Abstract
Biofilm formation in Burkholderia cenocepacia has been shown to rely in part on acylhomoserine lactone-based quorum sensing. For many other bacterial species, it appears that both the initial adherence and the later stages of biofilm maturation are affected when quorum sensing pathways are inhibited. In this study, we examined the effects of mutations in the cepIR and cciIR quorum-sensing systems of Burkholderia cenocepacia K56-2 with respect to biofilm attachment and antibiotic resistance. We also examined the role of the cepIR system in biofilm stability and structural development. Using the high-throughput MBEC assay system to produce multiple equivalent biofilms, the biomasses of both the cepI and cepR mutant biofilms, measured by crystal violet staining, were less than half of the value observed for the wild-type strain. Attachment was partially restored upon providing functional gene copies via multicopy expression vectors. Surprisingly, neither the cciI mutant nor the double cciI cepI mutant was deficient in attachment, and restoration of the cciI gene resulted in less attachment than for the mutants. Meanwhile, the cciR mutant did show a significant reduction in attachment, as did the cciR cepIR mutant. While there was no change in antibiotic susceptibility with the individual cepIR and cciIR mutants, the cepI cciI mutant biofilms were more sensitive to ciprofloxacin. A significant increase in sensitivity to removal by sodium dodecyl sulfate was seen for the cepI and cepR mutants. Flow cell analysis of the individual cepIR mutant biofilms indicated that they were both structurally and temporally impaired in attachment and development. These results suggest that biofilm structural defects might be present in quorum-sensing mutants of B. cenocepacia that affect the stability and resistance of the adherent cell mass, providing a basis for future studies to design preventative measures against biofilm formation in this species, an important lung pathogen of cystic fibrosis patients.
A wide array of bacteria, from environmental isolates to commensal organisms and opportunistic pathogens, appear to exist in their natural habitats as multicellular microbial communities rather than free-floating individual cells. Advantages to this mode of growth are thought to include cooperative degradation of complex nutrients, resistance to physical and chemical removal of cells, and a community-based regulation of gene expression (52). Understanding the factors involved in biofilm formation is crucial for developing ways of enhancing colonization, such as in the use of probiotics, or to prevent or remove adherent bacteria for effective treatment of biofilm-related infections (18, 44). The theory that a specific “biofilm phenotype” exists, resulting from differential gene regulation when cells are in a high-density biofilm, is gaining support. The cellular signaling pathways that control the transition from a planktonic cell to an adherent microbial community must be elucidated to fully understand the biofilm phenotype and subsequently develop novel antimicrobial strategies.
Recent studies have demonstrated the potential for biofilm formation in a variety of environmental and clinical isolates from the Burkholderia cepacia complex, on both abiotic surfaces (3, 14, 23, 55) and human airway epithelial cell cultures (47). Strains of the B. cepacia complex are organized into at least nine genetically distinct species (12, 13, 57, 58). Isolates classified as Burkholderia cenocepacia, particularly those containing the cable pilus gene and the B. cepacia epidemic strain marker, are thought to be more readily transmissible between patients with cystic fibrosis (CF), for reasons yet unknown (8, 34, 53).
Burkholder first identified B. cepacia in the 1950s as the causative agent of soft rot in onions (7), which was later isolated as a virulent pathogen in the lungs of CF patients (25). Colonization of the lungs of CF patients by B. cepacia complex can result in an unpredictable clinical presentation, including asymptomatic carriage, a progressive deterioration of lung function, or a rapid decline in health due to bacterial sepsis, causing death (referred to as the “cepacia syndrome”) (19). Circumstances resulting in such variations in clinical manifestations are poorly understood, although biofilm formation may be an important virulence factor in both establishing and maintaining the infection and in colonizing the surfaces of respiratory equipment as a source of transmission.
Quorum sensing is a cell-to-cell signaling mechanism used to regulate cellular processes in a cell density-dependent manner. Biofilm formation has been recognized to be a quorum-sensing-regulated phenomenon for an ever-increasing number of bacteria, including Streptococcus mutans (30, 39, 61, 63), Streptococcus gordonii (31), Staphylococcus aureus (60), Staphylococcus epidermidis (4, 59), Serratia liquefaciens (27), Vibrio cholerae (20, 56, 65), Salmonella spp. (43), Aeromonas hydrophila (32), Pseudomonas putida (51), and B. cenocepacia (23) and its fellow CF pathogen Pseudomonas aeruginosa (15). Mutations in the quorum-sensing systems of B. cenocepacia and P. aeruginosa appear to affect the latter stages of biofilm formation, particularly the maturation of dense adherent microcolonies into what have been classified as mushroom- and pillar-like three-dimensional structures (15, 23, 24, 49). Initial stages of attachment to the substratum do not appear to be affected, but the resultant biofilm structure of P. aeruginosa is denser, shallower, and more easily disaggregated (15). B. cenocepacia also shows reduced attachment and altered surface colonization patterns when quorum sensing has been disrupted (23); however, the downstream effects of this phenomenon on the properties of the biofilm have not been examined.
Multiple distinct quorum-sensing systems have been identified in the B. cepacia complex. The first identified pathway, cepIR, is a homologue of the lasIR/rhlIR systems of P. aeruginosa and has been shown to control the expression of a significant portion of the transcriptome (2) and proteome (45), including such virulence factors as proteases and iron acquisition machinery (28, 29, 36). Mutants with mutations in the cepIR system have decreased virulence in rat, plant, and nematode models of infection (1, 26, 50). The cepI gene encodes an autoinducer synthase, which is responsible for the production of N-hexanoyl-acylhomoserine lactone (C6-HSL) and N-octanoyl-acylhomoserine lactone (C8-HSL) signaling molecules. The signal concentration in the local environment increases in proportion to an increase in cell density, causing the signals to accumulate intracellularly and interact with the cepR gene product, the cognate transcription factor protein. Binding of the acylhomoserine lactone molecule is required for CepR to repress or activate the transcription of particular genes. Studies by Huber et al. (23) have indicated a requirement for cepIR in attachment to inert surfaces and the formation of mature biofilm structures.
