Abstract
The spatial organization of cells within bacterial aggregates on leaf surfaces was determined for pair-wise mixtures of three different bacterial species commonly found on leaves, Pseudomonas syringae, Pantoea agglomerans, and Pseudomonas fluorescens. Cells were coinoculated onto bean plants and allowed to grow under moist conditions, and the resulting aggregates were examined in situ by epifluorescence microscopy. Each bacterial strain could be localized because it expressed either the green or the cyan fluorescent protein constitutively, and the viability of individual cells was assessed by propidium iodide staining. Each pair of bacterial strains that was coinoculated onto leaves formed mixed aggregates. The degree of segregation of cells in mixed aggregates differed between the different coinoculated pairs of strains and was higher in mixtures of P. fluorescens A506 and P. agglomerans 299R and mixtures of P. syringae B728a and P. agglomerans 299R than in mixtures of two isogenic strains of P. agglomerans 299R. The fractions of the total cell population that were dead in mixed and monospecific aggregates of a gfp-marked strain of P. agglomerans 299R and a cfp-marked strain of P. agglomerans 299R, or of P. fluorescens A506 and P. agglomerans 299R, were similar. However, the proportion of dead cells in mixed aggregates of P. syringae B728a and P. agglomerans 299R was significantly higher (13.2% ± 8.2%) than that in monospecific aggregates of these two strains (1.6% ± 0.7%), and it increased over time. While dead cells in such mixed aggregates were preferentially found at the interface between clusters of cells of these strains, cells of these two strains located at the interface did not exhibit equal probabilities of mortality. After 9 days of incubation, about 77% of the P. agglomerans 299R cells located at the interface were dead, while only about 24% of the P. syringae B728a cells were dead. The relevance of our results to understanding bacterial interactions on leaf surfaces and the implications for biological control of pathogenic and other deleterious microorganisms is discussed.
Leaf surfaces are colonized by a variety of microorganisms, and many factors have been suggested to be involved in dictating the composition and organization of such communities. While evidence of direct interactions among particular populations has been provided by studies of the biological control of plant-pathogenic fungi and bacteria (1, 11, 19) and by studies of coexistence of particular epiphytic bacterial strains (33), the factors determining interactions between epiphytic bacteria are poorly understood. The efficacy of biological control of foliar disease, especially under variable field conditions, is often inconsistent (1, 13, 15). While antibiosis, hyperparasitism, and resource competition are believed to play essential roles in such interactions, how spatially and temporally variable such interactions are between populations remains unclear. Incomplete biological control by applied antagonists has been proposed to be due, at least in part, to spatial isolation of pathogen and antagonist (13).
While it has been shown that coexistence between two or more bacterial populations can be mediated through nutritional resource partitioning (25, 33), recent reports describing the spatial segregation of the majority of epiphytic bacteria into aggregates suggest that coexistence could also be achieved through spatial separation of individual populations (21, 23, 27). It seems likely that, since the majority of epiphytic bacteria are located within a few large aggregates (21, 23, 24), the opportunity for interactions would be limited to only a few sites on the leaf surface and to only a subset of cells in the population, irrespective of whether coexistence between individual populations is mediated through resource partitioning or spatial segregation. Since several bacterial and fungal species occur within individual cell aggregates on leaves (26), a complex pattern of interactions between these species, even on such small scales, is possible.
Studies of the location of bacterial cells within mixed-species biofilms found in aquatic environments revealed substantial spatial organization of individual populations (31). A given bacterial species usually occurred in discrete locations within the biofilm. The complex organizational structures of biofilms may result from several factors acting together, such as the creation of nutritional gradients resulting in growth differentiation, chemotactic movements within the community, establishment of syntrophic relationships, and excretion of regulatory communication signals leading to construction of new organizational forms (31). The spatial organization of bacterial aggregates in the phyllosphere, however, has not been described and might be influenced additionally by the topography and spatial heterogeneity in nutrients and water availability of the leaf surface environment. In an attempt to test the extent to which cells are spatially partitioned within aggregates on leaf surfaces, we established different pair-wise mixtures of three different bacterial species commonly found on leaves, Pseudomonas syringae, Pantoea agglomerans, and Pseudomonas fluorescens, by coinoculation and examined the spatial structures of the resulting aggregates in situ by epifluorescence microscopy. To differentiate the bacteria, each expressed either the green or the cyan fluorescent protein constitutively. Interactions among the strains were assessed by determining the viability of individual cells within aggregates by staining with propidium iodide. Our objective was to determine if the spatial organization of bacterial cells within aggregates on leaves was influenced by the bacterial strains constituting an aggregate. Since we found that different bacterial strains exhibited different degrees of spatial segregation, we also then estimated the fraction of cells of the individual populations in direct contact with each other. The relevance of our results to understanding the ecology of bacterial interactions on leaf surfaces and the implications for biological control of pathogenic and other deleterious microorganisms is discussed.
MATERIALS AND METHODS
Bacterial strains and culture media.
