Abstract
We analyzed the usefulness of rpoA, recA, and pyrH gene sequences for the identification of vibrios. We sequenced fragments of these loci from a collection of 208 representative strains, including 192 well-documented Vibrionaceae strains and 16 presumptive Vibrio isolates associated with coral bleaching. In order to determine the intraspecies variation among the three loci, we included several representative strains per species. The phylogenetic trees constructed with the different genetic loci were roughly in agreement with former polyphasic taxonomic studies, including the 16S rRNA-based phylogeny of vibrios. The families Vibrionaceae, Photobacteriaceae, Enterovibrionaceae, and Salinivibrionaceae were all differentiated on the basis of each genetic locus. Each species clearly formed separated clusters with at least 98, 94, and 94% rpoA, recA, and pyrH gene sequence similarity, respectively. The genus Vibrio was heterogeneous and polyphyletic, with Vibrio fischeri, V. logei, and V. wodanis grouping closer to the Photobacterium genus. V. halioticoli-, V. harveyi-, V. splendidus-, and V. tubiashii-related species formed groups within the genus Vibrio. Overall, the three genetic loci were more discriminatory among species than were 16S rRNA sequences. In some cases, e.g., within the V. splendidus and V. tubiashii group, rpoA gene sequences were slightly less discriminatory than recA and pyrH sequences. In these cases, the combination of several loci will yield the most robust identification. We can conclude that strains of the same species will have at least 98, 94, and 94% rpoA, recA, and pyrH gene sequence similarity, respectively.
Vibrios are gram-negative, usually motile rods, are mesophilic and chemoorganotrophic, and have a facultatively fermentative metabolism (5). They are generally able to grow on marine agar and on the selective medium thiosulfate-citrate-bile salt-sucrose agar and are mostly oxidase positive. Vibrios belong to the Gammaproteobacteria according to 16S rRNA gene sequence analysis. These bacteria are found abundantly in aquatic habitats and in association with eukaryotes. Associations established by vibrios range from mutualistic, e.g., Vibrio fischeri-bobtail squid (26), to pathogenic, e.g., V. cholerae-humans (45). Probiotic Vibrio strains for fish and shellfish have also been documented (44).
The current family Vibrionaceae comprises the genera Enterovibrio (2 species), Grimontia (1 species), Photobacterium (7 species), Salinivibrio (1 species), and Vibrio (64 species). The novel species Photobacterium rosenbergii and Enterovibrio coralii have recently been proposed to encompass isolates associated with coral bleaching (41). Several new Vibrio species, mainly in the phylogenetic neighborhood of V. harveyi, V. halioticoli, V. splendidus, V. tubiashii, and V. fluvialis, have been described in the last few years, with V. neonatus, V. ezurae (28), and V. ponticus (22) being the most recent ones. V. harveyi, V. splendidus, and V. tubiashii are frequently associated with disease in different species of fish and shellfish worldwide, while the V. halioticoli group comprises species that are potentially mutualist to abalones (28). These organisms may be promising probionts for abalone rearing.
Accurate identification of vibrios at the family and genus levels is obtained by 16S rRNA gene sequencing, whereas identification at the species and strain levels requires the application of genomic analyses, including DNA-DNA hybridization, repetitive extragenic palindromic PCR, and amplified fragment length polymorphism (AFLP) analysis (40). These techniques are essential for reliable species identification, because several vibrios have nearly identical 16S rRNA sequences and similar phenotypic features (10). Unfortunately, their use is restricted to a few reference laboratories. Interlaboratory comparisons of fingerprint patterns are difficult. The sequencing of housekeeping genes is emerging as an alternative to overcome this problem. In addition, this type of data may improve the current pragmatic definition of bacterial species (28a). In silico whole-genome analysis applied to the Gammaproteobacteria identified a set of 203 genes that are most valuable for inferring bacterial phylogeny (18a). According to Gevers et al. (8), 135 of these genes are not conserved outside the Gammaproteobacteria and thus may not be appropriate for phylogenetic studies on a broader taxonomic scale.