In the present study, it was our aim to investigate the contribution of the cepIR quorum-sensing system of the epidemic CF isolate B. cenocepacia K56-2 in attachment of the cells and subsequent development of a mature and functional biofilm. We have investigated the effect of quorum sensing in relation to both antibiotic resistance and anionic detergent challenge. The results presented here indicate that the attachment ability of the cepIR quorum-sensing mutants is affected and that they are unable to form structurally sound biofilms comparable to those formed by the wild-type strain.
Baldwin et al. (5) recently identified a second quorum-sensing system within the B. cenocepacia pathogenicity island that is associated with epidemic strains. This system, designated cciIR, is also involved in the regulation of virulence factors (5, 35), and a K56-2 cciI mutant was shown to be less virulent in a rat chronic infection model (5). The CciI autoinducer synthase produces C6-HSL and smaller amounts of C8-HSL. The cciIR system is positively regulated by CepR, and CciR negatively regulates the transcription of cciIR and cepI (35). Mutation of the cciR gene, but not the cciI gene, resulted in a significant defect in adherence. Biofilms formed by a double acylhomoserine lactone synthase mutant of B. cenocepacia (cepI cciI), although not hindered in attachment, were more sensitive to ciprofloxacin. Therefore, quorum-sensing pathways may present suitable therapeutic targets to interfere with B. cepacia complex biofilm formation and compromise the stability and resistance of the resulting biofilm.
MATERIALS AND METHODS
Bacterial strains and growth media.
The bacterial strains used in this study are listed in Table 1. Burkholderia cenocepacia K56-2 (formerly B. cepacia genomovar III) is an epidemically transmitted CF isolate (28). The genome of this strain contains both the B. cepacia epidemic strain marker and the cable pilus gene (33). All strains were subcultured twice from frozen stocks on solid tryptic soy agar (BDH, VWR International, Edmonton, Alberta, Canada) supplemented with 200 μg ml−1 of tetracycline (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) and/or 100 μg ml−1 of trimethoprim (Sigma) when required. Biofilms were grown in tryptic soy broth (TSB) (BBL Microbiology Systems, Cockeysville, MD) as described below. Cation-adjusted Mueller-Hinton broth (Becton Dickinson, VWR International) was used as the diluent for antibiotic susceptibility challenges.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description or genotypea | Reference |
|---|---|---|
| B. cenocepacia strains | ||
| K56-2 | Epidemic cystic fibrosis respiratory isolate, genomovar III, cblA+, BCESM+ | 28 |
| K56-I2 | cepI::tmp derivative of K56-2; Tpr | 28 |
| K56-R2 | cepR::Tn5-OT182 derivative of K56-2; Tcr | 28 |
| K56-2 cciI | cciI::tp derivative of K56-2 | 5 |
| K56-2 cciR | cciR::tp derivative of K56-2 | 35 |
| K56-2 cepI cciIa | ΔcepI cciI::tp derivative of K56-2 with a spontaneous mutation affecting ornibactin synthesis | 35 |
| K56-2 cepI cciIb | ΔcepI cciI::tp derivative of K56-2 | 35 |
| K56-2 cepR cciIR | cepR::tp ΔcciR derivative of K56-2 | 35 |
| Plasmids | ||
| pHKT1 | pBBR1MCS-3 containing 1.6-kbp KpnI/SstI gfp fragment from pASV2; Tcr | 54 |
| pHKT2 | pBBR1Tp containing 1.6-kbp KpnI/XbaI gfp fragment from pASV2; Tpr | 54 |
| pUCP28T | Broad-host-range vector; Tpr | 48 |
| pUCP26 | Broad-host-range vector; Tcr | 62 |
| pSLA3.2 | pUCP28T with 3.2-kb SphI fragment containing cepI; Tpr | 28 |
| pSLR100 | pUCP28T with 1.65-kb KpnI-SphI fragment from pSLA3.2 containing cepR; Tpr | 28 |
| pSLS225 | pUCP26 with 1.5-kb SphI-KpnI fragment from pSLA3.2 containing cepI; Tcr | 29 |
| pRM4.3 | pCR2.1TOPO with 4.3-kb XhoI fragment containing cciIR; Kmr | 35 |
| pRM164 | pUCP26 with 850-bp PCR amplified cciI fragment; Tcr | 35 |
| pRM165 | pUCP26 with 1.1-kb NcoI-ApaLI fragment from pRM4.3; Tcr | This paper |
| pRM166 | pUCP26 with 1.6-kb SphI-KpnI fragment from pSLA3.2; Tcr | This paper |
cblA, cable pilus gene; BCESM, Burkholderia cepacia epidemic strain marker; Tp, trimethoprim; Tc, tetracycline; Ap, ampicillin; Km, kanamycin.
DNA manipulations.
DNA manipulations were performed using standard techniques as described by Sambrook et al. (46). Restriction endonucleases and T4 DNA polymerase were purchased from Invitrogen (Burlington, Ontario, Canada). T4 DNA ligase was purchased from New England Biolabs (Mississauga, Ontario, Canada). Plasmids were introduced into B. cenocepacia by electroporation using a Gene Pulser (Bio-Rad, Richmond, CA) as previously described (17).
Cloning of cciR and cepR.
A 1.1-kb NcoI/ApalI fragment containing cciR was cloned from pRM4.3 (35) into pUCP26 (62), and the resulting plasmid was designated pRM165. A 1.6-kb SphI/KpnI fragment containing cepR was cloned from pSLA3.2 (28) into pUCP26 (62), resulting in pRM166.
Biofilm growth conditions.