The origins and characteristics of Pseudomonas syringae pv. syringae B728a, Pseudomonas fluorescens A506, and Pantoea agglomerans 299R have been previously reported (4, 18, 32). Cultures were stored at −80°C in 15% (vol/vol) glycerol in 10 mM potassium phosphate buffer, pH 7.0, and were routinely grown on King's medium B. Plasmid DNA was isolated from Escherichia coli DH5α(pKT-trp) (12) by using a QIAGEN DNA isolation kit (QIAGEN Inc., Valencia, CA) and was transferred into P. syringae B728a (22) and P. fluorescens A506 by electroporation using standard procedures (30). Plasmid pKT-trp consists of a green fluorescent protein marker gene (gfp) driven by the trp promoter from Salmonella enterica serovar Typhimurium (12). P. agglomerans 299R(CFP) carries plasmid pWM1009, which consists of a cyan fluorescent protein marker gene (cfp) fused to a consensus Campylobacter promoter sequence (20), yielding constitutive expression in P. agglomerans. P. agglomerans 299R(GFP) carries plasmid pFRU48, which consists of a gfp marker fused to the promoter of fruR and yielding constitutive expression of gfp in P. agglomerans.
Plant inoculation and growth conditions.
All experiments were conducted with 2-week-old, greenhouse-grown bean plants (Phaseolus vulgaris cv. Bush Blue Lake 274) incubated under controlled conditions in the laboratory. Mixtures of two bacterial strains were applied with an artist's airbrush by spraying the plants with an equal mixture of P. agglomerans 299R(GFP) and 299R(CFP), P. syringae B728a(pKT-trp) and P. agglomerans 299R(CFP), or P. fluorescens A506(pKT-trp) and P. agglomerans 299R(CFP) with a combined cell concentration of 107 CFU/ml in phosphate buffer, resulting in the deposition of ca. 105 bacteria per leaf. Drop size and inoculum concentration were calibrated so that cells arrived on the leaf surface as solitary cells. The plants were then kept in a moist chamber maintained at close to 100% relative humidity at 22°C for the duration of the experiment (up to 9 days). Experiments were conducted exclusively under high relative humidity conditions in order to avoid the presence of dead cells within aggregates resulting from desiccation stress (23).
To determine any differences in the growth and survival of genetically marked bacterial strains on plants, the plants were inoculated with equal numbers of each member of an isogenic strain pair, as described above, and the ratio of the two strains in the mixture was measured daily for up to 3 days. Likewise, the relative behaviors of a marked strain and its parental strain in mixtures with another species were determined on plants inoculated with equal numbers of cells of a genetically marked strain and a test strain (pair 1) as well as a nonmarked strain and the same test strain (pair 2); tests of the similarity of the ratio of strains in pair 1 and pair 2 were then done. The ratio of each strain in a particular pair of tests was measured daily for up to 3 days. Cells were recovered by dilution plating of leaf washings, as in other studies (22), onto King's medium B containing rifampin. The strains in a given mixture were distinguished from each other by visualization of the expression of fluorescent proteins in the marked strains. The ratio of each strain pair was measured on five individual replicate leaves for each sampling time for each mixture.
Sample preparation for microscopy.
The frequency and size of monospecific and mixed-strain aggregates, as well as the viability of bacterial cells in each aggregate, were determined directly in situ by epifluorescence microscopy. For each treatment, three leaves were randomly selected, and measurements were obtained from the upper leaf surface. Six segments of approximately 1 by 1 cm were randomly cut from each leaf and placed on top of 100 μl of melted water agar (1%) on a microscope slide in order to stabilize the leaf segments and to ensure a flat surface for microscopic observations. After solidification of the agar (in about 20 s), 20 μl of a solution of propidium iodide (10 μg/ml) in Aqua-Poly/Mount (Polysciences Inc., Warrington, PA) was placed on the center of a coverslip, which was then gently pressed down onto the leaf segment. The mountings were kept in the dark at room temperature for 10 min and then observed by epifluorescence microscopy. For each experiment, the viability of cells in the inoculum was determined prior to inoculation of bacteria onto leaves. Suspensions of inoculated cells were adjusted to a concentration of 108 CFU/ml and stained with propidium iodide (10 μg/ml) for 10 min in the dark at room temperature. Five microliters of each suspension was mixed with 5 μl of Aqua-Poly/Mount, deposited onto a slide, and immediately observed under a microscope. At least 20 random fields of view, containing a total of at least 400 cells, were observed per slide, and the total numbers of dead (red) and living cells were enumerated.
Epifluorescence microscopy.
Samples were observed by epifluorescence microscopy using an Axiophot Zeiss microscope equipped with a 10×/0.30-, 20×/1.30-, 40×/0.75-, or 100×/1.30-numerical-aperture Plan Neofluar objective (Zeiss Inc., Oberkochen, Germany). A CFP/yellow fluorescent protein 51017 filter set (Chroma Technology Corp., Brattleboro, VT) was used to visualize green and cyan fluorescence in the same field of view and to count the total number of GFP-marked and CFP-marked cells present in each aggregate. Dead (red) cells were visualized using a separate filter set for rhodamine (excitation filter, 526 to 566 nm; dichroic mirror, 580 nm; long-pass emission filter, 590 nm). In addition to the rhodamine filter set, a 650-nm short-pass filter (Melles-Griot, Irvine, CA) was used to improve the quality of images by partially blocking the strong red autofluorescence of the leaf. Images were captured with an Optronics DEI-750 video camera, transferred to a personal computer platform, and processed with Corel Photopaint software (Corel Co., Ottawa, Canada). Processed images were obtained by combining two images of the same field of view captured by using the filter sets for CFP and GFP and for propidium iodide. On the resulting image, live cells appeared either cyan or green, and dead cells of these strains appeared purple or orange, respectively.
Characterization of monospecific and mixed bacterial aggregates.