Different loci, e.g., 23S rRNA (21), gapA (23), gyrB (20), hsp60 (18), and recA (30), have been used for phylogenetic studies and the identification of Vibrionaceae species. So far, these genes (except for recA) have only been examined in a very limited number of species and strains. Thompson et al. (30) analyzed the recA sequences of most vibrios, but mainly using type strains, making it difficult to draw conclusions about the use of this gene as an identification marker. Alternative phylogenetic markers should fulfill several criteria, as put forward by Zeigler (46): (i) the genes must be widely distributed among genomes, (ii) the genes must be present as a single copy within a given genome, (iii) the individual gene sequences must be long enough to contain sufficient information but short enough to allow sequencing in a convenient way (900 to 2,250 nucleotides [nt]), and (iv) the sequences must predict whole-genome relationships with acceptable precision and accuracy that correlate well with the 16S rRNA data and with whole-genome similarities measured by, e.g., DNA-DNA hybridization. A combination of in silico analyses and recent experimental studies of different bacteria, including Bacillus, Proteobacteria, lactic acid bacteria, Mycobacterium, and Mycoplasma, suggested that the RNA polymerase alpha subunit gene (rpoA), recA, and the uridylate kinase gene (pyrH) fulfill these requisites and could therefore be used for identification purposes (8, 18a, 46).
For the present study, we analyzed the rpoA, recA, and pyrH gene sequences of 192 well-documented Vibrionaceae strains comprising all currently known species (except for Photobacterium profundum, V. agarivorans, V. calviensis, V. ruber, V. ponticus, and V. salmonicida). We also included 16 presumptive Vibrio isolates associated with healthy and bleached corals. In order to determine the intraspecies variation among the three loci, we included at least three representative strains each of 37 Vibrionaceae species. The aim of this study was to evaluate the application of different genetic loci as phylogenetic and identification markers, both individually and as concatenated elements. The multigene-based phylogeny roughly confirmed the 16S rRNA gene-based grouping obtained in previous studies and revealed new interesting relationships among different Vibrio species.
MATERIALS AND METHODS
The strains used for this study are listed in Table 1. All strains included in this study are deposited in the BCCM/LMG Bacteria Collection or the Research Collection at Ghent University (Ghent, Belgium). A detailed list of these strains can be found elsewhere (31, 40). This study also included fresh isolates associated with bleached and healthy corals of different species collected in 2002 in Australia and the United States. Isolates R-21409, R-21422, and R-21431 originated from different bleached Pachyseris speciosa colonies (Magnetic Island, Australia). R-21410 originated from a healthy Montipora capitata sample, and isolates R-21416 and R-21432 originated from a healthy P. speciosa sample, both in Kaneohe Bay (United States). R-21415, R-21419, and R-21433 were isolated from healthy Merulina ampliata samples (Magnetic Island, Australia), and R-21413 was isolated from bleached M. ampliata coral (Magnetic Island, Australia). R-21427 and R-21434 originated from bleached Acropora millepora coral (Davies Reef, Australia). R-21426, R-21435, and R-21439 were isolated from healthy A. millepora (Davies Reef, Australia), Pocillopora damicornis (Davies Reef, Australia), and Barabattoia amicorum (Magnetic Island, Australia) samples, respectively. R-23286 was isolated from a bleached Montipora sp. sample (Magnetic Island, Australia) in 2003.
TABLE 1.