Biofilms were formed on the pegs of the MBEC assay system (MBEC BioProducts Inc., Edmonton, Alberta, Canada), which was described by Ceri et al. (9). Briefly, the device consists of 96 conical pegs attached to a plastic lid that rests on top of a trough containing a 1.0 McFarland bacterial suspension diluted 1:30 in broth. The device is placed on a rocking table at 35°C and 95% humidity for 24 h. For growth curve analysis, biofilms were formed and CFU per peg determined, as previously outlined for single-species biofilms by Tomlin et al. (55), except that only 22 ml of the diluted inoculum was used in the trough according to the manufacturer’s directions.
Crystal violet staining of cellular matter.
The following procedure has been adapted for use with the MBEC system from methods originally developed by Christiansen (10, 11). Biofilms were formed on the MBEC system by placing the device lid in a 96-well microtiter plate. Inoculation suspensions of each strain were prepared in TSB and normalized to an optical density of 0.4 at 600 nm, and 5 μl of the suspension was inoculated into 145 μl of TSB. The device was incubated for 24 h as described above, after which the lid was removed and rinsed briefly to remove loose biomass in an MBEC trough containing double-distilled water. The lid was air dried for 10 min, stained with 1% crystal violet (Sigma) in a trough for 1 min, and rinsed three times in separate troughs containing double-distilled water. The stained pegs were decolorized with 175 μl of 100% methanol in a microtiter plate for 1 min. The quantity of crystal violet was measured using a Wallac Victor2 Multilabel counter (Perkin-Elmer Life Sciences, Woodbridge, Ontario, Canada) set to measure absorbance at 540 nm.
Antibiotic susceptibility testing.
MIC, minimum bactericidal concentration (MBC), and minimal biofilm eradication concentration were determined using methods outlined in Ceri et al. (9). The following antibiotics were screened: amikacin, azithromycin, aztreonam, cefepime, ceftazidime, chloramphenicol, ciprofloxacin, colistin, erythromycin, gentamicin, imipenem, meropenem, piperacillin, rifampin, ticarcillin, and tobramycin (Sigma). Stock solutions containing 5.12 mg ml−1 were made for each antibiotic in sterile double-distilled water and stored at −70°C until required.
SDS detachment assay.
The sodium dodecyl sulfate (SDS) detachment assay method was adapted from a similar study using P. aeruginosa PAO1 quorum-sensing mutants grown on a glass coverslip flow cell (15). Biofilms were grown on the MBEC assay system for 24 h as described above. Two pegs were removed from both the first and last columns of pegs and the CFU per peg determined to serve as pretreatment controls. The MBEC lid was transferred to a 96-well microtiter plate containing 225 μl of sterile 0.9% (wt/vol) sodium chloride to rinse off any loose biomass. The lid was then placed in a second microtiter plate containing 200 μl of sterile 0.2% (wt/vol) SDS (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada) in columns 1 through 6 and 200 μl of sterile double-distilled water as a control in the remaining columns. The plate was incubated at 35°C, and two pegs were removed for each treatment at 3, 8, and 24 h during the course of the assay, rinsed in sterile 0.9% (wt/vol) sodium chloride, and assayed for CFU per peg as described above.
Flow cell biofilms.
Three-chamber polycarbonate glass coverslip once-through flow cells were constructed as described elsewhere (54). Briefly, B. cenocepacia strains harboring the appropriate green fluorescent protein expression vector (Table 1) were subcultured twice from frozen stocks on selective-low salt (1.5 g ml−1) pH 7 Luria-Bertani (LB-1) agar (54) and grown up overnight in 5 ml of LB-1 broth containing 50 μg ml−1 of either tetracycline or trimethoprim. The chambers were inoculated and the flow cell maintained over 72 h exactly as described by Tomlin et al. (54). Cells were buffered during microscopy by using 300 μl of phosphate buffer (0.1 M, pH 7) (64). A Leica DMR epifluorescence microscope equipped with a fluorescein isothiocyanate filter, a focus motor (MAC5000/MAC2 Z Joystick; Ludl Electronic Products Ltd., Hawthorne, NY) controlled by Openlab 3.0.9 software (Improvision, Lexington, MA), and a charge-coupled-device camera (Q-Imaging Retiga EX; Quorum Technologies Inc, Guelph Ontario, Canada) was used to obtain three-dimensional image stacks of the biofilms, using a 63× oil immersion lens. Image stacks (512 by 512 pixels per slice) were deconvolved with the volume deconvolution module of Openlab 3.0.9, using the following parameters: z slice = 1 mm, nearest-two-neighbor analysis, and removal = 0.6399. Scion Image 4.0 Beta (Scion Corp.) and Microsoft Excel software were used to quantify and analyze the image stacks. Biovolume was calculated as the average percent area of active pixels over the slices of the stack multiplied by the stack depth (micrometers). Stack depth was determined by the number of slices in the stack multiplied by the slice depth (micrometers). Substratum coverage was calculated as the percent area of active pixels recorded in the slice at the coverslip surface. Stacks were averaged using Scion Image and adjusted with Adobe Photoshop 5.0 (Adobe Systems Canada, Ottawa, Ontario, Canada).
Statistical analysis.
All statistical analyses were completed using SigmaStat V2.03 analysis of variance (ANOVA) followed by Tukey all-pairwise multiple-comparisons tests for normally distributed data. Otherwise, ANOVA on ranks was performed, followed by Dunn’s multiple-comparison tests. For all statistical analyses, P < 0.05 was considered statistically significant. All colony count data underwent log10 transformation before analysis to achieve a normal distribution of the data.
RESULTS
Role of the cepIR system of cell attachment and growth in a biofilm.