The entire surface of each leaf segment was extensively scanned at different magnifications in order to locate aggregates of different sizes. Images of each monospecific aggregate or of mixed aggregates, containing 32 cells or more, that were encountered were captured at a final magnification of ×500 or ×1,250, depending on the actual size of the aggregate. The total number and viability of bacterial cells in each monospecific aggregate was determined. For each treatment, up to 42 mixed-strain aggregates were characterized as follows. The total number of clusters formed by each strain, the number and viability of bacterial cells in each cluster, and the number and viability of bacterial cells of a given strain in contact with the other strain (at the interface between two clusters) were determined. A segregation index, SI = (R1/C1) + (R2/C2), was used to quantitatively describe the degree of segregation of two given strains at a given time within an aggregate. C1 and C2 correspond to the number of clusters formed by strains 1 and 2, respectively, within the aggregate, and R1 and R2 are the fraction of cells represented by strain 1 and strain 2, respectively, within the aggregate as a whole, e.g., R1 = N1/(N1 + N2), where N1 and N2 are the numbers of cells of strain 1 and strain 2, respectively, within a given aggregate. SI values ranged from 0 (cells randomly distributed) to 1 (cells highly clustered) (Fig. 1).
FIG. 1.
Examples of SI values and the corresponding spatial distribution of cells within aggregates. Bacterial cells of strain 1 and strain 2 are represented by individual squares in white and gray, respectively. SI = (R1/C1) + (R2/C2), where C1 and C2 correspond to the numbers of clusters formed by strains 1 and 2, respectively, within the aggregate, and R1 and R2 are the proportions of cells of strain 1 and strain 2 in an aggregate.
Statistics.
Data management and computation were performed using Microsoft Excel software (Microsoft Co., Redmond, WA). Descriptive statistical analysis and analysis of variance of the arcsine square root-transformed measurements of the proportion of living cells were all performed with the software Statistica (Statsoft Inc., Tulsa, OK). A representative subset of segregation index values of aggregates, which did not differ in number of cells per aggregate and relative fractions of strains 1 and 2, was used to compare segregation indices from different strain pairs.
RESULTS
Relative behavior of genetically marked bacterial strains.
The genetically marked bacterial strains used in this study to facilitate visualization on leaf surfaces by fluorescence microscopy appeared to behave the same as their parental strain in all cases when cultured as a single species or as a mixture of species on leaves. Strain behavior was determined by measuring the ratio of two isogenic strains as a function of time after inoculation in equal cell numbers of plants by culturing bacteria removed from leaves. No significant change in the ratio of P. agglomerans 299R(CFP) to P. agglomerans 299R, P. agglomerans 299R(GFP) to P. agglomerans 299R, P. fluorescens A506(pKT-trp) to P. fluorescens A506, or of P. syringae B728a(pKT-trp) to P. syringae B728a were observed after incubation on leaves under sequential moist and dry conditions (data not shown). The later result is consistent with an earlier report that that P. syringae B728a(pKT-trp) behaved similarly to B728a on plants (22). The interactions of marked strains with other species also did not appear to differ from that of their parental strain. No significant difference in the ratio of P. fluorescens A506(pKT-trp) to P. agglomerans 299R in mixtures with that of P. fluorescens A506 to P. agglomerans 299R in mixtures at a given sampling time was observed, and the ratios remained constant over a 3-day sampling period (data not shown). Likewise, no significant difference in the ratio of P. syringae B728a(pKT-trp) to P. agglomerans 299R in mixtures with that of P. syringae B728a to P. agglomerans 299R in mixtures at a given sampling time was observed, and the ratios remained constant over a 3-day incubation period (data not shown).
Relative fraction of mixed and monospecific aggregates.
Each pair of bacterial strains coinoculated onto leaves formed mixed aggregates (Fig. 2). For each such pair of strains, the species compositions of between 100 and 200 aggregates, having from 32 to over 10,000 cells per aggregate, were analyzed. After 5 days of incubation on bean leaves under moist conditions, mixed aggregates represented between 40.1% and 51.3% of the total number of aggregates observed (Table 1). The fraction of aggregated cells located in mixed aggregates ranged from about 20% to 66% of the total number of aggregated cells (Table 1).
FIG. 2.
Mixed aggregates formed by P. agglomerans 299R(GFP) and 299R(CFP) (A and B), P. fluorescens A506 and P. agglomerans 299R(CFP) (C and D), and P. syringae B728a and P. agglomerans 299R(CFP) (E and F) observed after 5 days of incubation on bean leaf surfaces under moist conditions. 299R(CFP) cells are represented in cyan, while all the other strains are represented in green. Magnification, ×500.
TABLE 1.
Fraction of aggregates formed on leaves after inoculation of pairs of bacterial strains that contained mixtures of the two strains and degree of segregation of those cells within such mixed aggregates
| Strain paired with P. agglomerans 299R(CFP) | % MAa | % of cells in MAb
|
No. of clusters/aggregate
|
% Cells in contact | SI
|
|||
|---|---|---|---|---|---|---|---|---|
| Strain 1 | Strain 2 | Range | Avg | Value | Range | |||
| P. agglomerans 299R(GFP) | 51.3 Ac | 66.2 A | 66.2 A | 2-27 | 8.9 A | 24.7 A | 0.381 B | 0.080-1 |
| P. fluorescens A506 | 44.0 A | 80.6 A | 18.7 B | 2-15 | 3.6 B | 13.1 B | 0.718 A | 0.137-1 |
| P. syringae B728a | 40.1 A | 55.1 A | 51.6 AB | 2-10 | 4.6 B | 13.2 B | 0.610 A | 0.211-1 |
Percentages of mixed aggregates (MA) relative to the total number of aggregates observed.