Gene sequence variation within species
| Species (strains) | % Sequence variation
|
||
|---|---|---|---|
| rpoA | recA | pyrH | |
| Enterovibrio norvegicus (LMG 19839T, LMG 19840, LMG 19842, R-3717) | 0.2 | 0.4 | 1.0 |
| E. coralii (LMG 22228T) | |||
| Grimontia hollisae (LMG 17719T, LMG 21416, LMG 21538) | 0.1 | 0.0 | 0.1 |
| Photobacterium angustum (LMG 8455T) | |||
| P. damselae subsp. damselae (LMG 7892T, LMG 10940, LMG 19445) | 0.1 | 0.1 | 1.0 |
| P. rosenbergii (LMG 22223T, LMG 22224, LMG 22225, LMG 22226, LMG 22227, R-21419) | 0.4 | 5.5 | 3.0 |
| P. leiognathi (LMG 4228T, LMG 10944, LMG 11221) | 0.2 | 1.0 | 2.0 |
| P. phosphoreum (LMG 4233T) | |||
| Salinivibrio costicola subsp. costicola (LMG 11651T) | |||
| Vibrio aerogenes (LMG 19650T) | |||
| V. aestuarianus (LMG 7909T) | |||
| V. alginolyticus (LMG 4409T, LMG 2174, LMG 4407, LMG 19993, R-14876) | 0.6 | 1.0 | 1.5 |
| V. anguillarum (LMG 4437T, LMG 10861) | 0.2 | 0.5 | |
| V. brasiliensis (LMG 20546T, LMG 20010) | 0.0 | 0.1 | |
| V. campbellii (LMG 11216T, LMG 11256, LMG 16835, LMG 20369, R-14899, R-14902, R-21413, R-21427, R-21434) | 0.4 | 3.0 | 2.0 |
| V. chagasii (LMG 21353T, LMG 13219, LMG 13237) | 0.1 | 2.0 | |
| V. cholerae (LMG 21698T, R-18244, R-18258, R-18297, R-18303, R-20544, R-20545, R-20546, R-20548) | 0.1 | 6.0 | 5.0 |
| V. cincinnatiensis (LMG 7891T) | |||
| V. coralliilyticus (LMG 20984T, LMG 10953, LMG 19270, LMG 20538, LMG 20548, LMG 21350, R-14978, R-14968, R-21432, R-23286) | 0.9 | 4.0 | 3.0 |
| V. cyclitrophicus (LMG 21359T, LMG 20001, R-14874) | 0.0 | 3.0 | |
| V. diabolicus (LMG 19805T) | |||
| V. diazotrophicus (LMG 7893T, LMG 11217, LMG 13218, LMG 20033, R-3706) | 0.5 | 1.6 | 1.5 |
| V. ezurae (LMG 19970T, LMG 19979, R-15766) | 0.0 | 0.4 | 0.5 |
| V. fischeri (LMG 4414T, LMG 11653) | 0.2 | 4.0 | 2.0 |
| V. fluvialis (LMG 7894T, LMG 11654, R-15090) | 0.2 | 1.8 | 1.0 |
| V. fortis (LMG 21557T, LMG 20547, R-15037, R-21409, R-21431) | 0.3 | 6.0 | 6.0 |
| V. furnissii (LMG 7910T, LMG 11655, LMG 11656, LMG 11758) | 0.2 | 0.7 | 1.5 |
| V. gallicus (LMG 21330T) | |||
| V. gazogenes (LMG 19540T) | |||
| V. halioticoli (LMG 18542T, LMG 19700, LMG 19963, LMG 19975) | 0.1 | 1.5 | 2.0 |
| V. harveyi (LMG 4044T, LMG 7890, LMG 11226, LMG 19643, LMG 19714, LMG 20977, R-14947, LMG 11659, LMG 20370, R-14913, R-21410, R-21426, R-21435, R-21439) | 2.0 | 4.2 | 1.5 |
| V. hepatarius (LMG 20362T) | |||
| V. hispanicus (LMG 13240T) | |||
| V. ichthyoenteri (LMG 19664T, R-3774, R-3789, R-3911) | 0.1 | 1.0 | 0.1 |
| V. kanaloaei (LMG 20539T, R-15010) | 0.0 | 0.2 | 0.1 |
| V. lentus (LMG 21034T, R-3884, R-3895, R-3912) | 0.2 | 2.6 | 1.5 |
| V. logei (LMG 19806T) | |||