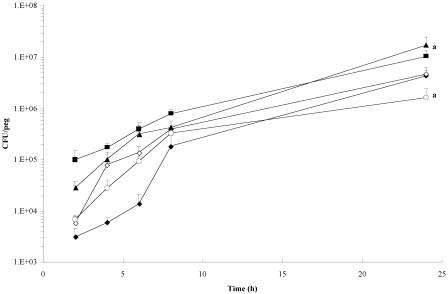
To determine the effect of quorum-sensing mutations on the initial stages of cell attachment and growth in a biofilm, B. cenocepacia K56-2, K56-I2, and K56-R2 and the mutant strains complemented with the wild-type genes were cultured on the pegs of the MBEC assay system over the course of 24 h. As shown in Fig. 1, the 2-, 8-, and 24-hour biofilm CFU counts do not differ significantly between the wild-type and mutant strains. However, a significant difference in cell counts was observed between K56-R2 and this mutant complemented with cepR, K56-R2(pSLR100). In addition, the progressions of biofilm development in terms of the general trend of increase in cell numbers during the course of the experiment were similar between strains.
FIG. 1.
Biofilm growth curves of wild-type B. cenocepacia K56-2 (♦), K56-I2 (⋄), K56-R2 (○), K56-I2(pSLS225) (▪), and K56-R2(pSLR100) (▴). Data points represent the mean (± standard error of the mean) CFU/peg of four pegs over two separate trials (two pegs per trial) every 2 h for 8 h, with a final biofilm density count at 24 h. Significant differences in 24-h biofilm counts were observed between K56-R2 and its complemented counterpart K56-R2(pSLR100) (a, P < 0.05 by ANOVA and Tukey test).
Crystal violet staining of the biomass demonstrates attachment deficiency in cepI, cepR, cciR, and cepR cciIR mutants.
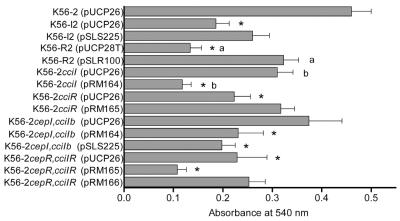
Biofilms of K56-2, K56-I2, and K56-R2 were grown on the MBEC assay system and stained with crystal violet as a semiquantitative measure of cell attachment to compare to colony counts. Indeed, the slight differences observed in 24-hour biofilm CFU counts seen in Fig. 1 are magnified and become more apparent using this method. As shown in Fig. 2, the crystal violet bound by both the K56-R2 and K56-I2 biofilms was significantly less than that bound by the wild-type strain K56-2 (P < 0.05 by ANOVA and Dunn test). In both cases, the attachment deficiency could be at least partially restored upon providing the mutants with wild-type copies of the genes (Fig. 2). However, the biofilms formed by strain K56-I2(pSLS225) were not significantly denser than those formed by K56-I2.
FIG. 2.
Quantification of cellular matter of the quorum-sensing mutants and the complemented mutants represented by absorbance at 540 nm of crystal violet stain bound to 24-h biofilms cultured on the MBEC assay system. The values shown are the means ± standard errors of the means from three trials, assessing at least five pegs per strain per trial. The asterisks indicate strains that produced significantly less biomass than K56-2 (P < 0.05 by ANOVA and Dunn test). K56-R2(pSLR100) produced significantly greater biomass than K56-R2(pUCP28T) (a), and K56-2 cciI(pRM164) produced significantly less biomass than K56-2 cciI(pUCP26) (b) (P < 0.01 by ANOVA and Dunn test).
During this study, the cciIR quorum-sensing system located within the B. cenocepacia pathogenicity island was identified (5). Biofilms formed by the K56-2 cciI and K56-2 cepI cciIb mutants were equivalent to those formed by the K56-2 wild-type strain (Fig. 2); however, attachment was impaired in both the K56-2 cciR and K56-2 cepR cciIR strains. Attachment was restored to wild-type levels in K56-2 cciR(pRM165) but could not be significantly restored in the cepR cciIR mutant with the provision of cciR(pRM165) (Fig. 2). A significant decrease in adherence was noted upon providing the K56-2 cciI and K56-2 cepR cciIR strains with the functional cciI or cepI gene on pRM164 or pSLS225 and the cciR gene on pRM165, respectively (Fig. 2).
Antibiotic susceptibility is quorum-sensing dependent.
Extensive research has demonstrated that biofilms appear to be intrinsically less susceptible to antibiotics than planktonic cells (22). We investigated the possibility that the reduced susceptibility observed in biofilms is a result of the regulation of antibiotic resistance mechanisms by quorum-sensing signaling in B. cenocepacia. Biofilms of K56-2, K56-I2, and K56-R2 grown for 24 h on the MBEC assay system were treated with a wide range of antibiotics (see Materials and Methods). After 24 h of antibiotic challenge, there were no major differences in the planktonic (MIC and MBC) or biofilm (minimal biofilm eradication concentration) antibiotic susceptibilities observed between the wild-type and mutant strains (Table 2). Only the data for antibiotics that were efficacious towards planktonic cultures were presented. Greater concentrations of the antibiotics were required to kill (MBC) rather than just inhibit (MIC) planktonic B. cenocepacia. As expected, even higher concentrations of the antibiotics were required to kill the biofilm cells (minimal biofilm eradication concentration), although there was no evidence that the cepIR system was directly responsible for the increased resistance of the biofilms, since both the cepI and cepR mutants were as resistant as the wild-type strain.
TABLE 2.
Representative antibiotic susceptibility valuesa of B. cenocepacia K56-2, K56-I2, K56-R2, K56-2 cciI, K56-2 cciR, and K56-2 cepI cciIa planktonic (MIC and MBC) and biofilm (minimal biofilm eradication concentration) populations
| Antibiotic | MIC (μg/ml) for strain:
|
MBC (μg/ml) for strain:
|
Minimal biofilm eradication concn (μg/ml) for strain:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K56-2 | K56-I2 | K56-R2 | K56-2 cciI | K56-2 cciR | K56-2 cepI cciI | K56-2 | K56-I2 | K56-R2 | K56-2 cciI | K56-2 cciR | K56-2 cepI cciI | K56-2 | K56-I2 | K56-R2 | K56-2 cciI | K56-2 cciR | K56-2 cepI cciI | |
| Ceftazidime | 8 | 8 | 8 | 16 | 32 | 1 | 128 | 256 | 256 | 128 | 128 | 64 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Chloramphenicol | 32 | 32 | 32 | 64 | 64 | 32 | 512 | 512 | 512 | 512 | 1,024 | 1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Ciprofloxacin | 8 | 8 | 8 | 4 | 4 | 4 | 64 | 64 | 64 | 64 | 128 | 64 | 512 | 512 | 1,024 | 1,024 | 512 | 64 |
| Meropenem | 8 | 8 | 8 | 8 | 4 | 8 | 128 | 128 | 128 | 64 | 128 | 64 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
Values shown represent those from one trial; values in subsequent trials did not vary by more than one or two dilutions from the representative value.
