Abstract
The failure to reduce the Campylobacter contamination of intensively reared poultry may be partially due to Campylobacter resisting disinfection in water after their internalization by waterborne protozoa. Campylobacter jejuni and a variety of waterborne protozoa, including ciliates, flagellates, and alveolates, were detected in the drinking water of intensively reared poultry by a combination of culture and molecular techniques. An in vitro assay showed that C. jejuni remained viable when internalized by Tetrahymena pyriformis and Acanthamoeba castellanii for significantly longer (up to 36 h) than when they were in purely a planktonic state. The internalized Campylobacter were also significantly more resistant to disinfection than planktonic organisms. Collectively, our results strongly suggest that protozoa in broiler drinking water systems can delay the decline of Campylobacter viability and increase Campylobacter disinfection resistance, thus increasing the potential of Campylobacter to colonize broilers.
Campylobacter jejuni is recognized as one of the leading causes of food-borne disease in the developed world (2, 4, 39). Estimates suggest that in the United Kingdom and the United States 1.1 and 1%, respectively, of the populations are affected annually by Campylobacter infection, and substantial sums are lost due to clinical costs and lost working hours, e.g., between $1.3 and $6.2 billion in the United States alone (26, 35, 50). Campylobacteriosis usually involves a self-limited gastrointestinal illness lasting up to 1 week, after an incubation period of 1 to 7 days (20, 22, 58). More than 90% of campylobacteriosis cases are sporadic, with the consumption of (and cross-contamination from) undercooked poultry an identified risk factor (4, 12, 28, 32). An extensive Food Safety Authority survey, conducted between April and June 2001, found the United Kingdom national average of Campylobacter contamination to be 50% in finished raw chicken meat (including both whole and portioned samples) (24).
The management of infection in breeder flocks appears to have relatively little importance in the epidemiology of infection since most researchers have found no compelling evidence that campylobacters are transmitted vertically (12, 14, 21, 65, 67). It is most likely that chicks become colonized from environmental sources, e.g., unchlorinated drinking water (22). Intensively reared broiler chickens readily pick up C. jejuni from the environment and, since campylobacters have a wide range of hosts, there are many potential sources of infection (4, 21, 57). In the developed world, a variety of biosecurity measures, e.g., boot dips and hygiene barriers, are generally practiced on broiler farms (27, 56). However, despite these measures, broilers still have high levels of Campylobacter contamination, e.g., 105 to 109 CFU per g of intestinal contents (12, 48, 66). The presence of Campylobacter in the intestinal tract implicates ingestion of a contaminated source (45). Neither feeds nor fresh litter seem to be likely sources of Campylobacter. Commercial feeds are dried, are pelleted, contain little moisture (8 to 10%), tend to be pasteurized, and are air blown into silos (45, 57). The litter used on farms is generally wood shavings which are dry and resinous (being mainly softwood) and normally come directly from sawmills (57).
Drinking water has sometimes been found to be a significant source of infection (12). Viable, nonculturable organisms in water may be important in C. jejuni transmission, but efforts to infect day-old hatched chicks have been variable, and the significance of this source is still under review. In addition to free suspensions, bacteria in water systems also exist attached to sediment or in biofilms on submerged surfaces, where these communities usually consist of bacteria, fungi, algae, and protozoa with high grazing activity (73). Aquatic biofilms may harbor potential human pathogens, e.g., C. jejuni, and promote their survival through a variety of mechanisms such as uptake by protozoa, resulting in protection from disinfection (73). King et al. (37) demonstrated that C. jejuni, when ingested by the protozoan Tetrahymena pyriformis, was more than 50 times more resistant to free chlorine (1 mg per liter, pH 7.0 at 25°C) than freely suspended C. jejuni (37). The potential and significance of protozoa to act as transfer vehicles for Campylobacter to infect intensively reared poultry is unclear. This is because there is currently very little information on the identity of eukaryotic microbes in poultry drinking water systems and of possible interactions between protozoa and Campylobacter. We describe here a combination of culture-based and molecular techniques to assess the potential of waterborne protozoa to act as vehicles for the Campylobacter infection of intensively reared poultry.
MATERIALS AND METHODS
Broiler house biosecurity.
All broiler houses contained between 10,000 and 20,000 cock Cob breed 500 broilers. One-day-old chicks were delivered to the farms and placed on litter consisting of wood shavings coming directly from sawmills. Pasteurized feed was supplied by an automatic auger system with feed pans, and water was provided through nipple drinker systems. Between each flock, litter was removed from houses, which were then cleaned and disinfected. Broilers fed and drank when their houses were lit for 24 h when they were 1 day old, and when broilers were 7 days old, lighting was reduced to 4 h per day. All of the houses sampled in this investigation had a demarcation zone between the outside and inside of the house “marked” by a stepover bench. Each broiler house had separate overalls and Wellington boots (inside and outside). A disinfectant boot dip with a 1:20 dilution of Virudine (DuPont, Sudbury, Suffolk, United Kingdom), an iodophor disinfectant with 2.8% iodine and 28% phosphoric acid activity, which was supposed to be replenished weekly, were present outside each broiler house. All of the water systems in houses were supposed to be disinfected by farmers between every two flocks of broilers using a 1:600 dilution of Virudine, where the diluted disinfectant was pumped though the water system, left overnight, and then removed. Farmers performed daily checks of the houses, recording a variety of information, e.g., age, broiler weight, cumulative mortality, feed delivery, water meter readings, and house temperature. Members of the poultry industry performed audits before the broilers arrived and when they were 4 weeks old, where the presence of biosecurity measures, e.g., the presence of a hygiene barrier, was assessed and recorded.
Microbiological analysis.
Feces/bedding and cloacal swab samples were taken from a single broiler house at five different farms. Fifteen cloacal swabs were taken from each broiler house, immediately placed into 5 ml of Preston broth (Oxoid, Basingstoke, Hampshire, United Kingdom), and transported to the laboratory at 4°C, where they were microaerophilically incubated for 48 h at 37°C (63). Each set of enrichment broths was pooled and filtered (0.6-μm pore size; Whatman, Middlesex, United Kingdom), and the flowthrough was centrifuged (9,300 × g for 5 min) (63). Most of the supernatant was discarded, the pellet was briefly vortexed in the remaining 1 ml of supernatant, and 0.1 ml of the suspension was incubated microaerophilically using CampyGen gas packs (Oxoid) in 3.5-liter gas jars (Oxoid) for 24 h at 42°C on Preston agar plates (Oxoid). Individual colonies were streaked for purity onto Preston agar plates and grown microaerophilically for 24 h at 42°C. Gram staining and biochemical testing using Mast ID Camp Identification Systems (Mast Diagnostics, Bootle, United Kingdom) were performed to check for vibroid morphology and for hippurate hydrolysis and indoxyl acetate and urease activity, respectively.
Water analysis.
Four of the five farms in this investigation used water directly from mains supply, and the remaining farm used a bore-hole source. Where possible, 510-ml water samples were taken from six places in the broiler drinking water systems (Fig. 1). Nipples were cleaned with 70% alcohol before water collection. An aliquot of water sample (10 ml) was screened microscopically for the presence of protozoa. Water samples were filtered by using nitrocellulose membranes (0.2-μm pore size; Whatman). DNA was then extracted from the filters by using DNA SPIN Kits for Soil (Bio 101, Anachem, Bedfordshire, United Kingdom) and processed according to the manufacturer's instructions.
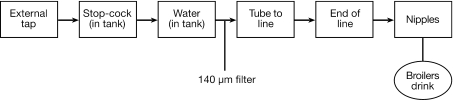
FIG. 1.
Flow diagram of drinking water systems in the broiler houses of intensively reared poultry. The broilers drink the water, obtained from main water supplies, at the nipples.
PCR detection of Campylobacter and protozoa.
All DNA extractions were performed using DNA SPIN Kits for Soil and processed according to the manufacturer's instructions. Bovine serum albumin (BSA) was obtained from Ambion, Cambridgeshire, United Kingdom. Unless stated otherwise, all other components used to perform PCR amplifications, purifications, and cloning were obtained from Invitrogen, Paisley, United Kingdom. DNA extractions were performed on Campylobacter colonies from the Preston agar plates. DNA was also extracted from samples of feces and bedding material and from water filtrates. Approximately 1 g of feces/bedding was taken from each of five locations randomly chosen within each broiler house. These were mixed in a sterile container and then placed on ice. DNA extractions were performed on 500 mg of each of these feces/bedding samples. The presence of C. jejuni and protozoa from environmental systems were determined by using the Winters et al. (78) seminested PCR system and the Einsele et al. (18) eukaryotic rRNA PCR system, respectively (Table 1). The seminested PCR amplifications for C. jejuni were performed in volumes of 100 μl containing 2.0 μl of DNA extracts, 50 pmol of each primer, 2.5 U of Taq DNA polymerase, 1.0 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 500 ng of BSA μl−1, and 200 μM concentrations of each deoxynucleoside triphosphate (78). For the first round of PCR, together with the primers WIN1 and WIN2 (Table 1), a protocol consisting of 1 cycle of 94°C for 3 min, followed by 40 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, with a final 3 min at 72°C was used (78). PCR product from the first round was used as the DNA template for the second round, together with primers WIN1 and WIN3 (Table 1). Twenty-four cycles were used in the second round with the conditions being the same, except for a lower annealing temperature (53°C) (78). For each of the three C. jejuni isolates a 122-bp amplicon from the Cj0343c gene was generated by using the Winters et al. (78) seminested PCR system (Table 2).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) | Annealing position | Reference |
|---|---|---|---|
| WIN1 | AAA TAA AGT TAG AGG TAG AAT GT | C. jejuni NCTC 11168, coordinates 66984 to 67006 in genome; Cj0343c gene | 78 |
| WIN2 | GGA TAA GCA CTA GCT AGC TGA T | C. jejuni NCTC 11168, coordinates 67140 to 67121 in genome; Cj0343c gene | 78 |
| WIN3 | GCA CGC CTA AAC CTA TAG C | C. jejuni NCTC 11168, coordinates 67105 to 67087 in genome; Cj0343c gene | 78 |
| EIN1 | ATT GGA GGG CAA GTC TGG TG | T. pyriformis, coordinates 537 to 557 in small subunit of rRNA | 18 |
| EIN2 | CCG ATC CCT AGT CGG AT AG | T. pyriformis, coordinates 998 to 1024 in small subunit of rRNA | 18 |
TABLE 2.