| V. mediterranei (LMG 11258T, LMG 11663, LMG 16836, LMG 19703T, R-14988, R-14989, R-14990, R-21415) | 0.2 | 0.4 | 4.0 |
| V. metschnikovii (LMG 11664T, LMG 4416, LMG 4426, R-22290) | 0.0 | 3.2 | 3.5 |
| V. mimicus (LMG 7896T, R-20564, R-20565, R-20568) | 0.1 | 2.5 | 3.0 |
| V. mytili (LMG 19157T) | |||
| V. natriegens (LMG 10935T) | |||
| V. navarrensis (LMG 15976T) | |||
| V. neonatus (LMG 19972T, LMG 19976, LMG 19978) | 0.1 | 1.8 | 2.0 |
| V. neptunius (LMG 20536T, LMG 20613, R-1575) | 0.2 | 0.6 | 1.0 |
| V. nereis (LMG 3895T) | |||
| V. nigripulchritudo (LMG 3896T) | |||
| V. ordalii (LMG 13544T, LMG 10951, R-15101, R-15107) | 0.0 | 0.7 | 2.0 |
| V. orientalis (LMG 7897T) | |||
| V. pacinii (LMG 19999T, LMG 13245, R-15016) | 0.1 | 1.1 | 1.5 |
| V. parahaemolyticus (LMG 2850T, LMG 11670, RIMD 2210633) | 0.3 | 6.0 | 6.0 |
| V. pectenicida (LMG 19642T, LMG 20549, LMG 20550) | 0.0 | 0.0 | 1.5 |
| V. pelagius (LMG 3897T, LMG 19995) | 0.1 | 0.5 | |
| V. penaeicida (LMG 19663T) | |||
| V. pomeroyi (LMG 20537T, R-14805) | 0.2 | 2.2 | 1.5 |
| V. proteolyticus (LMG 3772T, R-15065) | 0.0 | 0.0 | 0.5 |
| V. rotiferianus (LMG 21460T, R-14935, R-21416, R-21422, R-21433) | 0.0 | 0.6 | 0.5 |
| V. rumoiensis (LMG 20038T, LMG 20039) | 0.0 | 0.3 | 0.5 |
| V. scophthalmi (LMG 19158T, LMG 20023, R-15029) | 0.0 | 0.8 | |
| V. splendidus (LMG 19031T, R-14789, LMG 16748, LMG 16751, LMG 16752) | 0.8 | 6.0 | 1.0 |
| V. superstes (LMG 21323T) | |||
| V. tapetis (LMG 19706T, LMG 19704, LMG 19705) | 0.0 | 0.0 | 1.0 |
| V. tasmaniensis (LMG 20012T, R-14842, R-14846) | 0.0 | 0.6 | 1.0 |
| V. tubiashii (LMG 10936T, LMG 16851, R-14825) | 1.0 | 3.3 | 3.7 |
| V. vulnificus (LMG 13545T, R-15063, CMCP6) | 0.3 | 1.3 | 2.4 |
| V. wodanis (K16, K26) | 0.1 | 0.3 | 1.1 |
| V. xuii (LMG 21346T, LMG 20011, R-15053) | 0.1 | 1.7 | 1.2 |
Bacterial genomic DNAs were extracted according to the methodology described by Pitcher et al. (24). PCRs were performed essentially as described previously (30). The sequences of the primers used for amplification and sequencing are listed in Table 2. These primers were designed using 17 gene sequences of each locus of Vibrio cholerae (n16961_o1), V. parahaemolyticus (o3k6_rimd2210633), V. vulnificus (cmcp6), Escherichia coli (cft073, o157h7_edl933, o157h7_rimd0509952, and k12_mg1655), Shigella flexneri (2a_2457t and 2a_301), Salmonella enterica (ct18 and ty2_typhi 3), S. enterica serovar Typhimurium (lt2sgsc1412_atcc700720), Yersinia pestis (co92 and kim), Haemophilus influenzae (rd), Pasteurella multocida (pm70), and Shewanella oneidensis (mr1), which originated from publicly available data from whole-genome sequencing projects. All of the primers specifically amplified the target fragments of all currently known strains of vibrios.