The cciI, cciR, and cepI cciIa mutants were subsequently analyzed for changes in susceptibility to the four antibiotics to which the planktonic B. cenocepacia cells were sensitive (Table 2). As described above, the planktonic and biofilm cells were subjected to doubling dilutions of the antibiotics, beginning from 1,024 μg ml−1 as the greatest concentration tested. A significant change is considered to be a shift in the sensitivity of more than two doubling dilutions in either direction. There were no notable changes in the sensitivities of either the cciI or the cciR mutant planktonic and biofilm cultures compared to the K56-2 wild-type strain (Table 2). However, the double signal mutant (cepI cciIa) was more sensitive to ceftazidime than the wild type during the planktonic phase of growth (a drop from between 8 and 32 μg ml−1 to 1 μg ml−1) and to ciprofloxacin as a biofilm (a drop from between 512 and 1,024 μg ml−1 to 64 μg ml−1).
Quorum-sensing mutant biofilms are structurally deficient.
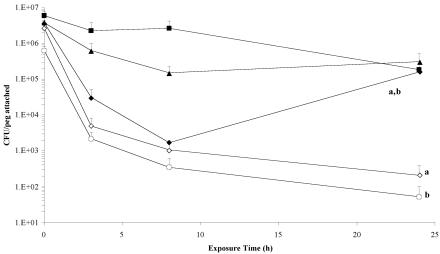
In a study of biofilm formation in P. aeruginosa, Davies et al. (15) demonstrated that lasI signaling mutants of PAO1 were readily detached from a glass coverslip substratum when subjected to treatment with 0.2% (wt/vol) sodium dodecyl sulfate. We were able to test this phenomenon in B. cenocepacia by treating the MBEC lid in a 96-well microtiter plate containing 0.2% SDS or double-distilled water and determining the number of the attached cells by using colony counts on sonicated pegs. By 3 h of treatment, the number of cells attached in the K56-I2 and K56-R2 biofilms drastically dropped by more than 2 log units (Fig. 3). The wild-type K56-2 strain also decreased by approximately 2 log units in this time. However, K56-2 was able to recover during 8 to 24 h of treatment in 0.2% SDS, while the mutant cells continued to detach from the peg. When the cepI and cepR genes were reintroduced into the mutants, the K56-I2(pSLS225) and K56-R2(pSLR100) strains did not demonstrate any appreciable detachment at any time during the course of treatment (Fig. 3). Indeed, they were more resistant to the biofilm-dispersing properties of SDS than the wild-type K56-2 strain. Curiously, no significant detachment of mutant biofilm cells compared to the wild type was observed after 24 h of treatment in the double-distilled water control treatment, except for K56-R2 (data not shown; P < 0.05 by ANOVA). Resistance to detachment was fully restored with the reintroduction of the cepR gene.
FIG. 3.
Comparative analysis of cell detachment of B. cenocepacia K56-2 (♦), K56-I2 (⋄), K56-R2 (○), K56-I2(pSLS225) (▪), and K56-R2(pSLR100) (▴) during a 24-hour treatment with 0.2% (wt/vol) SDS. Data points represent the mean (± standard error of the mean) CFU/peg of four pegs over two separate trials (two pegs per trial) every 2 h for 8 h, with a final biofilm density count at 24 h. By the end of the 24-hour time course, there were significantly fewer cells attached in the K56-I2 and K56-R2 biofilms than in those of the wild-type K56-2 and complemented K56-I2(pSLS225) and K56-R2(pSLR100) strains (a and b, P < 0.05 by ANOVA and Tukey test).