Strain 1, strain listed in column 1; strain 2, P. agglomerans 299R(CFP).
Mean values followed by the same letter do not differ (P = 0.05) based on Duncan's multiple-range test.
Spatial organization of cells within mixed aggregates.
The degree of clustering of mixed aggregates differed between the different pairs of strains coinoculated onto bean leaves (Table 1). Subsets of our data in which the numbers of cells per aggregate, as well as the ratios of the two strains within the aggregates, did not differ were used to compare the spatial organizations of mixed aggregates. The average fraction of cells of a given strain within an aggregate ranged from 0.44 to 0.56, and the average number of cells per aggregate ranged from 224 ± 40 to 394 ± 88 (means ± standard errors) in different pair-wise mixtures. SI values were highly variable between aggregates of a given mixture (Table 1) and ranged from 0.080 to 1, 0.137 to 1, and 0.211 to 1 for mixed aggregates of P. agglomerans 299R(GFP) and 299R(CFP), P. fluorescens A506 and P. agglomerans 299R, and P. syringae B728a and P. agglomerans 299R, respectively (Table 1). The average number of clusters per aggregates also varied greatly in a given strain pair (Table 1).
After 5 days of incubation, the average SI value of mixed aggregates of P. agglomerans 299R(GFP) and 299R(CFP) was significantly smaller (P < 0.01) than such measures of segregation for P. fluorescens A506 and P. agglomerans 299R or P. syringae B728a and P. agglomerans 299R (Table 1). No significant differences were observed between the SI values of a mixture of P. fluorescens A506 and P. agglomerans 299R and that of P. syringae B728a and P. agglomerans 299R. For each combination of two strains, average SI values of aggregates consisting of 512 cells or fewer did not differ significantly and ranged from 0.454 to 0.689 (0.566 ± 0.118). However, average SI values of aggregates consisting of 512 cells or more were significantly higher (P < 0.01) in mixed aggregates formed by P. fluorescens A506 and P. agglomerans 299R and by P. syringae B728a and P. agglomerans 299R (0.807 ± 0.172 and 0.824 ± 0.275, respectively) than in mixed aggregates formed by P. agglomerans 299R(GFP) and 299R(CFP) (0.112 ± 0.024). The average percentage of cells of one strain in contact with cells of the other strain (interface between clusters) for mixed aggregates of P. agglomerans 299R(GFP) and 299R(CFP) was significantly higher (P < 0.001) than that of mixed aggregates of P. fluorescens A506 and P. agglomerans 299R and of P. syringae B728a and P. agglomerans 299R (Table 1).
Viability of cells in monospecific and mixed bacterial aggregates.
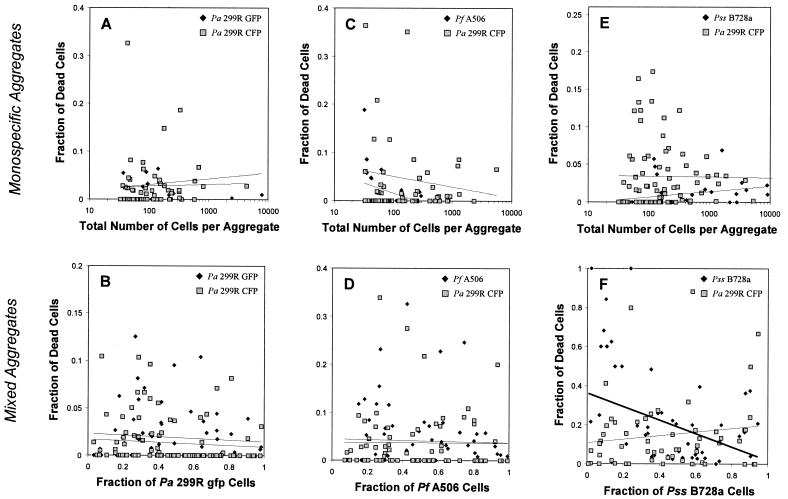
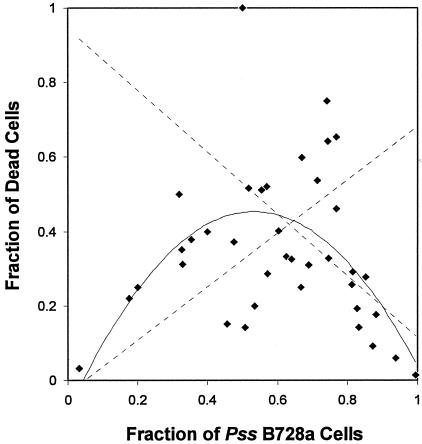
The relative fractions of the total cell population that were dead in mixed and monospecific aggregates of P. agglomerans 299R(GFP) and 299R(CFP) (Fig. 3A and B) or of P. fluorescens A506 and P. agglomerans 299R (Fig. 3C and D) did not differ significantly. However, the proportion of dead cells in mixed aggregates of P. syringae B728a and P. agglomerans 299R was significantly higher (13.2% ± 8.2%) than the fraction of dead cells in monospecific aggregates (1.6% ± 0.7%) of these two strains. The fractions of cells of P. syringae B728a and P. agglomerans 299R in mixed aggregates that were dead were 16.0% ± 10.1% and 10.7% ± 7.0%, respectively, compared with 0.5% ± 0.3% and 2.8% ± 1.5%, respectively, in monospecific aggregates. The fraction of dead cells of strain 299R increased, while the fraction of dead cells of strain B728a decreased, as a function of the increasing proportion of B728a cells in an aggregate (Fig. 3E and F). The fraction of cells of both B728a and 299R in mixed aggregates that were dead increased over time, and after 7 days of incubation, regression analysis revealed a significant positive correlation between the fraction of dead cells of one strain with the increasing proportion of the other strain in mixed aggregates (Fig. 4). The maximum proportion of dead cells in mixed cell aggregates of this pair of strains occurred when the strains were nearly equally represented in the mixture (Fig. 5).
