp expression in E. coli (8). To test the effect of pH on Anabaena sp. strain L-31 kdp expression, the cells were inoculated in BG-11/K0 or BG-11/0.05 medium at pH 6.0 or 7.0, and the total RNA was isolated at the end of 16 h. When the RNA was probed with the kdpA2 probe, no increase in kdp2 due to the pH shift (7.0 to 6.0) over that at pH 7.0 was observed (Fig. 6A). Expression of kdp1 was not detected, even at pH 6.0 (data not shown). The presence of a combined nitrogen source in the medium is known to regulate the expression of several genes, e.g., nif, het (18, 34), and several osmoresponsive proteins (19) in Anabaena spp. However, the addition of 17 mM NaNO3 during growth did not affect expression of kdp1 or kdp2 operons in Anabaena sp. strain L-31 (data not shown). Heat shock is known to alter the expression of several stress responsive genes in bacteria. Anabaena sp. strain L-31 cells grown in BG-11/K0 were exposed to 42°C for 30 min, and the RNA was isolated. Heat shock decreased the kdpA2 expression under K+ limitation, whereas no expression was seen in cells grown with 5 mM K+ (Fig. 6B). Heat shock did not influence the kdp1 expression (data not shown).
FIG. 6.
Effect of different environmental stresses on Anabaena sp. strain L-31 kdp2 expression. (A) Anabaena sp. strain L-31 cells were inoculated in BG-11 medium with K+ concentrations and pH as indicated. (B) Anabaena sp. strain L-31 cells were subjected to heat shock (HS) at 42°C for 30 min in the presence or absence of 5 mM K+. (C) Anabaena sp. strain L-31 cells grown in BG-11/K0.1 were filtered onto filter paper disks and desiccated under illumination for the specified time intervals. The total RNA (4 μg) from each of the above samples (A, B, and C) was hybridized to the kdpA2 or kdpA1 probe as denoted.
Cyanobacteria are periodically exposed to desiccation in the natural environment. Recently, kdp2 operon from Anabaena sp. strain 7120 was reported to be induced in response to desiccation stress (24). The response of Anabaena sp. strain L-31 kdp operons to desiccation stress was also analyzed. Anabaena sp. strain L-31 cells grown in BG-11/K5 were inoculated in BG-11/K0.1 medium, wherein no kdp expression was observed (Fig. 4A). After 16 h of incubation in this medium, the cells were filtered (0.45-μm pore size; Millipore) and allowed to desiccate under light for 1 and 3 h. The total RNA was isolated and hybridized to the kdpA2 probe. No expression was observed in the BG-11/K0.1 control, whereas kdp2 expression was clearly observed in the RNA obtained from desiccated Anabaena sp. strain L-31 (Fig. 6C). No expression was detected when the kdpA1 probe was hybridized to the RNA isolated from either desiccated or nondesiccated Anabaena sp. strain L-31 cells (Fig. 6C).
DISCUSSION
We earlier demonstrated the presence KdpB polypeptides in three strains of Anabaena (including L-31) and their regulation by K+ with the E. coli KdpB antiserum (3, 6). Here we report on the Anabaena sp. strain L-31 kdp operon’s organization and its transcriptional activation in response to various environmental conditions. Nucleotide sequencing and Southern hybridization analysis demonstrated the presence of two distinct kdp operons in Anabaena sp. strain L-31. The genome of another filamentous cyanobacterium, Anabaena sp. strain PCC 7120 (21), also possesses two kdp operons very similar to those of strain L-31, while two unicellular cyanobacteria, Synechocystis sp. strain PCC 6803 (22) and Gloeobacter violaceous PCC 7421, possess only one kdp operon (www.kazusa.or.jp/cyano/gloeobacter). Recently, the importance of Kdp for K+ uptake in absence of Ktr system has been suggested for Synechocystis sp. strain PCC 6803 (11).
In E. coli, the kdpFABC operon is transcriptionally controlled by the products of the adjacent kdpDE operon (33). In response to appropriate stimuli, the KdpD (a transmembrane sensor kinase) phosphorylates the cytosolic response regulator KdpE, and the KpdE∼P binds to the kdpFABC promoter and activates its transcription (20). The corresponding cyanobacterial genes appear to be distinctly different. Anabaena sp. strain L-31 kdp1, Anabaena sp. strain 7120 kdp1, and the Synechocystis sp. strain PCC 6803 kdp operon all show the presence of a naturally short kdpD ORF downstream of the kdpC and lack kdpE. Interestingly, the kdp2 operon, which is the major kdp operon induced in Anabaena sp. strain L-31, has neither a kdpD-like nor a kdpE-like ORF downstream of the kdpC gene. The cyanobacterial KdpD resembles only the KdpD N-terminal domain of the full-length KdpD from other bacteria. What role the naturally short KdpD plays in cyanobacterial kdp regulation is not clear and remains to be investigated. We have earlier shown that Anabaena sp. strain L-31 KdpD, when fused with the E. coli KdpD C-terminal domain, results in a functional protein that can induce kdpFABC expression in vivo in E. coli and phosphorylate the purified E. coli KdpE in vitro (9).
kdpABC expression is tightly regulated and is induced at the transcriptional level in all bacterial strains where it has been monitored (15). Anabaena sp. strain L-31 kdp operons are no exception. Of the two kdp operons, the kdp2 is the major operon expressed in response to K+ limitation. Minor kdpA1 or kdpC1 expression could be detected only by a very sensitive RT-PCR technique (Fig. 3). Comparison of the DNA sequences upstream of the kdpA1and kdpA2 (presumed promoter regions) revealed no significant homology (data not shown). This suggests that the regulatory DNA (cis) elements present upstream of the two kdp operons may be different from each other.