Library of C. jejuni clones with sample collection data
| Clone name | Sample data | GenBank no. |
|---|---|---|
| 1A | Farm 1, feces/bedding | AY830861 |
| 1B | Farm 1, feces/bedding | AY830862 |
| 2A | Farm 1, tank water | AY830863 |
| 2B | Farm 1, tank water | AY830864 |
| 3A | Farm 4, feces/bedding | AY830865 |
| 3B | Farm 4, feces/bedding | AY830866 |
| 4A | Farm 4, end of line | AY830867 |
| 4B | Farm 4, end of line | AY830868 |
| 5A | Farm 4, tube to line | AY830869 |
| 5B | Farm 4, tube to line | AY830870 |
| 6A | Farm 4, tank water | AY830871 |
| 6B | Farm 4, tank water | AY830872 |
| 7A | Farm 5, feces/bedding | AY830873 |
| 7B | Farm 5, feces/bedding | AY830874 |
| 8A | Farm 5, end of line | AY830875 |
| 8B | Farm 5, end of line | AY830876 |
| 9A | Farm 5, tube to line | AY830877 |
| 9B | Farm 5, tube to line | AY830878 |
| 10A | Farm 5, stop-cock water (in tank) | AY830879 |
| 10B | Farm 5, stop-cock water (in tank) | AY830880 |
| 11S | Farm 1, cloacal swab | AY830881 |
| 12S | Farm 4, cloacal swab | AY830882 |
| 13S | Farm 5, cloacal swab | AY830883 |
Amplification reactions for the detection of eukaryotic rRNA were performed in final volumes of 100 μl containing 2.0 μl of DNA, 50 pmol of each primer, 2.5 U of Taq DNA polymerase, 3 mM MgCl2, 51 mM KCl, 20 mM Tris-HCl (pH 8.4), 200 μM concentrations of each dNTP, and 500 ng of BSA μl−1. Using EIN1 and EIN2 primers (Table 1), a PCR protocol consisting of 1 cycle of 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min, with a final 5 min at 72°C was used (18). The technique of “band-stab PCR” was used to obtain a single desired PCR product of 500 bp (8). All PCR product purifications were performed by using Concert Rapid PCR purification kits, and the products were examined by using agarose gel electrophoresis. Each gel was composed of 1.5% multipurpose agarose (Boehringer, Ingelheim, Germany), stained with 0.5 μg of ethidium bromide ml−1, subjected to 100 V for 90 min, and scanned with a White/UV Transilluminator UVP Transilluminator (Ultra-Violet Products, Cambridge, United Kingdom).
Sequencing PCR products.
All PCR products (detected from water and feces/bedding samples) for DNA sequencing were first cloned using Original TA cloning kits. Samples were prepared for sequencing using a BigDye Terminator V2.0 cycle sequencing kit (Applied Biosystems, Foster City, CA) ethanol precipitation as described in the kit protocol and an ABI/Hitachi 3100 Genetic Analyzer capillary action sequencer (ABI/Hitachi, Arcade, NY). The forward strands of the three isolated C. jejuni strains and two clones from each of the 10 C. jejuni environmental amplicons were sequenced; these sequences were then deposited at the GenBank database under accession numbers AY830861 to AY830883 (Table 2). Forward and reverse DNA strands were also sequenced from 34 random eukaryotic clones—3 from each external tap and tank sample and 2 from each nipple water sample—and were then deposited at the GenBank database under accession numbers AY837467 to AY837500 (Table 3). Sequences were then analyzed by using Chromas version 1.62 (Technelysium, Tewantin Qld, Australia), NCBI BLAST, and EMBL-EBI (European Bioinformatics Institute, Heidelberg, Germany) CLUSTAL W alignment. A phylogenetic tree of Campylobacter amplicons was constructed by using the neighbor-joining method based on all nucleotide sites, with corrections for multiple substitutions by the Jukes Cantor method in MEGA version 2.1 (The Pennsylvania State University, University Park, PA).
TABLE 3.
Library of eukaryotic clones with sample collection data
| Clone | Sample data | GenBank no. | Most closely related microbe, % similarity | Clone | Sample data | GenBank no. | Most closely related microbe, % similarity | |
|---|---|---|---|---|---|---|---|---|
| 1A | Farm 1, external tap | AY837467 | Trichosporon montevideense, 99% small subunit rRNA | |||||
| 1B | Farm 1, external tap | AY837468 | Trichosporon montevideense, 99% small subunit rRNA | |||||
| 1C | Farm 1, external tap | AY837469 | Trichosporon montevideense, 97% small subunit rRNA | |||||
| 2A | Farm 4, external tap | AY837470 | Phialocephala fortinii, 91% small subunit rRNA | |||||
| 2B | Farm 4, external tap | AY837471 | Botryotinia fuckeliana, 18S 99% rRNA | |||||
| 2C | Farm 4, external tap | AY837472 | Botryotinia fuckeliana, 99% 18S rRNA | |||||
| 3A | Farm 5, external tap | AY837473 | Scopulariopsis brevicaulis, 99% 18S rRNA | |||||
| 3B | Farm 5, external tap | AY837474 | Paecilomyces lilacinus, 98% 18S rRNA | |||||
| 3C | Farm 5, external tap | AY837475 | Debaryomyces hansenii var. fabryi, 98% 18S rRNA | |||||
| 4A | Farm 1, tank | AY837476 | Paraurostyla weissei 17S, 90% rRNA gene, | |||||
| 4B | Farm 1, tank | AY837477 | Uncultured alveolate clone LEMD251, 93% small subunit rRNA | |||||
| 4C | Farm 1, tank | AY837478 | Trichosporon montevideense, 99% small subunit rRNA | |||||
| 5A | Farm 2, tank | AY837479 | Paraphysomonas butcheri, 96% 18S rRNA | |||||
| 5B | Farm 2, tank | AY837480 | Eurotium herbariorum, 98% 18S rRNA gene | |||||
| 5C | Farm 2, tank | AY837481 | Eurotium herbariorum, 97% 18S rRNA gene | |||||
| 6A | Farm 3, tank | AY837482 | Hydnum rufescens 18S, 96% rRNA gene | |||||
| 6B | Farm 3, tank | AY837483 | Spumella oblique, 96% 18S rDNA gene | |||||
| 6C | Farm 3, tank | AY837484 | Scopulariopsis brevicaulis, 97% 18S rRNA gene, | |||||
| 7A | Farm 4, tank | AY837485 | Phialophora verrucosa, 99% 18S rRNA | |||||
| 7B | Farm 4, tank | AY837486 | Phialophora verrucosa, 98% 18S rRNA | |||||
| 7C | Farm 4, tank | AY837487 | Unidentified eukaryote clone LKM67, 97% 18S rRNA | |||||
| 8A | Farm 5, tank | AY837488 | Scopulariopsis brevicaulis, 97% 18S rRNA gene | |||||
| 8B | Farm 5, tank | AY837489 | Uncultured alveolate clone LEMD251, 95% small subunit rRNA | |||||
| 8C | Farm 5, tank | AY837490 | Uncultured alveolate clone LEMD251, 95% small subunit rRNA | |||||
| 9A | Farm 1, nipples | AY837491 | Trichosporon montevideense, 98% small subunit rRNA | |||||
| 9B | Farm 1, nipples | AY837492 | Aspidisca steini, 92% small subunit rRNA | |||||
| 10A | Farm 2, nipples | AY837493 | Eurotium herbariorum, 99% 18S rRNA gene | |||||
| 10B | Farm 2, nipples | AY837494 | Gibberella pulicaris, 99% 18S rRNA | |||||
| 10B | Farm 2, nipples | AY837494 | Gibberella pulicaris, 99% 18S rRNA | |||||
| 11A | Farm 3, nipples | AY837495 | Uncultured alveolate clone LEMD251, 94% small subunit rRNA | |||||
| 12A | Farm 4, nipples | AY837497 | Capronia coronata, 97% 18S rRNA | |||||
| 12B | Farm 4, nipples | AY837498 | Penicillium italicum, 94% 18S rRNA | |||||
| 13A | Farm 5, nipples | AY837499 | Cladosporium cladosporioides, 99% 18S rRNA | |||||
| 13B | Farm 5, nipples | AY837500 | Gibberella pulicaris, 96% 18S rRNA |
Internalization of Campylobacter by protozoa.
