Abstract
A four-step flavanone biosynthetic pathway was constructed and introduced into Saccharomyces cerevisiae. The recombinant yeast strain was fed with phenylpropanoid acids and produced the flavanones naringenin and pinocembrin 62 and 22 times more efficiently compared to previously reported recombinant prokaryotic strains. Microbial biosynthesis of the flavanone eriodictyol was also achieved.
Flavonoids are a diverse family of plant polyphenols that demonstrate great potential in the treatment of various human pathological conditions (1, 10). The health-promoting effects of flavonoids have stimulated significant research toward the elucidation of their biosynthetic networks, as well as the development of production platforms using well-characterized hosts (8, 19). Within this context, Hwang et al. recently described recombinant Escherichia coli strains that lead to the synthesis of flavanones, the common precursors of the vast majority of flavonoids (7).
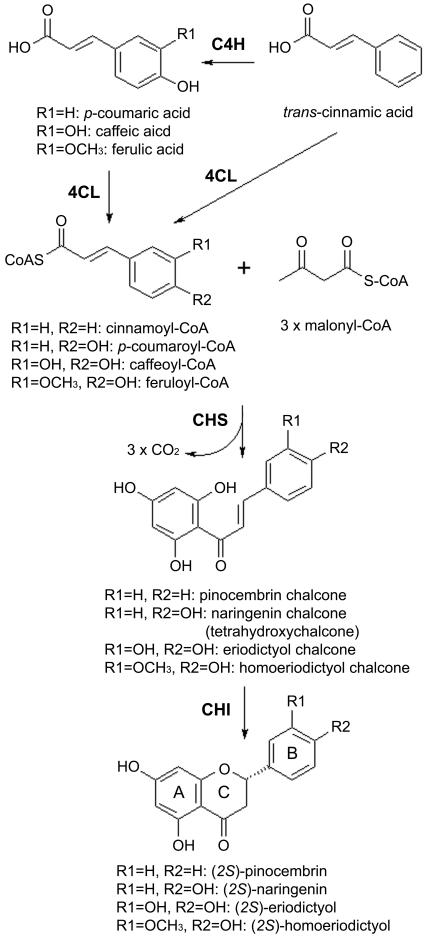
In plants, flavanone biosynthesis begins with the hydroxylation of cinnamic acid to p-coumaric acid by a membrane-bound P450 monooxygenase, cinnamate 4-hydroxylase (C4H) (Fig. 1). p-Coumaric acid is then activated to p-coumaroyl-coenzyme A (CoA) by 4-coumaroyl:CoA ligase. In the next step, three molecules of malonyl-CoA are sequentially added to one molecule of p-coumaroyl-CoA, yielding tetrahydroxychalcone, by the action of a polyketide synthase, chalcone synthase. Finally, chalcone isomerase converts the C15 compound tetrahydroxychalcone into (2S)-flavanones (Fig. 1) (5).
FIG. 1.
Flavanone biosynthesis in plants. Abbreviations: C4H, cinnamate 4-hydroxylase; 4CL, 4-coumaroyl:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase.
We present here the construction of a gene cluster that contains four plant-derived genes of the early flavonoid biosynthetic pathway that allows the conversion of phenylpropanoid acids into flavanones in Saccharomyces cerevisiae. This resulted in a substantial increase in the amount of flavanones produced compared to recombinant E. coli production and opens the possibility of producing several other flavonoid molecules whose biosynthesis requires the action of plant P450 monooxygenases.
Cloning of flavanone pathway in yeast.
Vector YEplac181 was utilized for cloning the flavanone biosynthetic gene cluster in S. cerevisiae. This cluster included four structural genes of heterologous plant origins: C4H cDNA from Arabidopsis thaliana (2), isolated from expressed sequence tag clone RAFL06-11-J16 (RIKEN BioResource Center) (14, 15); Pc4cL-2 cDNA from Petroselinum crispum (GenBank accession number AF233638) isolated from parsley young leaves by reverse transcription-PCR (RT-PCR) (9); CHI-A and chs cDNAs from Petunia × hybrida (GenBank accession numbers X14589 and X13225, respectively) isolated from petunia corolla also by RT-PCR (16, 17). We chose parsley 4-coumaroyl:CoA ligase-2 because this enzyme has been documented to accept caffeic acid, ferulic acid, and cinnamic acid with 28%, 66%, and 21% efficiency, respectively, in addition to its natural substrate, p-coumaric acid (9). We chose CHI-A because its previous cloning and expression in tomato plants led to a significant increase in the accumulation of flavanones (11).