FIG. 3.
Fractions of bacterial cells in monospecific aggregates (A, C, and E) and mixed aggregates (B, D, and F) that were dead after 5 days of incubation on leaves, as a function of the total number of cells per aggregate (monospecific aggregates) and as a function of the relative fraction of P. agglomerans (Pa) 299R(GFP) cells (B), P. fluorescens (Pf) A506 cells (D), and P. syringae pv. syringae (Pss) B728a cells (F) in mixed aggregates. Each point represents a single cell aggregate of at least 32 cells in size. Lines represent the linear regressions of the fraction of dead cells as a function of the total number of cells per aggregate in monospecific aggregates (A, C, and E) or the linear regressions of the fraction of dead cells as a function of the relative fraction of 299R(GFP) cells (B), A506 cells (D), or B728a cells (F) in mixed aggregates. All regressions were nonsignificant except for the negative correlation between the fraction of dead B728a cells and the increasing fraction of B728a cells within a given aggregate (y = −0.35x + 0.36, R2 = 0.148, P = 0.003) Bold line evident in panel F.
FIG. 4.
Fraction of cells of P. syringae pv. syringae (Pss) B728a (diamonds) and P. agglomerans 299R(CFP) (squares) in mixed aggregates after 7 days of incubation on leaves, as a function of the fraction of B728a cells in an aggregate. The solid line represents the significant positive correlation between the fraction of dead 299R(CFP) cells and the increasing fraction of B728a cells within a given aggregate (y = 0.172x − 0.03, R2 = 0.333, P < 0.001). The dashed line represents a significant negative correlation between the fraction of dead B728a cells and the increasing fraction of B728a cells within a given aggregate (y = −0.83x + 0.94, R2 = 0.326, P < 0.001).
FIG. 5.
Fraction of cells of either P. syringae pv. syringae (Pss) B728a or P. agglomerans 299R(CFP) that were dead in mixed aggregates of these two strains, after 7 days of incubation on leaves, as a function of the proportion of B728a cells in the aggregate. The solid line represents the polynomial regression of the fraction of dead cells as a function of proportion of B728a cells in mixed aggregates (y = −1.89x2 + 2.02x − 0.08, R2 = 0.305, P < 0.001).
Spatial distribution of dead cells within mixed aggregates.
While the spatial organizations of cells in mixed aggregates of P. syringae B728a and P. agglomerans 299R as well as P. fluorescens A506 and P. agglomerans 299R were similar, the fraction of dead cells was significantly higher in mixtures of strains B728a and 299R, and dead cells were preferentially found at the interface between cell clusters of these strains (Fig. 6). After 5 days of incubation, 19.7% ± 0.2% of the cells of either B728a or 299R that were in direct contact with each other were dead, while only 6.4% ± 0.1% of the rest of the cells in the aggregate were dead. The proportion of dead cells increased significantly with time, and by 7 days of incubation, 64.9% ± 7.4% (P = 0.005) of the cells of the two strains that were in contact with each other were dead, while only 15.2% ± 1.2% of the rest of the cells were dead. Cells of these two strains that were located at the interface did not exhibit equal probabilities of mortality. For example, after 5 days of incubation, 22.5% ± 0.3% of strain 299R cells located at the interface were dead, while a significantly lower proportion of strain B728a cells (13.3% ± 0.3%, P < 0.001) were dead. A similar trend was observed over time, and after 9 days of incubation, 77.1% ± 4.2% of 299R cells located at the interface were dead, while only 23.5% ± 8.7% of B728a cells were dead. Such differences in mortality between strains at the interfaces with each other were not observed for other strain pairs, and the proportions of dead cells located at an interface and in the rest of the aggregate were similar. For example, after 5 days of incubation, 0.7% of the cells located at the interface and 0.9% of the cells in the rest of aggregates of P. agglomerans 299R(GFP) and 299R(CFP) were dead. Likewise, 3.2% of cells in contact and 4.2% of more-distal cells in aggregates of P. fluorescens A506 and P. agglomerans 299R were dead. The proportion of dead cells in such aggregates did not increase significantly with time (data not shown).
FIG. 6.
Mixed aggregates formed by P. syringae pv. syringae B728a (green cells) and P. agglomerans 299R(CFP) (cyan cells) after 5 days (A) and 7 days (B and C) of incubation on bean leaves, showing the presence of dead cells at the interface between clusters. Dead cells of B728a are represented in orange, and dead cells of 299R(CFP) are represented in purple. Magnification, ×500 (A) and ×1,250 (B and C).