All of the following microscopy coculture assays were performed in triplicate, and the Campylobacter viability decline and disinfection assays were each performed in quadruplicate. T. pyriformis (CCAP 1630/14A) (Culture Collection of Algae and Protozoa, Oban, United Kingdom) and Acanthamoeba castellanii (CCAP 1501/10) were used in all in vitro experiments measuring bacterial-protozoan interactions. Protozoa were enumerated by using direct hemocytometer counts. A drop of 37% formaldehyde (Sigma, St. Louis, MO) was added to 1-ml aliquots of suspensions of T. pyriformis to suppress motility and to make the cells easier to count. Fluorescence microscopy used a Super High-Pressure Nikon Mercury Lamp (Kawasaki, Kanagawa, Japan) and B-2A and/or UV-1A Nikon filters, and images were recorded by using Kroma Scan (Kinetic Imaging, 2000, Nottingham, United Kingdom).
Establishing cocultures.
During this research, protozoa were grown for 3 days, starved, and then placed in low-nutrient conditions to replicate the low-nutrient conditions in water systems, minimize protozoa variation in feeding behavior, and increase protozoan ingestion of Campylobacter (29, 30, 68). A. castellanii organisms were grown for 3 days in 20 ml of proteose peptone glucose broth (CCAP) at 25°C to a population density of 106 cells ml−1. T. pyriformis organisms were grown for 3 days in 15 ml of proteose peptone yeast extract broth (CCAP) at 25°C to a population density of 7.5 × 105 cells ml−1. Cultures were gravity filtered by using membrane filters (0.8-μm pore size; Whatman) to remove broth. Cells were then resuspended in a 1:1 dilution of the appropriate culture broth and Page's ameba saline (PAS) solution (CCAP; total volume, 10 ml) and incubated at 25°C for 12 h. This was done to reduce the effects of osmotic shock to protozoa and to avoid cyst formation (16, 37). After 12 h, the cells were filtered again, as described above, resuspended in 5 ml of PAS solution, adjusted to a concentration of 108 cells ml−1, and incubated at 25°C for 12 h. Campylobacters were grown on Preston agar in microaerophilic conditions for 24 h at 42°C using CampyGen (Oxoid) gas packs in 3.5-liter anaerobic jars (Oxoid). Bacterial cells were resuspended in PAS, and optical densities were measured at 600 nm.
Determination of viability of internalized bacteria cells.
Live/Dead Baclight bacterial viability kits (Invitrogen) utilize SYTO 9 green-fluorescent nucleic acid stain and the red-fluorescent nucleic acid stain propidium iodide. Bacteria with intact membranes fluoresced green (alive), whereas bacteria with damaged membranes fluoresced red (dead). A. castellanii and T. pyriformis were prepared for coculture as previously described. C. jejuni NCTC 11351 (National Collection of Type Cultures), C. coli NCTC 11366 and C. jejuni subsp. jejuni (poultry isolate) were each grown for 24 h, suspended in 10 ml of phosphate-buffered saline (PBS; Oxoid), and adjusted to concentrations of 2.8 × 108 CFU ml−1 (optical density at 600 nm of 0.4). Each Campylobacter suspension was then stained by using Baclight according to the kit protocol. The Campylobacter suspensions were vortexed briefly in 10 ml of PBS and centrifuged for 1 min at 9,300 × g; the supernatant was then discarded. Each dyed bacterial pellet was then resuspended in 10 ml of PAS solution.
Cocultures of each protozoa and Campylobacter were obtained by adding by 2 ml of protozoan suspension and 0.36 ml of bacterial suspension to 17.64 ml of PAS solution, giving 1:1 ratios of protozoa to Campylobacter. The cocultures were incubated at 25°C for up to 24 h and were monitored at time intervals (1 h, 3 h, 6 h, 24 h, and 3 days) over this period. After the appropriate time period, 3 ml of coculture was gravity filtered, and each filter was carefully rinsed with 25 ml (5 by 5 ml) of PAS solution to remove Campylobacter from the surface of the protozoa. The invaded protozoa were then observed by using phase-contrast microscopy and fluorescence microscopy (×40 and ×100 Nikon lenses) with B-2A and U-1A filters (Nikon).
Distinguishing internal from external Campylobacter from a coculture.
A. castellanii and T. pyriformis were prepared for coculture with Campylobacter, as previously described. C. jejuni NCTC 11351, C. jejuni subsp. jejuni (poultry isolate), and C. coli NCTC 11366 were each grown for 24 h, suspended in 10 ml of PBS, and adjusted to a concentration of 2.8 × 108 CFU ml−1. Each Campylobacter suspension was centrifuged at 9,300 × g for 1 min, and the supernatant was removed. Bacterial pellets were stained with 50 μl of fluorescein-isothiocyanate (FITC)-labeled rabbit antibody to C. jejuni (AMS Biotechnology, Ltd., Abingdon, United Kingdom), vortexed briefly, and left in darkness for 15 min on ice. Each sample was centrifuged as described above, and the supernatant was removed. The stained bacterial pellets were rinsed in 10 ml of PBS, and the supernatant was discarded. The stained bacterial pellets (protected from light) were then resuspended in 10 ml of PAS solution and vortexed briefly. Cocultures of protozoa and Campylobacter were obtained by adding by adding 2 ml of protozoan suspension and 0.36 ml of bacterial suspension to 17.64 ml of PAS, giving 1:1 ratios of protozoa to Campylobacter. The cocultures were incubated at 25°C for up to 24 h and were monitored at time intervals (1 h, 3 h, 6 h, and 24 h) over this period. After the appropriate time period, 1 ml of coculture was removed, and 0.01 mg of DAPI (4′,6′-diamidino-2-phenylindole; Sigma) was added to the aliquot, which was then left to incubate at room temperature for 10 min in darkness. The invaded protozoa were then observed by using fluorescence microscopy using ×40 and ×100 phase-contrast lenses (Nikon) and B-2A (FITC) and UV-1A (DAPI) Nikon filters. As negative controls, 50 μl of FITC antibody was added to separate 20-ml suspensions of A. castellanii and T. pyriformis (107 cells ml−1) in PAS in the absence of bacteria. The negative protozoan controls fluoresced after staining with DAPI.
The survival of Campylobacter in cocultures.
Sonication (on ice) with a microtip probe for 10 s at 40 W appeared to have no effect on the viability of Campylobacter, and phase-contrast microscopy clearly confirmed that this treatment completely ruptured A. castellanii and T. pyriformis cells (37). A. castellanii and T. pyriformis were prepared for coculture with C. jejuni NCTC 11351, C. jejuni subsp. jejuni (poultry isolate), and C. coli NCTC 11366 as previously described. Each coculture was then incubated at 25°C, and at various time intervals (0 h, 3 h, 6 h, and then daily) 1 ml of coculture was removed and sonicated (on ice) for 10 s at 40 W; viable counts (24 h at 42°C on Preston agar) were then performed daily until viable cells were no longer obtained (37). As controls, each Campylobacter strain was grown for 24 h at 42°C, suspended in 10 ml of PAS solution, and adjusted to concentrations of 2.8 × 108 CFU ml−1. Concentrations of 107 Campylobacter CFU ml−1 were obtained by adding by 0.36 ml of the bacterial suspension to 19.64 ml of PAS solution. The bacterial suspensions were then incubated at 25°C. At various time intervals (0 h, 3 h, 6 h, and then daily), 1 ml of planktonic suspension was removed for each Campylobacter strain, and viable counts were then performed daily until viable cells were no longer obtained.
Campylobacter disinfection resistance studies.
Disinfection assays were mainly based on a previous resistance study by King et al. (37). A variety of preliminary experiments were conducted to verify the functionality of the system for analysis of bacterial viability in a protozoan model. First, it was confirmed that after exposure to 10% sodium thiosulfate (STS; Sigma) in PAS, no significant effects (P > 0.05) on the growth of Campylobacter, T. pyriformis, and A. castellanii were observed (data not shown). A 1:1,000 dilution of the disinfectant Virudine was found to kill planktonic Campylobacter but had no significant effect (P > 0.05) on the growth T. pyriformis and A. castellanii in PAS at 25°C (data not shown). This concentration was used for all subsequent disinfection experiments.
Cocultures of C. jejuni NCTC 11351, C. jejuni subsp. jejuni (poultry isolate), and C. coli NCTC 11366 with T. pyriformis and A. castellanii were each prepared as previously described, and each coculture was incubated for up to 24 h at 25°C. During the 24-h incubation period and after 3, 6, 9, 12, and 24 h, a 1:1,000 dilution of Virudine was used to kill planktonic C. jejuni; 2.2 ml of 1:100 Virudine was then added to each coculture for a contact time of 1 min. Then, 2.2 ml of sterile 10% STS (Sigma) was added to neutralize the disinfectant, and each sample was gravity filtered (0.8 μm) and rinsed with 7 ml of PAS. Serial dilutions of the filtrate were then performed to quantify the number of Campylobacter on the surface of protozoa that survived disinfection. Next, the filter was resuspended in 10 ml of PAS, 1 ml of coculture was removed and sonicated (on ice) for 10 s at 40 W, and viable Campylobacter counts (24 h at 42°C on Preston agar) were performed.
To examine the effect of the age of protozoa on Campylobacter disinfection resistance, coculture and disinfection assays were performed as previously described, except that before coculture incubation protozoa were grown 3, 6, and 9 days, and only a coculture time of 3 h was used.
Statistical analysis.
To determine whether values were significantly different (P < 0.05) between Campylobacter strains in a planktonic state compared to when they were in the presence of protozoa (cocultures), strain data were compared by using SPSS 11.0 software (SPSS, Inc., Chicago, IL) using the Bonferroni (one-way analysis of variance) multiple comparison test.