TABLE 2.
Amplification and sequencing primers for rpoA, recA, and pyrH
| Gene product | Primer | Sequence (5′→3′) | Position |
|---|---|---|---|
| Uridylate kinase (pyrH; 750 nt) | pyrH-02-R | GTRAABGCNGMYARRTCCA | 599 |
| pyrH-04-F | ATGASNACBAAYCCWAAACC | 1 | |
| RNA polymerase alpha chain (rpoA; 1,000 nt) | rpoA-01-F | ATGCAGGGTTCTGTDACAG | 1 |
| rpoA-03-R | GHGGCCARTTTTCHARRCGC | 951 | |
| rpoA-05-F | GCAGCDCGTGTWGARCARCG | 568 | |
| rpoA-06-R | CGYTGYTCWACACGHGCTGC | 568 | |
| RecA protein (recA; 1,300 nt) | recA-01-F | TGARAARCARTTYGGTAAAGG | 222 |
| recA-02-R | TCRCCNTTRTAGCTRTACC | 1040 | |
| recA-03-F | TYGGBGTGATGTTYGGTAACC | 767 | |
| recA-04-R | GGGTTACCRAACATCACVCC | 769 |
PCR mixtures were composed of 29.5 μl sterile MilliQ water, 5.0 μl PCR buffer (10×), 5.0 μl deoxynucleoside triphosphates (2 mM each), 2.5 μl forward primer rpoA-01-F (10 μM), 2.5 μl reverse primer rpoA-03-R (10 μM), 0.5 μl AmpliTaq DNA polymerase (1 U/μl), and 5.0 μl template DNA (0.01 μg/μl). PCRs were performed using a GeneAmp PCR System 9600 thermocycler (Applied Biosystems). The thermal program consisted of (i) 5 min at 95°C; (ii) 3 cycles of 1 min at 95°C, 2 min 15 s at 55°C, and 1 min 15 s at 72°C; (iii) 30 cycles of 35 s at 95°C, 1 min 15 s at 55°C, and 1 min 15 s at 72°C; and (iv) a final 7 min at 72°C. PCR products with the expected size and intensity were purified using the Nucleofast 96 PCR cleanup system (Macherey-Nagel, Germany). Purified PCR products were eluted in 30 to 200 μl sterile MilliQ water. Subsequently, 3.0 μl of purified PCR product was mixed with 1.0 μl ABI Prism Big Dye Terminator ready reaction mix, version 3.1 (Applied Biosystems), 3.0 μl sequencing primer (4 μM), 1.5 μl dilution buffer (5×), and 1.5 μl MilliQ water. The thermal program consisted of 30 cycles of 15 s at 96°C, 1 s at 35°C, and 4 min at 60°C. Sequencing products were purified using a Montage SEQ96 sequencing reaction cleanup kit (Millipore). Purified sequencing reactions were eluted in 20 μl of injection solution and mixed with 20 μl of deionized formamide. Subsequently, separation of the DNA fragments was obtained in an ABI PRISM 3100 genetic analyzer (Applied Biosystems). The time and voltage of sample injection were 20 s and 1.25 kV. Each run was performed at 50°C for 6,500 s at 0.1 mA and 12.2 kV. Raw sequence data were transferred to the Gene Builder module within Kodon package 2.03 (Applied Maths, Belgium), where consensus sequences were determined using the two to four reads. Consensus sequences were imported into BioNumerics 3.0 software (Applied Maths, Belgium), where a similarity matrix and phylogenetic trees were created based on the maximum parsimony and neighbor joining methods (27). Splits tree decomposition analysis was done using software available on the Internet (http://bibiserv.techfak.uni-bielefeld.de/splits/) (13), while the GC content, ratio of mean synonymous substitutions per synonymous site to mean nonsynonymous substitutions per nonsynonymous site (dS/dN), and Sawyer's test were calculated using the software package START obtained from http://pubmlst.org/software/analysis/start/ (15).