DISCUSSION
The degree of interaction between epiphytic bacterial species most likely depends on the spatial distribution of cells on the leaf surface (i.e., their respective habitats) as well as the spatial organization of cells within aggregates when the habitats are coincident (coaggregation occurs). While the proportion of cells of different bacterial species that come into contact with each other on a leaf will be a function of the population sizes of the different species, our results suggest that only a small fraction of the cells of different species are in contact even on heavily colonized leaves. For example, when P. syringae pv. syringae B728a was coinoculated with P. agglomerans 299R, only about 13% of the cells of the two species that were present even in mixed aggregates were in contact with each other. Knowing that mixed aggregates account for only about half of the aggregated cells and assuming that aggregated cells represented about half of the total number of cells on a leaf (21), we can estimate that only about 3% of the cells of these two species are actually in direct contact with each other. In an unpublished study, we found that when P. agglomerans 299R was first allowed to establish large populations on bean leaves and P. syringae B728a was subsequently inoculated onto these plants and allowed to grow for 5 more days under moist conditions, mixed aggregates represented only about 8% of the total number of aggregates observed, and about 0.2% and 1.9% of immigrant cells and residents cells, respectively, were in direct contact. The low proportion of cells of these two species in contact with each other may result from negative interactions between bacteria. We found, however, that when the isogenic P. agglomerans strains 299R(GFP) and 299R(CFP) were coinoculated, and no positive or negative interactions between the two strains are to be expected, about 8% of the cells of the two isogenic strains were in direct contact with each other. The process of enlargement of population sizes from relatively small numbers of immigrant cells apparently leads to the formation of a spatially “clonal” community where the nearest neighbors are most likely also direct descendants of the pioneer cell.
The significantly smaller fraction of mixed cell aggregates formed by P. syringae B728a and P. agglomerans 299R compared to those formed by the isogenic strains P. agglomerans 299R(GFP) and 299R(CFP) suggests that B728a and 299R have different colonization patterns on the leaf surface. In a recent study, we reported that P. syringae B728a forms aggregates preferentially at the base of glandular trichomes and in the grooves between cells associated with veins (21). While we observed that P. agglomerans 299R was able to form aggregates at these same sites, it also was able to form aggregates on top of undifferentiated plant epidermal cells. We hypothesize that such aggregates may have resulted from a modification of their microhabitat as a result of indole-3-acetic acid production, resulting in an increased rate of nutrient leakage from plant cells (5). Since mixed aggregates of P. syringae B728a and P. agglomerans 299R were found mostly at the base of glandular trichomes and were observed rarely on top of epidermal cells, negative interactions between these two strains in large 299R aggregates might have prevented B728a from colonizing these later sites. In contrast, the large majority of cells of P. fluorescens A506 (about 80%) were found in mixed aggregates with P. agglomerans 299R, suggesting that A506 might benefit from the presence of 299R. While we did not quantify the spatial distribution of mixed aggregates formed by these two strains, we often observed that A506 formed large aggregates on top of epidermal cells only when these cells were also colonized by 299R. Since no negative interactions were observed between these two strains, this observation supports our hypothesis that A506 benefits from the local modification of its microhabitat by coincident 299R cells.
While we observed a significant difference in the spatial organizations of mixed aggregates of the different pair-wise mixtures of strains on bean leaves, the variability in spatial organizations of the mixed aggregates formed by such strains precludes us from identifying traits that dictate such organization. The results instead suggest, as expected, that bacterial interactions per se are not the only mechanisms shaping the structure of epiphytic bacterial communities. The spatial organization of cells in mixed aggregates of P. syringae B728a and P. agglomerans 299R was similar to that of P. fluorescens A506 and P. agglomerans 299R. While the fraction of dead cells was significantly higher in the former, there were no significant differences between the respective SI values, the average numbers of microcolonies per aggregate, or the fractions of cells of the two strains in contact with each other. We conclude that negative interactions between strains do not strongly affect the spatial organization of cells within an aggregate. As hypothesized for biofilms formed in aquatic environments (31), the spatial organization of microorganisms and the various structural forms apparently result from differences in local substrate availability (29), as well as differential gene expression, that directly controls the spatial organization of bacteria (9).
As illustrated in Fig. 2, the topography of the leaf influences the shape of aggregates and therefore, indirectly, the spatial organization of bacterial cells within aggregates. For example, aggregates formed at the base of glandular trichomes were larger than and not as elongated as aggregates formed between the grooves of plant epidermal cells and therefore tended to have a larger proportion of cells of the two species in direct contact. While the source of nutrients on leaf surfaces remains unknown, we can assume that different leaf sites may differ quantitatively and qualitatively in the amount of nutrients available to bacteria (17, 25). Differences in local nutrient concentrations may promote heterogeneous growth activities of bacteria at particular sites. Such differences may favor growth of one bacterial species and not another and result in a more clustered organization or may promote metabolic interactions and therefore a closer coupling between two species (28). A qualitative change in nutrient availability or composition at a site, due to metabolic activity or microhabitat modification by a given strain, may induce structural changes in biofilms (34) and perhaps also on leaves. Since most epiphytic bacteria may have the ability to move in response to existing or resulting nutrient gradients in their vicinity, the spatial pattern of occurrence of bacteria on leaves may be dynamic, and further observations over longer periods of time may help us identify general rules that govern the spatial organization of epiphytic communities.