RESULTS
C. jejuni detected in poultry broiler drinking water.
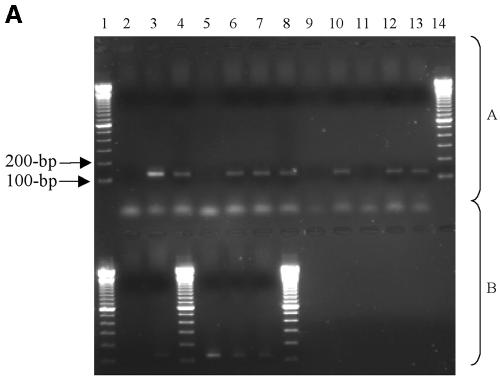
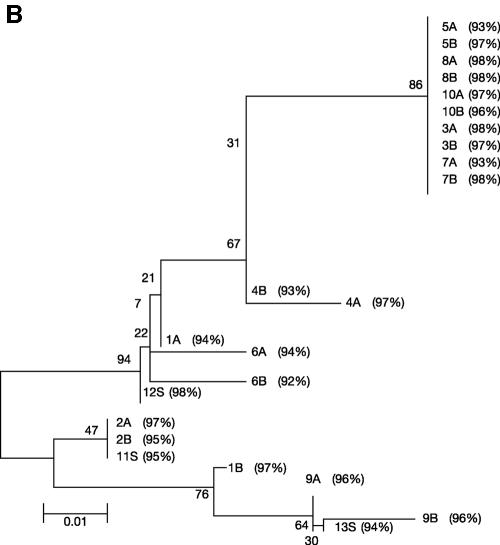
Strains of C. jejuni were recovered from broiler cloacal swabs, and this organism was also detected by seminested PCR in the drinking water and litter/feces on farms 1, 4, and 5 (Table 2). Identification was based on Gram staining and biochemical tests and confirmed by sequence analysis of seminested PCR products. No C. jejuni was detected in farms 2 and 3. In all instances, on farms where C. jejuni cultures were recovered from broilers a positive PCR result was also obtained from water analysis of the farm (Fig. 2A). All positive seminested 122-bp PCR C. jejuni products recovered from the various water samples were cloned and sequenced to confirm the specificity of the assay. Two clones of each amplicon were selected for sequencing (Table 2). The C. jejuni amplicons derived from cloacal swabs were not cloned. The homology studies of C. jejuni and sequence alignment data (Fig. 2B and C) illustrate that conserved regions of C. jejuni were amplified. In the dendrogram 2A, 2B, and 11S were grouped together, suggesting that the same strain of C. jejuni in broilers was also present in tank water from farm 1 (Fig. 2B). The distance between clones 1A and 1B in the dendrogram suggests that more than one C. jejuni strain was present in the feces/litter of farm 1. The C. jejuni amplicons all showed a high degree of homology to gene Cj0343c in C. jejuni subsp. jejuni NCTC 11168 (Fig. 2B). At farm 4 the same strain of C. jejuni detected in tank water (6A and 6B) may have colonized broilers (12S; Fig. 2B), and in farm 5 the same strain of C. jejuni that was detected in the tube to the line (9A and 9B) may have also colonized broilers (13S; Fig. 2B). Collectively, this genome-localized evidence suggests that the C. jejuni isolated from cloacal swabs taken from farms 1, 4, and 5 were the same as those isolated from water systems of these farms.
FIG. 2.
(A to C) Detection and analysis of C. jejuni DNA from the drinking water and feces/bedding and of isolates from cloacal swabs from five broiler farms. The primer system was based on that of the Winters et al. (78) seminested primer system for the Cj0343c gene in C. jejuni, with positive detection resulting in the presence of 122-bp amplicons. (A) Agarose gel (1.5%) of C. jejuni PCR amplicons detected from the drinking water and feces/bedding and C. jejuni isolated from cloacal swabs from five broiler farms. Positive results are indicated by 122-bp amplicons. (Block A) Lanes: 1, 100-bp DNA ladder; 2, negative control (no DNA); 3, positive control (C. jejuni NCTC 11351); 4, feces/bedding (farm 4); 5, nipples; 6, end of line; 7, tube to line; 8, tank water; 9, stop-cock water (in tank); 10, feces/bedding (farm 5); 11, nipples; 12, end of line; 13, tube to line; 14, 100-bp DNA ladder. (Block B) Lanes: 1, 100-bp DNA ladder; 2, tank water; 3, stop-cock water (in tank); 4, 100-bp DNA ladder; 5, swab (farm 1); 6, swab (farm 4); 7, 5 swab (farm 5); 8, 100-bp DNA ladder. (B) Phylogenetic tree of C. jejuni PCR amplicons detected from the drinking water, feces/bedding, and C. jejuni isolated from cloacal swabs from five broiler farms. The tree was constructed by using the neighbor-joining method based on all nucleotide sites, with corrections for multiple substitutions by the Jukes-Cantor method in MEGA version 2.1. The letter “S” represents cloacal swab isolates, and the percent values indicate the percent similarity to gene Cj0343c, coordinates 66984 to 67087, in the C. jejuni subsp. jejuni NCTC 11168 genome. (C) EMBL-EBI CLUSTAL W alignment of 13 DNA sequences from the Cj0343c gene in C. jejuni; 10 of the sequences were detected from broiler drinking water and feces/bedding, and 3 of the sequences were amplified from C. jejuni isolated from cloacal swabs, again represented by the letter “S.” The asterisk represents conserved sequences in all C. jejuni seminested PCR amplicons.
Detection of protozoa in all broiler drinking water systems.
The same water samples that were screened for the presence of Campylobacter were also analyzed for eukaryotic microbes. After collection of 510-ml samples, each sample was subjected to brief screening by bright-field light microscopy using simple wet mounts. The presence of flagellate and ciliate protozoa was confirmed, although further microscopic identification was not performed. Instead of using the highly skilled and potentially subjective process of protozoa microscopy identification (53, 54, 55), eukaryotic identification was based on the most powerful approach to explore microbial diversity, the analysis of cloned rRNA ribosomal gene sequences using the primer system of Einsele et al. (18, 47). Eukaryotic microorganisms were detected in all water and fecal/bedding samples from all farms, based upon the presence of a 500-bp band in all PCR samples (data not shown). All of these PCR products were cloned and sequenced (Table 3). A wide diversity of eukaryotic microbes were detected in broiler drinking water, i.e., protozoa, uncultured eukaryotic clones, and yeasts and molds (Table 3). Species of protozoa which were detected were the chrysophyte (two flagella) flagellates Paraphysomonas butcheri (5- to 20-μm body length; farm 2) and Spumella oblique (5- to 20-μm body length; farm 3), and the ciliates Paraurostyla weissei (farm 1) and Aspidisca steini (20- to 100-μm body length; farm 5), which are all generally reported to be widely distributed in freshwater (53, 54, 55). Aspidisca is commonly isolated from freshwater and are small, flattened cells, moving over immersed surfaces with their ventral mouths, usually consuming individually attached particles, e.g., bacteria (54, 55). The majority (73.5%) of clones were yeasts and molds, e.g., Trichosporon montevideense, which are generally distributed widely in nature, especially in soils (5, 17, 33, 34, 38, 40-42, 47, 59, 61, 69). The uncultured alveolate clone LEMD251 was detected from farms 1, 3, and 5, while an unidentified eukaryotic 18S rRNA clone LKM67 was detected in the tank of farm 4. Alveolates are a vast group of protozoa comprising of ciliates, dinoflagellates, and apicomplexa characterized by the possession of a system of abutting sacs (alveoli) underlying the cell surface (13, 46, 51). The most divergent of known eukaryotic lineages in phylogenetic trees is represented by anaerobic or aerotolerant organisms, the inhabitants of anoxic (low-oxygen) environments (7, 13). Broiler drinking water systems provide low-oxygen or anoxic habitats for microbes, explaining the detection of anoxic unculturable alveolate clones. Although reports of intracellular prokaryotes in rumen ciliate protozoa (anoxic environment) exist, the potential of anoxic uncultured alveolate clones to act as Campylobacter hosts is unknown but nevertheless may exist (9, 43, 44). Protozoa were detected in the water systems of all farms, thus indicating the potential for Campylobacter and protozoa to interact.
Internalization of Campylobacter in protozoa.
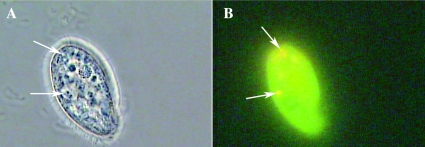
Campylobacter and protozoa were shown to coexist in poultry water systems. The potential interactions between the two were investigated in a series of in vitro experiments by using established model systems. T. pyriformis and A. castellanii are two types of bacterivorous protozoa commonly observed in surface water that can be grown axenically; studies have shown these bacteria to be subject to infection by bacteria, and both protozoa therefore provide good in vitro models (37, 62). In fluid media, axenically grown T. pyriformis and A. castellanii ingest nutrients through food vacuole formation (49, 74). In the presence of bacteria, T. pyriformis and A. castellanii contain digestive food vacuoles containing live bacteria, which for a period of time stay undamaged and can be observed microscopically (64). The use of light microscopy to study protozoa ingesting bacteria has the inherent difficulty of discriminating between bacteria that are bound to the external surface of cells and those that are internalized by them (15). An uncomplicated and inexpensive method for studying phagocytosis uses FITC-labeled bacteria and DAPI as a quenching agent, allowing the simultaneous viewing of intracellular and extracellular bacteria and providing the ability to discriminate between them (31, 60, 75). The bacterial viability assay clearly showed viable (green) and after longer incubation times (3 days) dead (red) Campylobacter inside food vacuoles of A. castellanii and T. pyriformis (Fig. 3 and 4, respectively). The final proof that Campylobacter were inside protozoa was provided by the DAPI/FITC method, wherein C. jejuni were clearly visible inside T. pyriformis (Fig. 5A to C). Internal Campylobacter were stained an extremely bright green color (FITC) (Fig. 5B), while external Campylobacter and protozoan DNA (especially in the nucleus) were stained blue with DAPI (Fig. 5C).