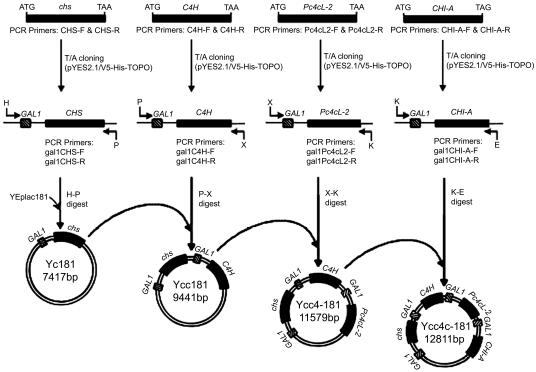
Cloning of the four-gene cluster in yeast was accomplished by following a cloning strategy that has previously been utilized successfully for E. coli (19). In the first step, each gene is cloned separately under the control of a species-specific promoter (in the present study, the strong S. cerevisiae GAL1 promoter). In the second step, each gene is amplified together with the promoter and is cloned using restriction digestions into the plasmid of choice (in the present study, S. cerevisiae plasmid YEplac181) (Fig. 2 and Table 1).
FIG. 2.
Schematic representation of the cloning strategy used for assembling plasmid Ycc4c181, carrying the four flavanone biosynthetic genes. Abbreviations for restriction enzymes: H, HindIII; P, PstI; X, XbaI; K, KpnI; E, EcoRI. The PCR and RT-PCR primer sequences used are presented in Table 1.
TABLE 1.
Primers used in this study and referred to in Fig. 1
| Primer | Sequence (5′-3′)a |
|---|---|
| C4H-F | ATGGACCTCCTCTTGCTGGAGAAGT |
| C4H-R | TTAACAGTTCCTTGGTTTCATAACGATTATGGAGTGG |
| Pc4cL2-F | ATGGGAGACTGTGTAGCACCCAAAG |
| Pc4cL2-R | TTATTTGGGAAGATCACCGGATGC |
| CHS-F | ATGGTGACAGTCGAGGAGTATCGTA |
| CHS-R | TTAAGTAGCAACACTGTGGAGGACA |
| CHI-A-F | ATGTCTCCTCCAGTGTCCGTTACTA |
| CHI-A-R | CTAGACTCCAATCACTGGAATAGTAGATTTCTCGG |
| gal1C4H-F | GGGGCTGCAGACGGATTAGAAGCCGCCGAG |
| gal1C4H-R | CCCCTCTAGATTAACAGTTCCTTGGTTTCATAACGATTATGGAGTGG |
| gal1Pc4cL2-F | GGGGTCTAGAACGGATTAGAAGCCGCCGAG |
| gal1Pc4cL2-R | CCCCGGTACCTTATTTGGGAAGATCACCGGATGC |
| gal1CHS-F | GGGGAAGCTTACGGATTAGAAGCCGCCGAG |
| gal1CHS-R | CCCCCTGCAGTTAAGTAGCAACACTGTGGAGGACA |
| gal1CHI-A-F | GGGGGGTACCACGGATTAGAAGCCGCCGAG |
| gal1CHI-A-R | CCCCGAATTCCTAGACTCCAATCACTGGAATAGTAGATTTCTCGG |
Underlining indicates restriction enzyme cleavage sites, corresponding to the primer description. Boldface indicates a start codon, and italics indicate a stop codon.
The successful application of this strategy in E. coli cannot be considered a precedent for success in yeast. This is because S. cerevisiae tends to be more recombination prone, and as a result, tandemly repeated copies of DNA (such as the 451-bp GAL1 promoter) have a higher probability of being looped out by excisional recombination (12). Therefore, the stability of the insertions must be carefully determined (3, 18). For that purpose, we grew the recombinant yeast strain for a maximum of 65 h at 30°C in leucine minimal selection medium (SC-Leu minimal medium) (4). Culture samples were taken every 24 h, and after plasmid isolation, the presence of each of the four genes together with the GAL1 promoter was tested by restriction digestions and PCRs. No recombination events that would lead to the loss of a gene(s) or promoters were observed during the 65-h time frame.
Exploring flavanone biosynthesis in S. cerevisiae.