The kdp2 operon is rapidly induced by transcriptional activation in response to K+ limitation and immediately repressed by addition of exogenous K+ (5 mM). Experiments with the transcription inhibitor rifampin showed kdp2 mRNA to have a half-life of 30 min in Anabaena sp. strain L-31 (data not shown). However, on addition of K+ very little kdp2 RNA was observed after 30 min (Fig. 4B). This suggests that not only is the kdp2 repressed by externally added K+ but also that the kdp2 mRNA is unstable in the presence of K+. We have earlier shown that the KdpB protein is degraded upon K+ repletion of K+-starved Anabaena cells (3). This correlates well with the need of Anabaena cells to express/repress and degrade the kdp2 operon products in response to changing environmental stimuli.
In E. coli and Salmonella enterica serovar Typhimurium the kdp operon has been shown to be induced by osmotic stress (particularly NaCl) (25, 16) and low pH (8). However, the Anabaena sp. strain L-31 kdp2 operon did not respond to increase in osmolarity mediated by PEG or sucrose, and exposure to NaCl in fact repressed it. Global-scale gene expression analysis of Synechocystis sp. strain PCC 6803 has also shown that the kdp is not induced in response to osmotic shock (23, 26). A shift in pH, heat shock, or the presence or absence of combined nitrogen in the medium also did not influence Anabaena sp. strain L-31 kdp expression.
Desiccation is known to markedly affect gene expression and gene regulation in bacteria (27). The kdp2 operon from Anabaena sp. strain L-31 was distinctly induced in response to desiccation. Katoh et al., using gene array technology, reported the manyfold induction of kdpA and kdpB genes of kdp2 operon (but not kdp1) from Anabaena sp. strain 7120 in response to desiccation stress (24). Desiccation stress increases the cellular requirement of K+ and the expression of KdpATPase encoded by the Anabaena sp. strain L-31 kdp2 operon may fulfill such requirement of K+.
The data presented here show that, of the two kdp operons present in Anabaena sp. strain L-31, only the kdp2 operon is predominantly expressed under K+ deficiency or during desiccation and is regulated at transcriptional level in a need-based manner. Unlike the enterobacterial kdp operons, Anabaena sp. strain L-31 kdp is unlikely to play a role in saline environments. It is more likely to contribute to survival during K+ starvation and the well-known desiccation tolerance of cyanobacteria.
Acknowledgments
The gene probes AB1 and AB2 used in this study were generated from the initial work on cloning and sequencing of Anabaena sp. strain L-31 kdp operons carried out by A.B. in the laboratory of K. Altendorf at the Department of Microbiology, University of Osnabrueck, Osnabrueck, Germany.
REFERENCES
- 1.Alahari, A., and S. K. Apte. 1998. Pleiotropic effects of potassium deficiency in heterocystous, nitrogen-fixing cyanobacterium, Anabaena torulosa. Microbiology 144:1557-1563. [DOI] [PubMed] [Google Scholar]
- 2.Alahari, A., and S. K. Apte. 2004. A novel potassium deficiency-induced stimulon in Anabaena torulosa. J. Biosci. 29:153-161. [DOI] [PubMed] [Google Scholar]
- 3.Alahari, A., A. Ballal, and S. K. Apte. 2001. Regulation of the potassium-dependent Kdp-ATPase expression in the nitrogen-fixing cyanobacterium Anabaena torulosa. J. Bacteriol. 183:5778-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altendorf, K., and W. Epstein. 1993. Kdp-ATPase of Escherichia coli. Cell. Physiol. Biochem. 4:160-168. [Google Scholar]
- 5.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipmann. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 6.Apte, S. K., and A. Alahari. 1994. Role of alkali cations (K+ and Na+) in cyanobacterial nitrogen fixation and adaptation to salinity and osmotic stress. Indian J. Biochem. Biophys. 31:267-279. [PubMed] [Google Scholar]
- 7.Apte, S. K., and R. Haselkorn. 1990. Cloning of salinity stress-induced genes from salt tolerant nitrogen-fixing cyanobacteruim Anabaena torulosa. Plant Mol. Biol. 15:723-733. [DOI] [PubMed] [Google Scholar]
- 8.Asha, H., and J. Gowrishankar. 1993. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as a signal for transcriptional control. J. Bacteriol. 181:4528-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Ballal, A., M. Bramkamp, H. Rajaram, P. Zimmann, S. K. Apte, and K. Altendorf. 2005. An atypical KdpD homologue from the cyanobacterium Anabaena sp. strain L-31: cloning, in vivo expression, and interaction with Escherichia coli KdpD-CTD. J. Bacteriol. 187:4921-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballal, A., R. Heermann, K. Jung, M. Gassel, S. K. Apte, and K. Altendorf. 2002. A chimeric Anabaena/Escherichia coli KdpD protein (Anacoli KdpD) functionally interacts with E. coli KdpE and activates kdp expression in E. coli. Arch. Microbiol. 178:141-148. [DOI] [PubMed] [Google Scholar]