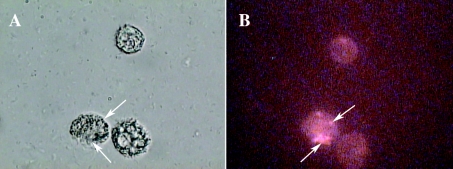
FIG. 3.
(A and B) Microscopy of T. pyriformis (CCAP 1630/14A) after 24 h of coculture with C. jejuni NCTC 11351 in PAS at 25°C. C. jejuni was stained with Baclight viability dye before coculture (1:1) with T. pyriformis. Magnification, ×100. The arrows indicate T. pyriformis vacuoles containing viable and intact C. jejuni. (A) Bright-field image of an intact T. pyriformis cell cocultured with C. jejuni. (B) Fluorescent image of viable (green) C. jejuni inside T. pyriformis.
FIG. 4.
(A and B) Microscopy of A. castellanii (CCAP 1501/10) after 3 days of coculture with C. jejuni NCTC 11351 in PAS at 25°C. C. jejuni was stained with Baclight viability dye before coculture (1:1) with A. castellanii. magnification, ×40. The arrows indicate A. castellanii vacuoles containing dead C. jejuni. (A) Bright-field image of an intact A. castellanii cell cocultured with C. jejuni. (B) Fluorescent image of dead (red) C. jejuni inside A. castellanii.
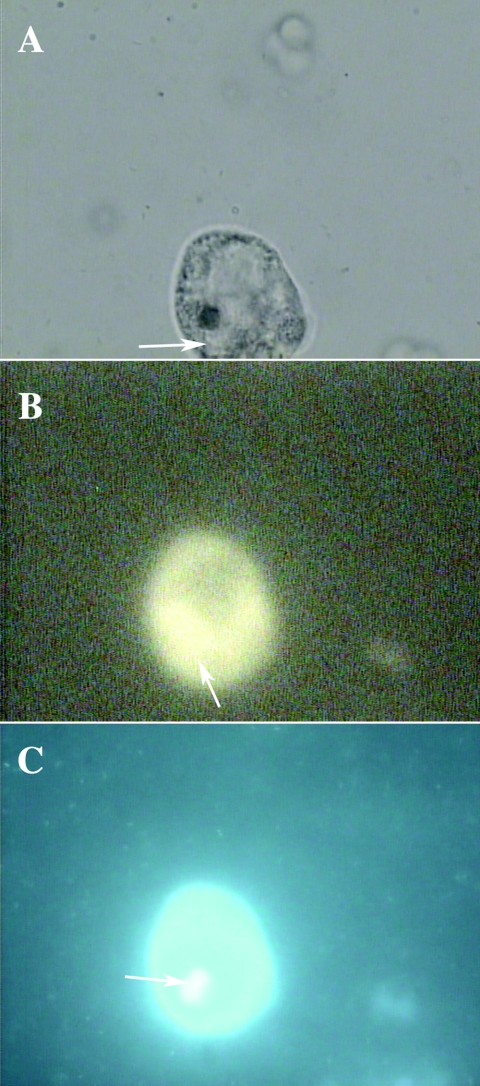
FIG. 5.
(A through C) Microscopy of T. pyriformis (CCAP 1630/14A) after 3 h of coculture with C. jejuni NCTC 11351 at 25°C. C. jejuni was stained with FITC-labeled rabbit antibody before coculture (1:1) with T. pyriformis. magnification, ×100. (A) Bright-field image of an intact T. pyriformis cell cocultured with C. jejuni. (B) Fluorescent image of C. jejuni inside T. pyriformis. The arrow indicates T. pyriformis vacuoles containing C. jejuni. (C) Intact T. pyriformis stained with DAPI after 3 h. DNA (especially in the nucleus) was stained blue with DAPI, and the arrow indicates a T. pyriformis nucleus. C. jejuni, also stained blue, can be seen outside around the intact T. pyriformis cell.
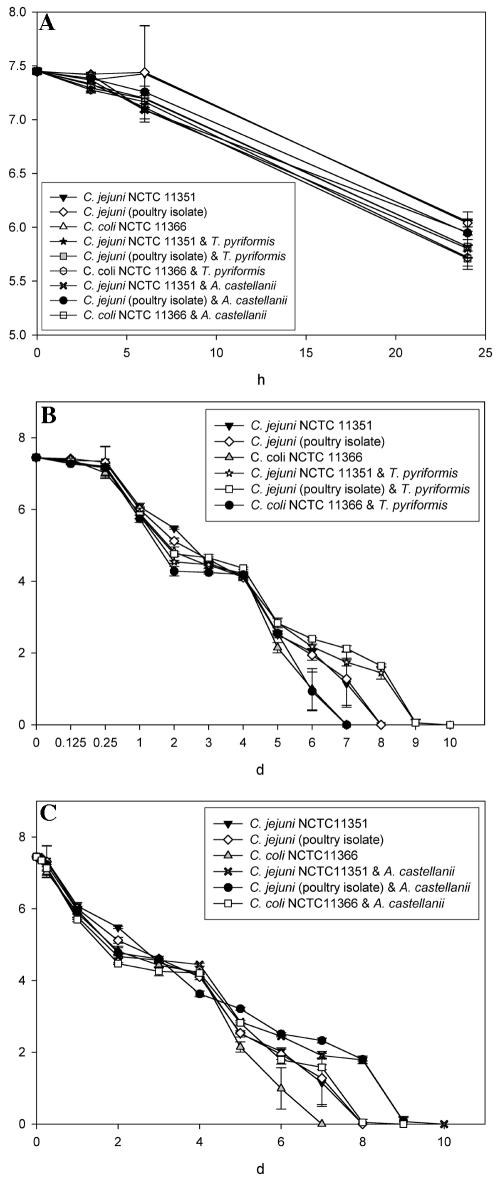
Delayed decline of Campylobacter viability in cocultures.
The effect of A. castellanii and T. pyriformis on the Campylobacter viability at 25°C in PAS was examined. When all strains of Campylobacter were incubated with protozoa, the number of recovered viable Campylobacter decreased (Fig. 6A to C). No significant (P > 0.05) differences were noted between Campylobacter strain viability during the first 24 h (Fig. 6A). During the first 3 h slightly more viable Campylobacter were recovered from planktonic suspensions than from suspensions of Campylobacter and protozoa (Fig. 6A). This may have been because the protozoa were feeding at a high rate after being starved. Initially fewer bacteria were recovered from cocultures containing T. pyriformis than A. castellanii, probably because T. pyriformis moves at a much higher rate and would be expected to be a more active bacterial predator (9, 37, 43, 44). After 5 days more notable differences existed between the Campylobacter viable counts (Fig. 6B and C). For example, after 5 days the number of viable C. jejuni organisms (poultry isolate) recovered from coculture with A. castellanii was significantly (P < 0.05) higher than the numbers of recovered planktonic C. jejuni NCTC 11351, C. jejuni (poultry isolate), and C. coli NCTC 11366 (Fig. 6C). After 5 days, significantly less (P < 0.05) planktonic C. coli NCTC 11366 and C. jejuni NCTC 11351 were recovered compared to the C. jejuni (poultry isolate) in coculture with T. pyriformis (Fig. 6B).
FIG. 6.
(A through C) Survival of Campylobacter jejuni NCTC 11351, C. coli NCTC 11366, and C. jejuni subsp. jejuni (poultry isolate) when cocultured with A. castellanii (CCAP 1501/10) and T. pyriformis (CCAP 1630/14A). Cocultures (1:1 ratio of Campylobacter to protozoa) and planktonic strains of Campylobacter were prepared in PAS solution and incubated at 25°C for up to 10 days, when viable Campylobacter were no longer obtained. The results are presented as average (performed in quadruplicate) viable Campylobacter recovered per ml from PAS, and error bars indicate the standard deviations. (A) Viablecounts of recovered Campylobacter during the first 24 h of incubation from cocultures with A. castellanii and T. pyriformis. (B) Viable counts of recovered Campylobacter during 10 days of incubation from cocultures containing T. pyriformis. (C) Viable counts of recovered Campylobacter during 10 days of incubation from cocultures containing A. castellanii.
The most important overall viability finding was that the C. jejuni strains both remained viable for significantly (P < 0.05) longer (an extra 36 h, i.e., from 7.75 to 9.25 days) when they were incubated with both strains of protozoa (Fig. 6B and C). The presence of T. pyriformis did not significantly increase (P > 0.05) the time required for a complete decline in C. coli viability. However, the decline of C. coli viability was significantly (P < 0.05) delayed during coculture with A. castellanii, i.e., 8 days (Fig. 6C). This suggests that the relationship between Campylobacter and protozoa is strain specific. Tezcan-Merdol et al. (72) found that the uptake and replication of different serovars of Salmonella enterica varied greatly within Acanthamoeba spp., suggesting that Acanthamoeba spp. can differentiate between serovars of salmonellae.
Increased Campylobacter disinfection resistance in cocultures.