Although bacterial gene expression on leaf surfaces might be influenced by many factors that differ within the microenvironment encountered on the leaf, several traits important in plant-microbe interactions have been shown to be regulated in a cell density-dependent manner via quorum-sensing mechanisms (2, 3, 8, 10, 16). Most plant-pathogenic bacteria, including species with an epiphytic phase such as Erwinia spp. and Pseudomonas syringae, produce N-acylated homoserine lactones (3, 7, 10) as a means of determining local cell abundance and coordinating cell density-dependent gene expression. Our preliminary studies reveal that aggregated cells of P. syringae produce N-acylated homoserine lactones on leaf surfaces, strongly suggesting that they may benefit from production of this signal molecule (unpublished data). We sometimes observed that the largest aggregates of P. syringae had a looser structure than the smaller aggregates (as illustrated in Fig. 2F and E, respectively), which might reflect the presence of a cell density-mediated exopolymeric matrix surrounding such cells. In addition, we observed that SI values of mixed aggregates formed by P. agglomerans 299R and P. syringae B728a were significantly higher in larger aggregates and that the fraction of dead cells of 299R in contact with B728a cells tended to increase with the increasing number of cells of P. syringae in the adjacent microcolony. We hypothesize that the loose structure of larger aggregates and the increasing segregation of cells with the increasing size of mixed aggregates formed by B728a and 299R, as well as the negative interactions observed between these two strains, may result from traits regulated in a cell density-dependent manner in P. syringae.
Our finding that a relatively small proportion of the total cells in a population are in direct contact, and hence maximally interacting, in a community may also explain the variable degree of coexistence of bacterial strains observed under different conditions with de Wit replacement design experiments (33). For example, Wilson and Lindow (33) found that P. agglomerans 299R exhibited variable levels of coexistence with P. syringae TLP2, ranging from a high degree of coexistence to a relatively low level of coexistence of P. syringae relative to P. agglomerans. While we did not examine the same P. syringae strain as that used by Wilson and Lindow (33), it is tempting to speculate that mixtures of these two species can be mutually inhibitory when in direct contact, and that under different environmental conditions such contact may be limiting and the coexistence observed may, in fact, be simply reflective of an environmentally mediated spatial segregation of the bacteria on the plant.
While the spatial organization of epiphytic bacterial populations had remained obscure until recently, the use of marker genes conferring the production of fluorescent proteins combined with propidium iodide as a viability stain has proven to be a valuable tool to study dual-species aggregates on leaf surfaces and has provided new insight into our understanding of bacterial interactions on leaf surfaces. Independently of the apparent complexity of the biological and environmental factors regulating the spatial structures of epiphytic communities, our study reveals that direct bacterial interactions on leaf surfaces is limited to only a few sites involving only a small fraction of the total bacterial population. While some interactions, such as nutrient acquisition via diffusion of soluble carbon sources, presumably could occur over large distances and not require actual cell-cell contact, studies using reporter gene fusions to environmentally responding genes have shown that even closely adjacent cells may sense different environmental conditions (6, 17, 20). Aspects of the physical environment of epiphytic cells, such as the wettability of the leaf surface and the availability of free moisture, could presumably modulate interactions that occur at a distance.
Spatial aggregation of bacteria on leaves may also explain the incomplete biological control of disease by applied antagonistic bacteria. A theoretical model developed by Johnson (13) suggests that incomplete biological control would be expected if a pathogen existed in refuges that were inaccessible to an applied biological control agent. Our results provide direct evidence of such spatial segregation, per se, of bacteria on the leaf. A recent theoretical contribution of Kinkel et al. (15) has addressed the implications of microbial dynamics across different spatial scales in a habitat such as a leaf in which resources are expected to be aggregated. Their modeling results suggest that at reasonable rates of immigration of bacterial cells to a leaf, while there could be a significant effect of competition by a superior competitor on the population density of an inferior competitor, there will be few sites on a leaf that are jointly colonized and that the majority of the population development for each strain in a mixture will be within sites that are singly colonized (15). Kinkel et al. further illustrate that when resources are highly localized on a leaf, frequent escape from competitive (and presumably also other antagonistic) interactions will strongly reduce the significance of interactions to microbial population dynamics on leaves. Their prediction of leaf colonization on small scales is a close match to our observations. Our results that illustrate the infrequent occurrence of mixed species aggregates on leaves and the infrequent contact of cells even within these mixed-species aggregates, together with assessments of resource availability made with whole-cell nutritional biosensors (6, 14, 17, 20), are all consistent with a model of bacterial colonization of leaves characterized by resource aggregation.
Acknowledgments
We thank Maria Brandl and William Miller from the U.S. Department of Agriculture in Albany for providing us with Pantoea agglomerans 299R(pWM1009) and Sergio Lucero from the University of California—San Francisco for his valuable mathematical insight.
This study was supported by grant 99-35303-8633 from the U.S. Department of Agriculture National Research Initiative, by grant DR-F603-86ER13518 from the Department of Energy, and by support from the Torrey Mesa Research Institute, Syngenta Research and Technology, San Diego, CA.