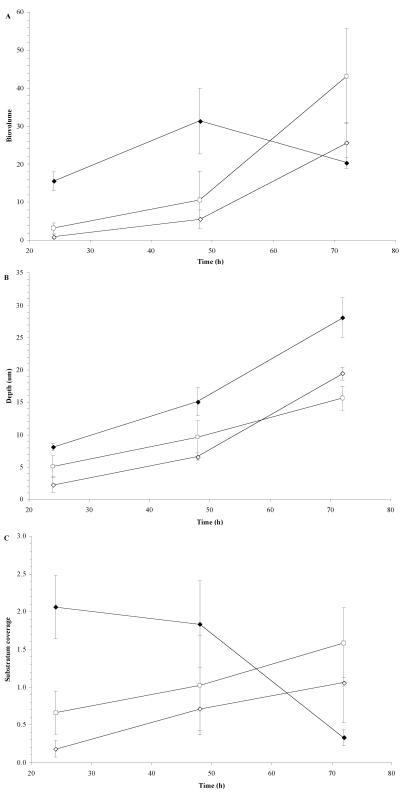
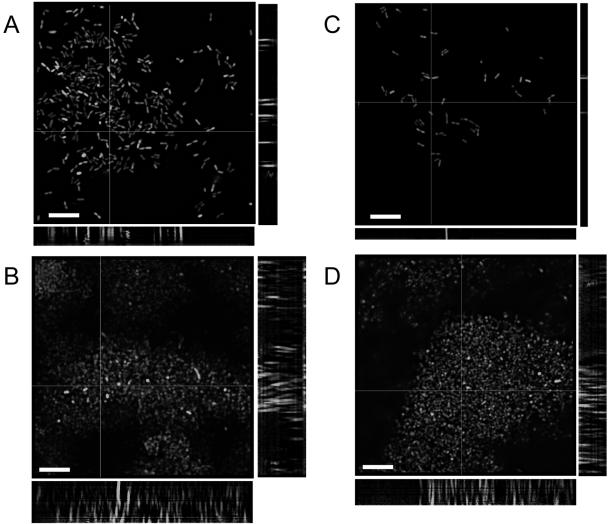
We were able to monitor the structural evolution of the wild-type and mutant biofilms over time by using a glass coverslip three-chamber polycarbonate flow cell. Cells were labeled for deconvolution epifluorescence microscopy using green fluorescent protein expression vectors developed and tested for use in B. cepacia complex biofilms (54). The biovolume, or the proportion of the biofilm comprised of cellular matter, was much slower to increase in both mutant biofilms over the first 48 h (Fig. 4A). However, by 72 h the biovolumes of the mutant biofilms had surpassed that of the wild type. The mutant biofilms were also shallower over the 72 h than the wild-type biofilm (Fig. 4B). However, the differences were not drastic, and the mutant biofilms did achieve a depth of 15 to 20 μm, compared to 25 to 30 μm for the wild-type K56-2. The development of the wild-type strain from the perspective of substratum coverage also differed (Fig. 4C). The cells of the wild-type strain accumulated on the surface of the coverslip, and over time as the biovolume increased (indicating an increased cellular content), the biofilm increased in depth and the cellular matter developed vertically into a more mature pillar-like structure (as indicated by a decrease in substratum coverage but an increase in depth and biovolume). Conversely, the mutant strains accumulated slowly but consistently on the coverslip surface over time as the biovolume and depth increased, indicating that cells were primarily accumulating on the surface and there was less structural development into more pillar-like structures. These data are reflected by the deconvolved epifluorescent images of B. cenocepacia K56-2 (Fig. 5A and B) and K56-R2 (Fig. 5C and D) flow cell biofilms at 24 and 72 h of incubation.
FIG. 4.
Characteristics of glass coverslip flow cell biofilms of B. cenocepacia K56-2 pHKT2 (♦), K56-I2 pHKT1 (⋄), and K56-R2 pHKT2 (○) as assessed by deconvolution epifluorescence microscopy. A) Biovolume was defined as the average percent area of active pixels over the slices of the stack multiplied by the stack depth (micrometers). B) Stack depth was defined as the number of slices in the stack multiplied by the slice depth (micrometers). C) Substratum coverage was defined as the percent area of active pixels recorded in the slice at the coverslip surface. All data points represent the means ± standard errors of the means from at least three separate trials, with three image stacks captured per time point per trial.
FIG. 5.
Deconvolved epifluorescence micrographs (stack average) of B. cenocepacia K56-2(pHKT2) flow cell biofilms at A) 24 h and B) 72 h and of B. cenocepacia K56-R2(pHKT2) flow cell biofilms at C) 24 h and D) 72 h. Slices were taken at 1.0-mm z-plane intervals using a Leica DMR epifluorescence microscope equipped with a focus motor at an exposure of 1.0 seconds and were volume deconvolved using Openlab 3.0.9 software. Side panels represent 1.0-μm slices in the x and y planes. Bars, 10 μm.
DISCUSSION
Previous work by Huber et al. (23) demonstrated that cepI and cepR mutants of B. cenocepacia H111 (CF isolate) will still attach to a glass flow cell surface, but the spatial organization of the cells in the biofilm is drastically altered. This finding is in agreement with a previous study using a lasI signaling mutant of P. aeruginosa PAO1, which also demonstrated attachment without the development of mushroom- and pillar-like three-dimensional structures that are usually observed in mature biofilms of this strain (15). Using cell attachment determined by viable colony counts as a measure of biofilm colonization, our data also suggest that initial attachment and proliferation are not affected by mutations in the cepIR quorum-sensing system of B. cenocepacia. The cepI and cepR mutant biofilm counts on the MBEC assay system did not differ significantly from those of the wild-type K56-2 at 24 h of incubation (Fig. 1). Davies et al. (15) also found no differences in cell attachment in P. aeruginosa quorum-sensing mutants growing in a minimal medium, and DeKievit et al. (16) and Shih and Huang (49), studying the same set of mutants, recorded differences only when the cells were grown in rich medium. Therefore, we decided to utilize additional methods to determine the cellular content of the biofilm.
We employed a second measure of colonization to confirm the initial findings by colony counts. Christensen et al. (10, 11) first developed the method of staining biofilms grown on plastic surfaces with crystal violet to determine colonization and exopolysaccharide production in Staphylococcus isolates. O’Toole and Kolter (41) later adapted the principles of this method to microtiter plate biofilms in which the bound crystal violet could be solubilized and quantified by measuring the absorbance of the dye. We have further adapted this method for use with the MBEC assay system by culturing and staining the biofilms on the pegs rather than the microtiter plate. The small variations in colony counts observed in Fig. 1 were amplified, and we were able to show more drastic and statistically significant differences in colonization between the cepR mutant K56-R2, the cepI mutant K56-I2, and the wild-type K56-2 as well as between K56-R2 and K56-R2(pSLR100). Huber et al. (23) demonstrated similar deficiencies in microtiter plate biofilms of both cepR and cepI mutants in B. cenocepacia H111 and showed an appreciable drop in crystal violet staining (approximately 50%) of both mutants.
Further studies were performed to determine if the cciIR system was involved in biofilm formation, particularly since we still observed appreciable attachment to the pegs in the absence of a functioning cepIR system. Biofilms formed by K56-2 cciIR mutants were also stained with crystal violet, and the quantity of biomass was compared to those of the K56-2 cepIR mutants and complemented strains. Not surprisingly, the cciI signal mutant, with residual production of C6- and C8-HSL provided by the functional copy of cepI, formed biofilms on the MBEC assay system that were statistically equivalent to those formed by the wild-type strain. Presumably, both the CepR and CciR transcriptional regulators are able to respond to the signal and activate or repress the appropriate genes involved in biofilm formation. However, we did not expect that the complemented cciI mutant would demonstrate a significant decrease in attachment compared to those of both the wild-type strain and the original cciI mutant.