When protozoa were 3 and 6 days old before coculturing, Campylobacter internalized by protozoa were significantly more resistant to disinfection (P < 0.05) than purely planktonic Campylobacter, which were all killed by the Virudine (Table 4 and Fig. 7). Significantly more (P < 0.05) internalized campylobacters survived when T. pyriformis and A. castellanii used in coculture were grown for 3 and 6 days than for 9 days, with maximal Campylobacter survival when protozoa were 3 days old (Table 4). After coculture with both T. pyriformis and A. castellanii, slightly more internalized C. jejuni subsp. jejuni (poultry isolate) survived disinfection than C. jejuni NCTC 11351 and C. coli NCTC 11366; however, these differences were not significant (P > 0.05) (Table 4).
TABLE 4.
Effect of age of protozoa during coculture on Campylobacter disinfection resistancea
| Protozoan age (days) | Location | Avg Campylobacter count (CFU/ml) ± SD
|
||
|---|---|---|---|---|
| C. coli NCTC 11366 | C. jejuni NCTC 11351 | C. jejuni (poultry isolate) | ||
| T. pyriformis | ||||
| 3 | Internal | (3.52 × 103) ± 285 | (4.28 × 103) ± 730 | (5.77 × 103) ± 999 |
| 3 | External | 0 ± 0 | 132 ± 18 | 313 ± 22 |
| 6 | Internal | (11.21 × 102) ± 134 | (3.25 × 103) ± 164 | (4.56 × 103) ± 644 |
| 6 | External | 5 ± 3 | 16 ± 10 | 33 ± 10 |
| 9 | Internal | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 9 | External | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| A. castellanii | ||||
| 3 | Internal | 269 ± 33 | 326 ± 32 | 375 ± 16 |
| 3 | External | 1 ± 1 | 3 ± 1 | 5 ± 1 |
| 6 | Internal | 113 ± 12 | 154 ± 24 | 172 ± 20 |
| 6 | External | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 9 | Internal | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 9 | External | 0 ± 0 | 0 ± 0 | 0 ± 0 |
T. pyriformis (CCAP 1630/14A) and A. castellanii (CCAP 1501/10) were each grown for 3, 6, and 9 days before being cocultured for 3 h at 25°C (1:1 ratio) with Campylobacter spp. in PAS solution. A 1:1,000 dilution of Virudine (1-min contact time) was then used to kill planktonic C. jejuni, followed by neutralization using STS, gravity filtration (0.8-μm pore size), rinsing and resuspension in PAS, sonication (10 s at 40 W), and counts of viable Campylobacter. Results are presented as the average (performed in quadruplicate) numbers of viable Campylobacter recovered ml of PAS.
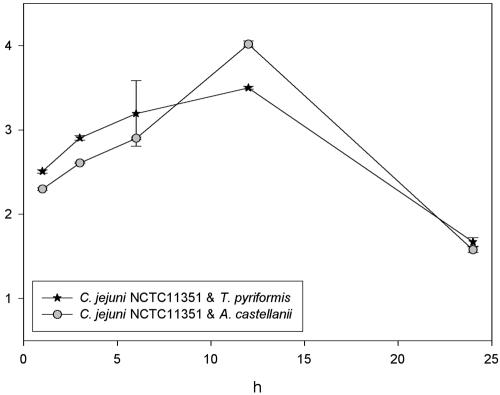
FIG. 7.
Effect of coculture time on the survival of C. jejuni NCTC 11351 during disinfection. Cocultures with C. jejuni and A. castellanii (CCAP 1501/10) and T. pyriformis (CCAP 1630/14A) (1:1 ratio of Campylobacter to protozoa) were prepared in PAS solution and incubated for up to 24 h at 25°C. After 3, 6, 9, 12, and 24 h, a 1:1,000 dilution of Virudine (1-min contact time) was used to kill planktonic C. jejuni, followed by neutralization with STS, gravity filtration (0.8 μm), rinsing and resuspension in PAS, sonication (10 s at 40 W), and viable Campylobacter counts. The results are presented as average (performed in quadruplicate) viable Campylobacter recovered per ml of PAS, and error bars indicate the standard deviations.
After 3 h of coculture time, significantly less (P < 0.05) internalized C. jejuni NCTC 11351 survived disinfection within A. castellanii than T. pyriformis (Fig. 7). However, after 6 h of coculture time, the amount of viable C. jejuni was not significantly different between both kinds of protozoa (P > 0.05) (Fig. 7). Maximum survival of internalized C. jejuni NCTC 11351 occurred after 12 h of coculture time with both protozoa, with significantly more (P < 0.05) C. jejuni surviving within A. castellanii. After 24 h of coculture time the amount of recovered viable C. jejuni NCTC 11351 had significantly dropped (P < 0.05), and the disinfection survival of internalized C. jejuni was no longer significantly different (P > 0.05) between T. pyriformis and A. castellanii. Collectively, these data suggest that the age or growth phase of protozoa and the interaction time (coculture time) greatly affects internalized Campylobacter disinfection resistance.
DISCUSSION
Protozoa detected in broiler drinking water systems.
Despite the presence of disinfectant, low temperatures, and flow regimes in drinking water distribution systems, the growth and persistence of bacteria in is well documented, and the diversity of protozoa and number of organisms are usually a function of the amount of available organic matter, including bacterial load (11, 53). Poultry drinking water systems allow sediment accumulation on pipe walls and tanks (Fig. 1), partly explaining the wide diversity of eukaryotes that was detected in the water systems (11, 53). The central role of protozoa in aquatic food webs as major grazers of phytoplankton and bacteria is firmly established, and species of protozoa detected in water systems were the flagellates Paraphysomonas butcheri and Spumella oblique (ingest bacteria) and the ciliate Aspidisca steini, which are frequently isolated from freshwater sources (23, 54, 55, 71, 73). Broiler drinking water contained a variety of protozoa; thus, it is concluded that poultry drinking water systems have strong potential to support and facilitate biological interactions between bacteria and protozoa.
Campylobacter and protozoan epidemiology.
The strains of C. jejuni present in water systems also appeared to colonize broilers as on farms where C. jejuni was detected in water; the organism was also found in broilers and feces/bedding. Previous farm epidemiology studies looking at a variety of potential reservoirs have shown that common strains can be found across a number of hosts. On et al. (50), using pulsed-field gel electrophoresis, found evidence of identical C. jejuni clones infecting humans, poultry, and cattle. Water plays an important role in the ecology of C. jejuni, and it can enter drinking water distribution systems through the fecal contamination of untreated ground or surface water, treatment failure, or distribution system failure (4, 25). Providing water is of low turbidity, standard chlorination procedures are normally sufficient to prevent the spread of planktonic campylobacters along water mains (12, 25, 79). Protozoa and C. jejuni were both detected in the drinking water systems of intensively reared poultry, highlighting the strong potential of protozoan-Campylobacter interactions. The detection of protozoa, including heterotrophic flagellates which are characteristically phagotrophic and are quantitatively the most important consumers of other microbes, means that such protozoa in the water supplies of broiler farms would probably ingest Campylobacter (3, 6, 23, 70).
Campylobacter internalization and viability decline.
The implications to the poultry industry of our in vitro coculture assays are greatly increased by the detection of ciliates closely related to T. pyriformis and by the usage of A. castellanii, the most commonly used protozoa in in vitro coculture assays. The studies reported here provide new evidence of the ability of Campylobacter to survive in the presence of protozoa. The three microscopic methods used to examine cocultures provided images where Campylobacter was clearly observed within vacuoles of A. castellanii and T. pyriformis. The viability coculture study, which examined the effects of the presence of protozoa toward Campylobacter viability, revealed new data with important implications for the broiler industry. In in vitro conditions, the presence of protozoa can significantly (P < 0.05) delay the decline in C. jejuni viability for up to 36 h at temperatures at which broilers are reared (25°C), thus potentially increasing the risk of Campylobacter colonization of broilers. This could be because Campylobacter released from protozoa undergo phenotypic changes, becoming more resistant to low-nutrient conditions and temperatures at which their decrease in viability is more rapid than at lower temperatures, e.g., 4°C (10, 15). When the broilers from the farms were 17 days old they were given a vaccine for infectious bursal disease in their drinking water. For this vaccine to be effective, the chlorine must be removed from the water. This method of administration of the infectious bursal disease vaccine could actually infect broilers with C. jejuni. Campylobacter would be expected to be periodically released from the protective environment of protozoa. However, during this vaccination process, the now planktonic Campylobacter would be in unchlorinated drinking water; thus, there would be a higher potential for infecting broilers. The timing with which the vaccine is administrated may be crucial because broilers are 17 days old and soon after this age, i.e., 3 weeks, they start to be infected with Campylobacter.
In broiler water supplies many other factors would affect Campylobacter viability. For this research, 1:1 ratios of protozoa to Campylobacter in cocultures were used. In water systems the concentrations of protozoa and Campylobacter would vary between farms and also within the different sections of water drinking systems in the same farm. In addition to nonliving organic matter, many varieties of eukaryotic and prokaryotic microbes also exist in a planktonic state and/or within biofilms. There would also be much greater variation in the physiological status, e.g., age and nutrient availability, of microbes in broiler drinking water. Further studies examining the effects of coculturing different Campylobacter strains (and other bacteria) with T. pyriformis, A. castellanii, and flagellates in cocultures, with variations in the ages of the bacteria and protozoa, would prove interesting.
Campylobacter disinfection resistance.