- 10.Behrens, M., W. Schreiber, and P. Durre. 2001. The high-affinity K+-translocating complex from Clostridium acetobutylicum consists of 6 subunits. Anaerobe 7:159-169. [Google Scholar]
- 11.Berry, S., B. Esper, I. Karandashova, M. Teuber, I. Elanskaya, M. Roegner, and M. Hagemann. 2003. Potassium uptake in the unicellular cyanobacterium Synechocystis sp. strain 6803 mainly depends on a Ktr-like system encoded by slr1509 (ntpJ). FEBS Lett. 548:53-58. [DOI] [PubMed] [Google Scholar]
- 12.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castenholz, R. W. 1988. Culturing methods for cyanobacteria. Methods Enzymol. 167:68-93. [Google Scholar]
- 14.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, and D. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 15.Epstein, W. 2003. The roles and regulation of potassium in bacteria. Prog. Nucleic Acids Res. Mol. Biol. 75:293-320. [DOI] [PubMed] [Google Scholar]
- 16.Frymier, J. S., T. D. Reed, S. A. Fletcher, and L. N. Csonka. 1997. Characterization of transcriptional regulation of the kdp operon of Salmonella typhimurium. J. Bacteriol. 179:3061-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gassel, M., T. Mollenkamp, W. Puppe, and K. Altendorf. 1999. The KdpF subunit is a part of the K+-translocating Kdp complex of the Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 274:37901-37907. [DOI] [PubMed] [Google Scholar]
- 18.Haselkorn, R. 1992. Developmentally regulated gene arrangements in prokaryotes. Annu. Rev. Genet. 26:113-130. [DOI] [PubMed] [Google Scholar]
- 19.Iyer, V., T. Fernandes, and S. K. Apte. 1994. A role of osmotic-stress induced proteins in the osmotolerance of a nitrogen-fixing cyanobacterium, Anabaena sp. strain L-31. J. Bacteriol. 176:5868-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung, K., and K. Altendorf. 2003. Stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli, p. 53-58. In P. Dürre and B. Friedrich (ed.), Regulatory networks in prokaryotes. Horizon Scientific Press, Wymondham, United Kingdom.
- 21.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, et al. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213; 227-253. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko., T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, et al. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 23.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate expression of different set of genes in Synechocystis PCC 6803. Biochem. Biophys. Res. Commun. 290:339-348. [DOI] [PubMed] [Google Scholar]
- 24.Katoh, H., R. K. Asthana, and M. Ohmori. 2004. Gene expression in the cyanobacterium Anabaena sp. PCC7120 under desiccation. Microb. Ecol. 47:164-174. [DOI] [PubMed] [Google Scholar]
- 25.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marin, K., Y. Kanesaki, D. A. Los, N. Murata, I. Suzuki, and H. Hagemann. 2004. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 136:3290-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potts, M. 1994. Desiccation tolerance in prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stumpe, S., A. Schlösser, M. Schleyer, and E. P. Bakker. 1996. K+ circulation across the procaryotic cell membrane: K+-uptake systems, p. 473-499. In W. N. Konings, H. R. Kaback, and J. S. Lolkema (ed.), Handbook of biological physics, vol. 2. Elsevier Science BV, Amsterdam, The Netherlands. [Google Scholar]
- 29.Suelter, C. H. 1970. Enzymes activated by monovalent cations: patterns and predictions for the enzyme-catalyzed based reactions are explored. Science 168:789-795. [DOI] [PubMed] [Google Scholar]
- 30.Sugiura, A., K. Hirokawa, K. Nakashima, and T. Mizuno. 1994. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol. Microbiol. 14:929-938. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, J. 1970. Absence of the pigments of photosystem II of photosynthesis in heterocysts of a blue-green alga. Nature 228:181-183. [DOI] [PubMed] [Google Scholar]
- 32.Walderhaug, M. O., E. D. Litwack, and W. Epstein. 1989. Wide distribution of homologs of Escherichia coli Kdp K+-ATPase among gram-negative bacteria. J. Bacteriol. 171:1192-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein. 1992. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolk, C. P. 1991. Genetic analysis of cyanobacterial development. Curr. Opin. Genet. Dev. 1:336-341. [DOI] [PubMed] [Google Scholar]