REFERENCES
- 1.Andrews, J. H. 1992. Biological control in the phyllosphere. Annu. Rev. Phytopathol. 30:603-635. [DOI] [PubMed] [Google Scholar]
- 2.Barber, C. E., J. L. Tang, J. X. Feng, M. Q. Pan, T. J. G. Wilson, H. Slater, J. M. Dow, P. Williams, and M. J. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24:555-566. [DOI] [PubMed] [Google Scholar]
- 3.Beck von Bodman, S., and S. K. Farrand. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177:5000-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl, M. T., and S. E. Lindow. 1996. Cloning and characterization of a locus encoding an indolepyruvate decarboxylase involved in indole-3-acetic acid synthesis in Erwinia herbicola. Appl. Environ. Microbiol. 62:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandl, M. T., and S. E. Lindow. 1997. Environmental signals modulate the expression of an indole-3-acetic acid biosynthetic gene in Erwinia herbicola. Mol. Plant-Microbe Interact. 10:499-505. [DOI] [PubMed] [Google Scholar]
- 6.Brandl, M. T., B. Quiñones, and S. E. Lindow. 2001. Heterogeneous transcription of an indoleacetic acid biosynthetic gene in Erwinia herbicola on plant surfaces. Proc. Natl. Acad. Sci. USA 98:3454-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe. Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 8.Clough, S. J., K.-E. Lee, M. A. Schell, and T. P. Denny. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:3639-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 208:295-298. [DOI] [PubMed] [Google Scholar]
- 10.Dumenyo, C. K., A. Mukherjee, W. Chun, and A. K. Chatterjee. 1998. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur. J. Plant Pathol. 104:569-582. [Google Scholar]
- 11.Fravel, D. R. 1988. Role of antibiosis in the biological control of plant diseases. Annu. Rev. Phytopathol. 26:75-91. [Google Scholar]
- 12.Hallman, J., A. Quadt-Hallmann, W. G. Miller, R. A. Sikora, and S. E. Lindow. 2001. Endophytic colonization of plants by the biological control agent Rhizobium etli G12 in relation to Meloidogyne incognita infection. Phytopathology 91:415-422. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, K. B. 1994. Dose-response relationships and inundative biological control. Phytopathology 84:780-784. [Google Scholar]
- 14.Joyner, D. C., and S. E. Lindow. 2000. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 146:2435-2445. [DOI] [PubMed] [Google Scholar]
- 15.Kinkel, L. L., M. R. Newton, and K. J. Leonard. 2002. Resource aggregation in the phyllosphere: implications for microbial dynamics across spatial scales, p. 317-340. In S. E. Lindow, E. I. Hecht-Poinar, and V. J. Elliot (ed.), Phyllosphere microbiology. APS Press, St. Paul, Minn.
- 16.Koiv, V., and A. Mae. 2001. Quorum sensing controls the synthesis of virulence factors by modulating rsmA gene expression in Erwinia carotovora subsp. carotovora. Mol. Genet. Genomics 265:287-292. [DOI] [PubMed] [Google Scholar]
- 17.Leveau, J. H. J., and S. E. Lindow. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 98:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loper, J. E., and S. E. Lindow. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449-1454. [Google Scholar]
- 19.Loper, J. E., and S. E. Lindow. 1993. Roles of competition and antibiosis in suppression of plant diseases by bacterial biological agents, p. 144-145. In R. D. Lumsden and J. L. Vaughn (ed.), Pest management: biologically based technologies. Proceedings of the Beltsville Symposium XVIII. American Chemical Society, Washington, D.C.
- 20.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monier, J.-M., and S. E. Lindow. 2004. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 70:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monier, J.-M., and S. E. Lindow. 2003. Pseudomonas syringae responds to the environment on leaves by cell size reduction. Phytopathology 93:1209-1216. [DOI] [PubMed] [Google Scholar]
- 23.Monier, J.-M., and S. E. Lindow. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. USA 100:15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris, C. E., M. B. Barnes, and R. J. C. McLean. 2002. Biofilms on leaf surfaces: implications for the biology, ecology and management of populations of epiphytic bacteria, p 138-154. In S. E. Lindow, E. I. Hecht-Poinar, and V. J. Elliot (ed.), Phyllosphere microbiology. APS Press, St. Paul, Minn.
- 25.Morris, C. E., and D. I. Rouse. 1985. Role of nutrients in regulating epiphytic bacterial populations, p. 63-82. In C. E. Windels and S. E. Lindow (ed.), Biological control on the phylloplane. American Phytopathological Society, St. Paul, Minn.
- 26.Morris, C. E., J.-M. Monier, and M.-A. Jacques. 1997. Methods for observing microbial biofilms directly on leaf surfaces and recovering them for isolation of culturable microorganism. Appl. Environ. Microbiol. 63:1570-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris, C. E., J.-M. Monier, and M.-A. Jacques. 1998. A technique to quantify the population size and composition of the biofilm component in communities of bacteria in the phyllosphere. Appl. Environ. Microbiol. 64:4789-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen, A. T., T. Tolker-Nielsen, K. B. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 29.Picioreanu, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 1998. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol. Bioeng. 58:101-116. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Tolker-Nielsen, T., and S. Molin. 2000. Spatial organization of microbial biofilm communities. Microb. Ecol. 40:75-84. [DOI] [PubMed] [Google Scholar]
- 32.Wilson, M., and S. E. Lindow. 1993. Effect of phenotypic plasticity on epiphytic survival and colonization by Pseudomonas syringae. Appl. Environ. Microbiol. 59:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, M., and S. E. Lindow. 1994. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl. Environ. Microbiol. 60:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, S. J. Caldwell, and D. E. Caldwell. 1994. Multicellular organization in a degradative biofilm community. Appl. Environ. Microbiol. 60:434-446. [DOI] [PMC free article] [PubMed] [Google Scholar]