We tested the ability of the recombinant yeast strain to synthesize flavanone compounds by feeding it with phenylpropanoid acids, such as cinnamic acid, p-coumaric acid, caffeic acid, and ferulic acid. In order to reduce cell growth inhibition, phenylpropanoid precursors were added every 13 h to the cultures in five equal doses, reaching a final concentration of 1 mM. In all cases, cultures were terminated after 65 h of incubation and flavonoid substances were extracted from the culture broth with an equal volume of ethyl acetate. They were further analyzed by reverse-phase high-performance liquid chromatography (HPLC) using an acetonitrile-water gradient, at a flow rate of 1.0 ml/min. The HPLC conditions were as follows: 10 to 40% for 10 min, 40 to 60% for 5 min, and 60 to 10% for 2 min. The retention times under these HPLC conditions for the standard authentic samples are presented in Table 2.
TABLE 2.
Retention times of standard (authentic) compounds used in the present study
| Compound | Retention time (min) |
|---|---|
| Pinocembrin | 16.3 |
| Naringenin | 12.8 |
| Eriodictyol | 11.1 |
| Homoeriodictyol | 12.9 |
When cinnamic acid was used as a precursor metabolite and galactose as the sole carbon source and inducer, a large amount (16.3 mg/liter) of the corresponding unhydroxylated flavanone pinocembrin accumulated in the medium. This is a 22-fold increase compared to the amount of pinocembrin produced by the most efficient E. coli recombinant strain (7). However, only a relatively low concentration of naringenin (0.2 mg/liter) was detected, demonstrating that although C4H was functionally expressed in yeast, it is still a rate-limiting step enzyme in the four-enzyme hybrid pathway. It is possible that increasing the activity of CPR1, the yeast P450 reductase that is required for P450 monooxygenase function through episomal overexpression, could lead to increased C4H activity (6). When p-coumaric acid was used as a precursor, a large amount of naringenin (28.3 mg/liter) accumulated in the culture, which is 62 times higher compared to the amount of naringenin produced by the most efficient recombinant E. coli strain. Similarly, when caffeic acid was used as a precursor, natural (2S)-eriodictyol was produced in significant amounts (6.5 mg/liter). This is the first time eriodictyol biosynthesis has been achieved through microbial fermentation. Finally, ferulic acid, which carries a methoxy group on the aromatic ring, failed to be metabolized by the recombinant yeast strain. This result is in agreement with recent data obtained by Schroeder et al. demonstrating that O-methylations on the B ring of flavonoid substrates result in complete loss of enzymatic activity. It is therefore possible that B-ring methylations occur later in the complex flavonoid pathway to the end products (i.e., after flavanones have been synthesized) (13). The biotransformation results are summarized in Table 3.
TABLE 3.
Production of natural flavanones by recombinant yeast S. cerevisiae INVSCI harboring plasmid Ycc4c181a
| Acid converted | Production (mg/liter) of:
|
|||
|---|---|---|---|---|
| Pinocembrin | Naringenin | Eriodictyol | Homoeriodictyol | |
| Cinnamic acid | 16.3 | 0.2 | NAb | NA |
| p-Coumaric acid | NA | 28.3 | NA | NA |
| Caffeic acid | NA | NA | 6.5 | NA |
| Ferulic acid | NA | NA | NA | 0 |
Cultures were grown in SC-Leu minimal medium with galactose as a carbon source and inducer.
NA, not applicable.
In conclusion, we describe the biosynthesis of milligram quantities of flavanone substances from an S. cerevisiae recombinant strain that carries a plant-derived gene cluster. Since many flavonoid substances are formed through the action of P450 monooxygenases that cannot be readily expressed in E. coli, our success in producing a variety of flavanone skeletons from recombinant yeast will allow us to proceed in the future with the biosynthesis of several other high-value flavonoid molecules, such as genistein and quercetin.
Acknowledgments
This work was supported by a research grant from the National Science Foundation (BES-0331404) to M. A. G. Koffas.