Biofilms of the cciR mutant were significantly deficient compared to those of the wild-type strain. At first glance one would expect the cciR mutant to be at least as proficient in attachment as the wild-type strain, since repression of cepI would be removed. However, this mutant produces more C6-HSL than the aforementioned cciI mutant (35); it is possible that this signal is less efficient at binding and activating the CepR transcriptional regulator than C8-HSL, thus providing some inhibition of the activation of cepIR-regulated genes. A similar phenomenon is observed in P. aeruginosa, in which the 3-oxododecanoyl-HSL signal synthesized by the lasI synthase can compete with the rhlI-derived butyroyl-HSL signal for binding to RhlR (42). Successful binding of the 3-oxododecanoyl-HSL to RhlR prevents transcription of the RhlRI-regulated rhamnolipid genes (42).
The most surprising result was that obtained with the cciI cepIb mutant. With no functional signal, one would expect to see very little biomass on the pegs if quorum sensing played a role in attachment, as was suggested by a decrease in biomass with both the cepI and cepR mutants. When the cepI cciIb mutant was complemented with the cciI gene, a reduction in attachment was observed, similar to that observed for the cciI mutant discussed earlier. This may be due to the abundance of C6-HSL signal in the complemented strain available for activation of the CciR protein, which possibly also is an inefficient activator of the CepR protein. However, we also observed a significant lack of biofilm formation when the double mutant was complemented with the cepI gene. In this case, signals would be free to interact with both CciR and CepR.
As expected, the cepR cciIR mutant was significantly impaired in attachment compared to the K56-2 wild type. Restoration of this phenotype was not accomplished by adding cciR in trans, which can be explained by the fact that functional CepR is required for expression of cciIR (37). Provision of the cepR gene partially restored attachment, although only slightly more adherence than for the double mutant was evident.
In an manner analogous to that for studies of P. aeruginosa (15, 49), we sought to determine the effect of quorum-sensing mutations on the resistance and stability of the biofilm. If the later stages of biofilm development and maturation were affected by the lack of a functional quorum-sensing system, it was predicted that we would see values of antibiotic resistance in biofilms that were similar to those of planktonic cultures, particularly if the genes responsible for antibiotic resistance were quorum-sensing regulated. We saw no considerable changes in the planktonic (MIC and MBC) or biofilm (minimal biofilm eradication concentration) antibiotic susceptibility values against the multiple agents tested (Table 2). These results are in contrast to a previous study in P. aeruginosa showing that the lasI signaling mutant and lasI rhlI double mutant of PAO1 were more susceptible to kanamycin than the wild-type strain (49). In addition, resistance to trimethoprim, ciprofloxacin, and chloramphenicol in B. cenocepacia is mediated at least in part by the efflux pump CeoA OpcM (40), of which the CeoA putative periplasmic linker protein is regulated by cepIR (2). It was not surprising that the cepIR mutants were not more susceptible, due to residual signal production provided by the cciIR system. Although the cepI cciI mutant did not demonstrate a compromised ability to form biofilms, the biofilms were more sensitive to killing after treatment with ciprofloxacin; however, the reduction in efficacy of the antibiotic is not of clinical significance.
Nevertheless, when we treated B. cenocepacia K56-2 biofilms with 0.2% sodium dodecyl sulfate as a biofilm dispersant, the cepI and cepR mutants were significantly impaired in their ability to remain intact after 24 h of treatment (Fig. 3). Even the K56-R2 biofilms subjected to sterile double-distilled water as a control showed significant detachment. This deficiency was restored in the reconstituted mutant strains K56-I2 and K56-R2 to a level of stability even greater over the course of the experiment than what was observed with the wild-type strain. Deconvolution epifluorescence microscopy of green fluorescent protein-labeled biofilms grown on a glass coverslip flow cell indicated that the mutant biofilms were developmentally delayed over 72 h of growth compared to the wild-type biofilm (Fig. 4 and 5). In general, the mutant biofilms contained less biomass, were thinner, and covered less of the substratum than the wild-type strain over the first 48 h. The mutant biofilm did recover in biomass, but much of the biomass remained at the level of the substratum, indicating a lack of structural development into vertical three-dimensional structures. Davies et al. (15) also saw similar flat and densely packed biofilms in the lasI mutant of P. aeruginosa, and the biofilm was readily dislodged by treatment with 0.2% SDS, suggesting that the lack of biofilm maturation results in structural instability. Davies et al. (15) suggested that the densely packed and flat lasI mutant biofilm might result from a lack of exopolysaccharide distribution between cells. Brown et al. (6) demonstrated that the exopolysaccharide layer on the surface of planktonic P. aeruginosa cells existed as a thin, dense layer closely associated with the cell surface, while that of biofilm cells was projected from the cell surface to connect with adjoining cells and fill the intervening space. Davies et al. (15) speculated that quorum sensing might play a role in the described transition of the exopolysaccharide layer, although that question has yet to be answered.
The results suggest that the cepR and cepI mutants of B. cenocepacia K56-2 are particularly impaired in attachment, the ability to maintain cells within the biofilm, and structural resistance to mechanical and chemical insults. The observed decrease in cell attachment may actually result from loss of bacteria during the rinsing process while preparing pegs for cell counts and crystal violet staining rather than from deficient attachment. However, scanning electron micrographs of untreated K56-I2 and K56-R2 did not show any noticeable detachment from the MBEC pegs during processing (data not shown). Regardless, both the cepI and the cepR mutants were significantly destabilized in the presence of a biofilm-dispersing detergent, the cepR mutant detached in significant amounts even in the control water treatment, and the biofilms were shown to be structurally and temporally impaired.