To our knowledge this is the first report of Campylobacter within protozoa demonstrating resistance to a disinfectant widely used in the poultry industry. Other novel areas of this research included examinations of the effects of the growth phase of the protozoa used in coculture and of the coculture time on Campylobacter disinfection resistance. Campylobacter in the presence of T. pyriformis and A. castellanii was significantly more (P < 0.05) resistant to disinfection when the protozoa used for coculture were grown for 3 and 6 days. The presence of cellulose in the cyst walls of Acanthamoeba spp. is a unique factor that may contribute to their disinfectant resistance, providing a physical barrier protecting them from extremes in pH and temperature, desiccation, anoxia, and antibiotics and disinfectants (76). The LuxS gene of C. jejuni 11168 produces the functional signal autoinducer 2 (AI-2), which is responsible for quorum sensing (19). As well as physical protection from external stresses, bacteria are densely packed in biofilms and/or protozoa. When Campylobacter were inside protozoa in food vacuoles, signaling molecules, e.g., AI-2, could have been present at higher concentrations than in equal numbers of there planktonic counterparts, potentially resulting in increased stress resistance. The exposure of protozoa to free iodine residuals may disrupt lysosomal hydrolase activity, delaying bacterial digestion (37). Undigested, viable bacterial cells may remain inside A. castellanii and T. pyriformis due to hydrolase disruption for up to 24 h after chlorine exposure, which killed planktonic bacteria (37).
King et al. (37) performed the only other major study examining the survival of bacterial pathogens within T. pyriformis and A. castellanii during disinfection. Similarities in protocol procedure include the use of 24-h-old bacterial strains, 1:1 amounts of protozoa for coculture (both at 104 ml−1), and neutralization of a halogen disinfectant with STS (1, 0.5, 0.25, and 0.125 mg of chlorine) after a 1-min contact period. However, King et al. did not differentiate between surviving external and internal bacteria, and the site of bacterial carriage was not unequivocally proven. Other established protozoan-bacterial relationships include Helicobacter pylori, one of the world's leading pathogens, colonizing ca. 60% of the global population (52, 77). The major mode of transmission of H. pylori remains unknown, and the finding of bacterial DNA in water samples, together with the high infection rate in developing countries suggests that environmental factors, e.g., interaction with amebas, could be involved in its transmission (77). The cocultivation of H. pylori with A. castellanii circumvented the bacterial requirements for precise microaerophilic conditions and a large supply of nutrients in order to grow, with a 100-fold increase of bacterial counts after 7 days (77). The putative dependence of H. pylori on free-living amebas in nature could be important with respect to transmission and prevalence, as has already been shown for Legionella pneumophila (1, 36, 37, 64, 76, 77). The potential for waterborne protozoa to act as vehicles for the Campylobacter infection of broilers is greatly increased by reports of H. pylori growing within Acanthamoeba, which was originally assigned taxonomically into the genus Campylobacter.
In conclusion, C. jejuni and a variety of protozoa were detected in broiler houses. In vitro, the presence of T. pyriformis and A. castellanii can significantly delay the decline of Campylobacter viability and significantly increase Campylobacter resistance to industrial disinfection. Collectively, these findings strongly suggest that the presence of protozoa and their interaction with Campylobacter in the water supplies of intensively reared poultry greatly increases the potential of broilers being colonized with Campylobacter. Viable pathogenic bacteria residing in protozoa present a new challenge in terms of disease control and sanitation of contaminated water sources since disinfectant efficiency is based upon planktonic tests.
REFERENCES
- 1.Ahearn, D. G., and M. M. Gabriel. 1997. Contact lenses, disinfectants and Acanthamoeba keratitis. Adv. Appl. Microbiol. 43:35-56. [DOI] [PubMed] [Google Scholar]
- 2.Allos, B. M. 2001. Campylobacter jejuni Infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 3.Alongi, D. M. 1991. Flagellates of benthic communities: characteristics and methods of study, p. 57-75. In D. J. Patterson and J. Larsen (ed.), The biology of free-living heterotrophic flagellates: the systematics association, special vol. 45. Clarendon Press, Oxford, England. [Google Scholar]
- 4.Altekruse, S. F., D. L. Swerdlow, and N. J. Stern. 1998. Microbial food-borne pathogens: Campylobacter jejuni. Vet. Clin. N. Am. Food Anim. Pract. 14:31-40. [PubMed] [Google Scholar]
- 5.Bacon, C. W., J. K. Porter, W. P. Norred, and J. F. Leslie. 1996. Production of fusaric acid by Fusarium species. Appl. Environ. Microbiol. 62:4039-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, J., P. A. Lambert, and M. R. Brown. 1993. Influence of intra-amoebic and other growth conditions on the surface properties of Legionella pneumophila. Infect. Immun. 61:3503-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard, C., A. G. Simpson, and D. J. Patterson. 2000. Some free-living flagellates (protista) from anoxic habitats. Ophelia 52:113-142. [Google Scholar]
- 8.Bjourson, A. J., and J. E. Cooper. 1992. Band-stab PCR: a simple technique for the purification of individual PCR products. Nucleic Acids Res. 20:4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet, J. L., P. Bogaerts, and J. Bohatier. 1999. Biological treatment of whey by Tetrahymena pyriformis and impact study on laboratory-scale wastewater lagoon process. Chemosphere 38:2979-2993. [DOI] [PubMed] [Google Scholar]
- 10.Brown, M. R., and J. Barker. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46-50. [DOI] [PubMed] [Google Scholar]
- 11.Camper, A., M. Burr, B. Ellis, P. Butterfield, and C. Abernathy. 1999. Development and structure of drinking water biofilms and techniques for their study. J. Appl. Microbiol. 28:1S-12S. [DOI] [PubMed] [Google Scholar]
- 12.Corry, J. E., and H. I. Atabay. 2001. Poultry as a source of Campylobacter and related organisms. Symp. Ser. Soc. Appl. Microbiol. 30:96S-114S. [DOI] [PubMed] [Google Scholar]
- 13.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle, M. 1984. Association of Campylobacter jejuni with laying hens and eggs. Appl. Environ. Microbiol. 47:533-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drevets, D. A., and P. A. Campbell. 1991. Macrophage phagocytosis: use of fluorescence microscopy to distinguish between extracellular and intracellular bacteria. J. Immunol. Methods 142:31-38. [DOI] [PubMed] [Google Scholar]
- 16.Drozanski, W. 1978. Activity and distribution of bacteriolytic N-acetyl-muramidase during growth of Acanthamoeba castellanii in axenic culture. Acta Microbiol. Pol. 27:243-256. [PubMed] [Google Scholar]
- 17.Eichner, C. A., R. W. Erb, K. N. Timmis, and I. Wagner-Dobler. 1999. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl. Environ. Microbiol. 65:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. A. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signaling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 20.Endtz, H. P., C. W. Ang, N. van den Braak, B. Duim, A. Rigter, L. J. Price, D. L. Woodward, F. G. Rodgers, W. M. Johnson, J. A. Wagenaar, B. C. Jacobs, H. A. Verburgh, and A. van Belkum. 2000. Molecular characterization of Campylobacter jejuni from patients with Guillain-Barré and Miller Fisher syndromes, J. Clin. Microbiol. 38:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans, S. J. 1992. Introduction and spread of thermophilic campylobacters in broiler flocks. Vet. Rec. 131:574-576. [PubMed] [Google Scholar]
- 22.Fields, P. I., and D. L. Swerdlow. 1999. Campylobacter jejuni. Clin. Lab. Med. 19:489-504. [PubMed] [Google Scholar]
- 23.Finlay, B. J., C. Tellez, and G. Esteban. 1993. Diversity of free-living ciliates in the sandy sediment of a Spanish stream in winter. J. Gen. Microbiol. 139:2855-2863. [Google Scholar]
- 24.Food Standards Agency. 2001. UK-wide survey of Salmonella and Campylobacter contamination of fresh and frozen chicken on retail sale. [Online.] http://www.foodstandards.gov.uk/news/chikensum.htm.
- 25.Ford, T. E. 1999. Microbiological safety of drinking water: United States and global perspectives. Environ. Health Perspect. 107:191-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsythe, S. J. 2000. Food poisoning microorganisms, p. 146-148. In S. J. Forsythe (ed.), The microbiology of safe food. Blackwell Science Publishers, Abingdon, United Kingdom.