REFERENCES
- 1.Arts, I. C. W., and P. C. H. Hollman. 2005. Polyphenols and disease risk in epidemiologic studies. Am. J. Clin. Nutr. 81:317s-325s. [DOI] [PubMed] [Google Scholar]
- 2.BellLelong, D. A., J. C. Cusumano, K. Meyer, and C. Chapple. 1997. Cinnamate-4-hydroxylase expression in Arabidopsis—regulation in response to development and the environment. Plant Physiol. 113:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii, T., K. Kondo, F. Shimizu, H. Sone, J. Tanaka, and T. Inoue. 1990. Application of a ribosomal DNA integration vector in the construction of a brewer's yeast having α-acetolactate decarboxylase activity. Appl. Environ. Microbiol. 56:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology, p. 14-15. In J. N. Abelson and M. I. Simon (ed.), Methods in enzymology. Academic Press, San Diego, Calif.
- 5.Heller, W. 1986. Flavonoid biosynthesis: an overview, p. 25-42. In V. Cody, E. Middleton, and J. B. Harbone (ed.), Plant flavonoids in biology and medicine: biochemical, pharmacological, and structure-activity relationships, vol. 213. Alan R. Liss, New York, N.Y. [Google Scholar]
- 6.Hotze, M., G. Schroder, and J. Schroder. 1995. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 374:345-350. [DOI] [PubMed] [Google Scholar]
- 7.Hwang, E. I., M. Kaneko, Y. Ohnishi, and S. Horinouchi. 2003. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69:2699-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi, Y., M. Akita, K. Sakamoto, H. Liu, T. Shigeoka, T. Koyano, M. Kawamura, and T. Furuya. 1993. Large-scale production of anthocyanin by Aralia cordata cell suspension cultures. Appl. Microbiol. Biotechnol. 40:215-218. [Google Scholar]
- 9.Lozoya, E., H. Hoffmann, C. Douglas, W. Schulz, D. Scheel, and K. Hahlbrock. 1988. Primary structures and catalytic properties of isoenzymes encoded by the 2,4-coumarate-CoA ligase genes in parsley. Eur. J. Biochem. 176:661-667. [DOI] [PubMed] [Google Scholar]
- 10.Middleton, E., Jr., C. Kandaswami, and T. C. Theoharides. 2000. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 52:673-751. [PubMed] [Google Scholar]
- 11.Muir, S. R., G. J. Collins, S. Robinson, S. Hughes, A. Bovy, C. H. Ric De Vos, A. J. van Tunen, and M. E. Verhoeyen. 2001. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 19:470-474. [DOI] [PubMed] [Google Scholar]
- 12.Romanos, M. A., C. A. Scorer, and J. J. Clare. 1992. Foreign gene-expression in yeast—a review. Yeast 8:423-488. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder, G., E. Wehinger, R. Lukacin, F. Wellmann, W. Seefelder, W. Schwab, and J. Schroder. 2004. Flavonoid methylation: a novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry 65:1085-1094. [DOI] [PubMed] [Google Scholar]
- 14.Seki, M., P. Carninci, Y. Nishiyama, Y. Hayashizaki, and K. Shinozaki. 1998. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15:707-720. [DOI] [PubMed] [Google Scholar]
- 15.Seki, M., M. Narusaka, A. Kamiya, J. Ishida, M. Satou, T. Sakurai, M. Nakajima, A. Enju, K. Akiyama, Y. Oono, M. Muramatsu, Y. Hayashizaki, J. Kawai, P. Carninci, M. Itoh, Y. Ishii, T. Arakawa, K. Shibata, A. Shinagawa, and K. Shinozaki. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296:141-145. [DOI] [PubMed] [Google Scholar]
- 16.Vantunen, A. J., S. A. Hartman, L. A. Mur, and J. N. M. Mol. 1989. Regulation of chalcone flavanone isomerase (Chi) gene-expression in Petunia-Hybrida—the use of alternative promoters in corolla, anthers and pollen. Plant Mol. Biol. 12:539-551. [DOI] [PubMed] [Google Scholar]
- 17.Vantunen, A. J., R. E. Koes, C. E. Spelt, A. R. Vanderkrol, A. R. Stuitje, and J. N. M. Mol. 1988. Cloning of the 2 chalcone flavanone isomerase genes from Petunia-Hybrida—coordinate, light-regulated and differential expression of flavonoid genes. EMBO J. 7:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, X. H., Z. J. Wang, and N. A. DaSilva. 1996. G418 selection and stability of cloned genes integrated at chromosomal delta sequences of Saccharomyces cerevisiae. Biotechnol. Bioeng. 49:45-51. [DOI] [PubMed] [Google Scholar]
- 19.Yan, Y., J. Chemler, L. Huang, S. Martens, and M. A. Koffas. 2005. Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 71:3617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]