Efforts have been directed to understanding the quorum-sensing systems of pathogens, with the hope of interrupting cell-to-cell signaling (for example, by using acylhomoserine lactone analogues) (21, 37, 38), to increase the efficacy of current antibiotic therapies. Our results indicate that disrupting the B. cenocepacia quorum-sensing systems make the bacteria more susceptible to antibiotics during the biofilm state of growth; however, the change observed with the biofilm model employed may not be clinically relevant Yet, we have demonstrated a decrease in the overall structural stability of the biofilm when the cepIR quorum-sensing system has been inactivated, which may allow for more efficacious removal of bacteria in the lungs by mechanical means, such as the daily regimen of postural drainage and percussion therapy used with CF patients.
Acknowledgments
This work was generously supported by funding from an NSERC operating grant (to H.C.) and by the Special Initiative Program in memory of Michael O’Reilly funded by the Canadian Cystic Fibrosis Foundation (to H.C., P.A.S., and D.G.S.). G.R. was supported by a postdoctoral fellowship from the Canadian Institute of Health Research, K.L.T. was funded by NSERC and Alberta iCORE graduate fellowships and a graduate research assistantship from the University of Calgary, and R.J.M. was funded by the Alberta Heritage Foundation for Medical Research.
We thank Doug Muench for the use of his epifluorescence microscope and J. R. Lawrence and G. Swerhone of the National Hydrology Research Institute for providing the specifications for the polycarbonate flow cells.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Bakri, A. G., P. Gilbert, and D. G. Allison. 2004. Immigration and emigration of Burkholderia cepacia and Pseudomonas aeruginosa between and within mixed biofilm communities. J Appl. Microbiol. 96:455-463. [DOI] [PubMed] [Google Scholar]
- 4.Balaban, N., A. Giacometti, O. Cirioni, Y. Gov, R. Ghiselli, F. Mocchegiani, C. Viticchi, M. S. Del Prete, V. Saba, G. Scalise, and G. Dell’Acqua. 2003. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J. Infect. Dis. 187:625-630. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. L., H. C. Aldrich, and J. J. Gauthier. 1995. Relationship between glycocalyx and povidone-iodine resistance in Pseudomonas aeruginosa (ATCC 27853) biofilms. Appl. Environ. Microbiol. 61:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 8.Burns, J. L. 2001. Burkholderia cepacia—a transmissible cystic fibrosis pathogen. J. Pediatr. 139:618-619. [DOI] [PubMed] [Google Scholar]
- 9.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 13.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 14.Conway, B. A., V. Venu, and D. P. Speert. 2002. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. J. Bacteriol. 184:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 16.DeKievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frangolias, D. D., E. Mahenthiralingam, S. Rae, J. M. Raboud, A. G. F. Davidson, R. Wittmann, and P. G. Wilcox. 1999. Burkholderia cepacia in cystic fibrosis: variable disease course. Am. J. Respir. Crit. Care Med. 160:1572-1577. [DOI] [PubMed] [Google Scholar]
- 20.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 21.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 22.Hogan, D., and R. Kolter. 2002. Why are bacteria refractory to antimicrobials? Curr. Opin. Microbiol. 5:472-477. [DOI] [PubMed] [Google Scholar]
- 23.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 24.Huber, B., K. Riedel, M. Kothe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 25.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 26.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 27.Labbate, M., S. Y. Queck, K. S. Koh, S. A. Rice, M. Givskov, and S. Kjelleberg. 2004. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvikovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutants is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 33.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. K. Wong, G. F. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 35.Malott, R., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73:4982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malott, R., and P. Sokol. 2003. Cell-cell signaling mechanisms of the Burkholderia cepacia complex. Recent Res. Dev. Infect. Immun. 1:277-292. [Google Scholar]
- 37.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 38.Manefield, M., T. B. Rasmussen, M. Henzter, J. B. Andersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119-1127. [DOI] [PubMed] [Google Scholar]
- 39.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nair, B. M., K. Cheung, Jr., A. Griffith, and J. L. Burns. 2004. Salicylate induces an antibiotic efflux pump in Burkholderia cepacia complex genomovar III (B. cenocepacia). J. Clin. Investig. 113:464-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 42.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prouty, A. M., J. C. Van Velkinburgh, and J. S. Gunn. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 45.Riedel, K., C. Arevalo-Ferro, G. Reil, A. Gorg, F. Lottspeich, and L. Eberl. 2003. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 24:740-750. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning a laboratory manual :, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schwab, U., M. Leigh, C. Ribeiro, J. Yankaskas, K. Burns, P. Gilligan, P. Sokol, and R. Boucher. 2002. Patterns of epithelial cell invasion by different species of the Burkholderia cepacia complex in well-differentiated human airway epithelia. Infect. Immun. 70:4547-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 49.Shih, P. C., and C. T. Huang. 2002. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 49:309-314. [DOI] [PubMed] [Google Scholar]
- 50.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 51.Steidle, A., M. Allesen-Holm, K. Riedel, G. Berg, M. Givskov, S. Molin, and L. Eberl. 2002. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ. Microbiol. 68:6371-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 53.Sun, L., R. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia cepacia) causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 54.Tomlin, K. L., S. R. Clark, and H. Ceri. 2004. Green and red fluorescent protein vectors for use in biofilm studies of the intrinsically resistant Burkholderia cepacia complex. J. Microbiol. Methods 57:95-106. [DOI] [PubMed] [Google Scholar]
- 55.Tomlin, K. L., O. P. Coll, and H. Ceri. 2001. Interspecies biofilms of Pseudomonas aeruginosa and Burkholderia cepacia. Can. J. Microbiol. 47:949-954. [PubMed] [Google Scholar]
- 56.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 58.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 59.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 60.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 61.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida, A., and H. K. Kuramitsu. 2002. Streptococcus mutans biofilm formation: utilization of a gtfB promoter-green fluorescent protein (PgtfB::gfp) construct to monitor development. Microbiology 148:3385-3394. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]