- 27.Gibbens, J. C., S. J. S. Pascoe, S. J. Evans, R. H. Davies, and A. R. Sayers. 2001. A trial of biosecurity as a means to control Campylobacter infection of broiler chickens. Prev. Vet. Med. 48:85-99. [DOI] [PubMed] [Google Scholar]
- 28.Hald, B., E. Rattenborg, and M. Madsen. 2001. Role of batch depletion of broiler houses on the occurrence of Campylobacter spp. in chicken flocks. Lett. Appl. Microbiol. 32:253-256. [DOI] [PubMed] [Google Scholar]
- 29.Hatzis, C., F. Srienc, and A. G. Fredrickson. 1993. Feeding heterogeneity in ciliate populations effects of culture age and nutritional status. Biotechnol. Bioeng. 43:371-380. [DOI] [PubMed] [Google Scholar]
- 30.Hatzis, C., P. J. Sweeney, F. Srienc, and A. G. Fredrickson. 1993. Determination of cellular rate distributions in microbial cell populations: feeding rates of ciliated protozoa. Biotechnol. Bioeng. 42:284-294. [DOI] [PubMed] [Google Scholar]
- 31.Hed, J. 1986. Methods for distinguishing ingested from adhering particles. Methods Enzymol. 132:198-204. [DOI] [PubMed] [Google Scholar]
- 32.Hendricks, R. A., E. A. Boyle, C. L. Kastner, and D. Y. Fung. 2000. Compilation of intervention methods and conditions and ingredient limits, for controlling Campylobacter jejuni in meat and poultry products. J. Rapid Methods Aut. Mic. 8:285-305. [Google Scholar]
- 33.Issakainen, J., J. Jalava, E. Eerola, and C. K. Campbell. 1997. Relatedness of Pseudallescheria, Scedosporium, and Graphium pro parte based on SSU rDNA sequences. J. Med. Vet. Mycol. 35:389-398. [PubMed] [Google Scholar]
- 34.Jha, D. K., G. D. Sharma, and R. R. Mishra. 1992. Ecology of soil myorrhizal symbionts in degraded forests at two altitudes. Biol. Fert. Soils 12:272-278. [Google Scholar]
- 35.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiolology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 36.Kilvington, S., and J. Price. 1990. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68:519-525. [DOI] [PubMed] [Google Scholar]
- 37.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kokalis-Burelle, N., and R. Rodriguez-Kabana. 1994. Changes in populations of soil microorganism, nematodes, and enzyme activity associated with application of powdered pine bark. Plant Soil 162:169-175. [Google Scholar]
- 39.Kopecko, D. J., L. Hu, and J. M. Zaal. 2001. Campylobacter jejuni microtubule-dependent invasion. Trends Microbiol. 9:389-396. [DOI] [PubMed] [Google Scholar]
- 40.Korhonen, L. K., and P. J. Martikainen. 1991. Comparison of the survival of Campylobacter jejuni and Campylobacter coli in culturable form in surface water. Can. J. Microbiol. 37:530-533. [DOI] [PubMed] [Google Scholar]
- 41.Kunova, Z., and E. Piekova. 2002. Soil-borne micromycetes tolerant to amphotericin B. Biologia 57:345-349. [Google Scholar]
- 42.Kunova, Z., and E. Piekova E. 2002. Isolation of fluconazole-tolerant micromycetes onto different cultivation media. Folia Microbiol. 47:113-117. [DOI] [PubMed] [Google Scholar]
- 43.Liou, J. J., A. G. Fredrickson, and F. Srienc. 1998. Selective synchronization of Tetrahymena pyriformis cell populations and cell growth kinetics during the cell cycle. Biotechnol. Prog. 14:450-456. [DOI] [PubMed] [Google Scholar]
- 44.Lloyd, D., A. G. Williams, R. Amann, A. J. Hayes, L. Durrant, and J. R. Ralphs. 1996. Intracellular prokaryotes in rumen ciliate protozoa: detection by confocal laser scanning microscopy after in situ hybridization with fluorescent 16S rRNA probes. Eur. J. Protistol. 32:523-531. [Google Scholar]
- 45.Montrose, M. S., S. M. Shane, and K. S. Harrington. 1985. Role of litter in the transmission of Campylobacter jejuni. Avian Dis. 29:392-399. [PubMed] [Google Scholar]
- 46.Moreira, D., and P. Lopez-Garcia. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 47.Muyzer, G. 1999. DGGE/TGGE: a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 48.Newell, D. G. 2001. Animal models of Campylobacter jejuni colonization and disease and the lessons to be learned from similar Helicobacter pylori models. Symp. Ser. Soc. Appl. Microbiol. 30:57S-67S. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson, J. R. 1987. Structural aspects of digestion of Escherichia coli in Tetrahymena. J. Protozool. 34:1-6. [Google Scholar]
- 50.On, S. L. W., E. M. Nielsen, J. Engberg, and M. Madsen. 1998. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI \polymorphisms: evidence of identical clones infecting humans, poultry, and cattle, Epidemiol. Infect. 120:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orpin, C. G., and S. D. Mathiesen. 1986. Microcetus lappus gen. nov., sp. nov.: new species of ciliated protozoan from the bovine rumen. Appl. Environ. Microbiol. 52:527-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Toole, P. W., M. C. Lane, and S. Porwollik. 2000. Helicobacter pylori motility. Microbes Infect. 2:1207-1214. [DOI] [PubMed] [Google Scholar]
- 53.Patterson, D. J. 1998. Protozoan communities, p. 181-193. In D. J. Patterson (ed.), Free-living freshwater protozoa. John Wiley & Sons, New York, N.Y.
- 54.Patterson, D. J. 1998. Classification of protozoa, p. 19-21. In D. J. Patterson (ed.), Free-living freshwater protozoa, John Wiley & Sons, New York, N.Y.
- 55.Patterson, D. J. 1998. The key, p. 23-180. In D. J. Patterson (ed.), Free-living freshwater protozoa. John Wiley & Sons, New York, N.Y.
- 56.Pattison, M. 2001. Practical intervention strategies for Campylobacter. Symp. Ser. Soc. Appl. Microbiol. 30:121S-125S. [DOI] [PubMed] [Google Scholar]
- 57.Pearson, A. D., M. H. Greenwood, T. D. Healing, D. Rollins, M. Shahamat, J. Donaldson, and R. R. Colwell. 1993. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl. Environ. Microbiol. 59:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pebody, R. G., M. J. Ryan, and P. G. Wall. 1997. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33-R37. [PubMed] [Google Scholar]
- 59.Pedras, M. S., and P. W. Ahiahonu. 2002. Probing the phytopathogenic stem rot fungus with phytoalexins and analogues: unprecedented glucosylation of camalexin and 6-methoxycamalexin. Bioorgan. Med. Chem. 10:3307-3312. [DOI] [PubMed] [Google Scholar]
- 60.Premke, K., and H. Arndt. 2000. Predation on heterotrophic flagellates by protists: food selectivity determined using a live-staining technique. Arch. Hydrobiol. 150:17-28. [Google Scholar]
- 61.Ragazzi, A., S. Moricca, I. Dellavalle, and F. Mancini. 1995. Infection of cotton by Fusarium oxysporum f. sp. vasinfectum. Phytoparasitica 23:315-321. [Google Scholar]
- 62.Rodríguez-Zaragoza, S. 1994. Ecology of free-living amoebae. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 63.Rosef, O., G. Kapperud, and E. Skjerve. 1987. Comparison of media and filtration procedures for qualitative recovery of thermotolerant Campylobacter spp. from naturally contaminated surface water. Int. J. Food Microbiol. 5:29-39.
- 64.Schlimme, W., B. Baur, K. Hanselmann, and B. Jenni. 1995. An agarose slide method to determine the fate of bacteria within digestive vacuoles of protozoa. FEMS Microbiol. Lett. 133:169-173. [DOI] [PubMed] [Google Scholar]
- 65.Shane, S. M. 1992. The significance of Campylobacter jejuni infection in poultry: a review. Avian Pathol. 21:189-213. [DOI] [PubMed] [Google Scholar]
- 66.Shane, S. M. 2000. Campylobacter infection of commercial poultry. Rev. Sci. Tech. OIE 19:376-395. [DOI] [PubMed] [Google Scholar]
- 67.Shanker, S., A. Lee, and T. C. Sorrell. 1986. Campylobacter jejuni in broilers: the role of vertical transmission. J. Hyg. 96:153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steinert, M., M. M. Ott, P. C. Luck, E. Tannich, and J. Hacker. 1994. Studies on the uptake and intracellular replication of Legionella pneumophila in protozoa and in macrophage-like cells. FEMS Microbiol. Ecol. 15:299-307. [Google Scholar]
- 69.Stundl, U. M., D. Patzak, and F. Schauer. 2000. Purification of a soluble cytochrome P450 from Trichosporon montevideense. J. Basic Microbiol. 40:289-292. [DOI] [PubMed] [Google Scholar]
- 70.Takeda, N., and K. Sugiyama. 1993. Metabolism of biogenic monoamines in the ciliated protozoan Tetrahymena pyriformis. Comp. Biochem. Physiol. 106:63-70. [DOI] [PubMed] [Google Scholar]
- 71.Tamar, H. 1990. Halteria bifurcata Tamar, 1968, distribution and variation, and Halteria gradinella O.F. Müller, 1773. Arch. Protistenkd. 138:3-15. [Google Scholar]
- 72.Tezcan-Merdol, D., M. Ljungstrom, J. Winiecka-Krusnell, E. Linder, L. Engstrand, and M. Rhen. 2004. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 70:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas, C., H. Gibson, D. J. Hill, and M. Mabey. 2001. Campylobacter epidemiology: an aquatic perspective. J. Appl. Microbiol. 85:168S-177S. [DOI] [PubMed] [Google Scholar]
- 74.Verity, P. G. 1991. Feeding in planktonic protozoans: evidence for nonrandom acquisition of prey. J. Protozool. 38:69-76. [Google Scholar]
- 75.Wassenaar, T. M., M. Engelskirchen, S. Park, and A. Lastovica. 1997. Differential uptake and killing potential of Campylobacter jejuni by human peripheral monocytes/macrophages. Med. Microbiol. Immunol. 186:139-144. [DOI] [PubMed] [Google Scholar]
- 76.Winiecka-Krusnell, J., and E. Linder. 2001. Bacterial infections of free-living amoebae. Res. Microbiol. 152:613-619. [DOI] [PubMed] [Google Scholar]
- 77.Winiecka-Krusnell, J., K. Wreiber, A. von Euler, L. Engstrand, and E. Linder. 2002. Free-living amoebae promote growth and survival of Helicobacter pylori. Scand. J. Infect. Dis. 34:253-256. [DOI] [PubMed] [Google Scholar]
- 78.Winters, D. K., A. E. O'Leary, and M. F. Slavik. 1998. Polymerase chain reaction for rapid detection of Campylobacter jejuni in artificially contaminated foods. Lett. Appl. Microbiol. 27:163-167. [DOI] [PubMed] [Google Scholar]
- 79.World Health Organization. 1996. Guidelines for drinking-water quality, vol. 2: health criteria and other supporting information, 2nd ed., p. 2-3. World Health Organization, Geneva, Switzerland. [Google Scholar]