RESULTS AND DISCUSSION
We sequenced fragments of the rpoA (931 nt), recA (613 to 783 nt), and pyrH (443 nt) genes of 192 vibrios corresponding to 60 to 93% of the coding regions of these genes (EMBL accession no. AJ842347 to AJ842743). The GC contents of rpoA (46% ± 0.9%), recA (46.6% ± 1.9%), and pyrH (48.7% ± 0.9%) are within the average for the total genomes of vibrios (34-39). We compared the rpoA, recA, pyrH, and 16S rRNA pairwise similarities using Pearson's product-moment correlation coefficient. A significant correlation was obtained between 16S rRNA and the three loci (R = 0.81 for rpoA and 0.68 for recA and pyrH). The rpoA and 16S rRNA data had a linear relationship, whereas the pyrH and recA data were best fitted with a polynomial regression of the second degree. These significant correlations show that rpoA, recA, and pyrH are indeed phylogenetic markers in vibrios. The gene sequences of these three loci have high proportions of synonymous mutations (dS/dN = 29 for rpoA and recA and 39 for pyrH), suggesting that there is selection against amino acid changes in these genes (6).
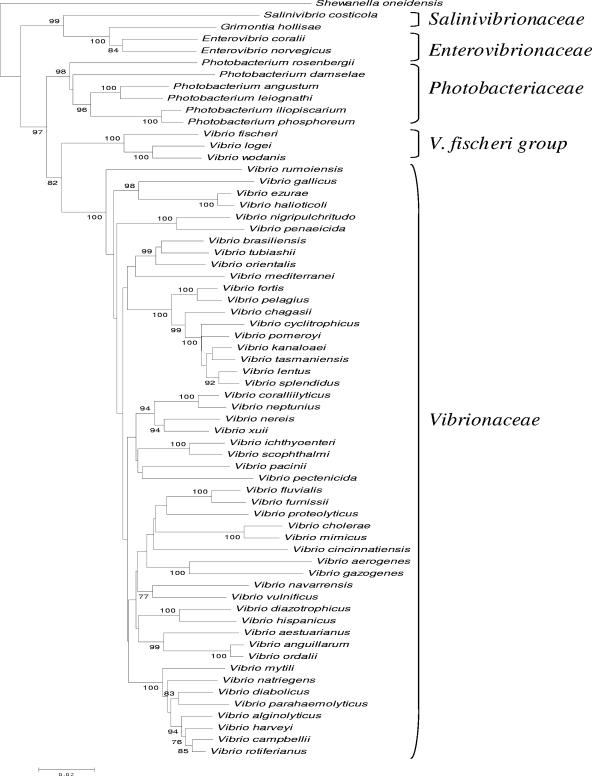
The families Vibrionaceae, Photobacteriaceae, Enterovibrionaceae, and Salinivibrionaceae were clearly separated from one another on the basis of the four loci (Fig. 1). The five currently known genera of vibrios were all differentiated on the basis of the gene sequences (Fig. 1). Enterovibrio strains had 95.4 to 100%, 88.6 to 100%, and 85.6 to 99.8% rpoA, recA, and pyrH gene sequence similarity, respectively. Grimontia hollisae and E. coralii had 96.2%, 85%, and 86% rpoA, recA, and pyrH sequence similarity, respectively. The new species E. coralii was tentatively allocated to the genus Enterovibrio on the basis of polyphasic taxonomic analysis (41), but we anticipate that this organism may be allocated to a new genus in future studies. Photobacterium species had 90.4 to 100%, 80 to 100%, and 79 to 99.6% rpoA, recA, and pyrH sequence similarity, respectively. Strains of the genus Vibrio showed the highest gene sequence variation, with ca. 19% variation for rpoA and 27% variation for recA and pyrH. V. logei, V. fischeri, and V. wodanis grouped apart from the other genus members, suggesting that Vibrio species are polyphyletic. The two squid symbionts had 96.6, 84, and 86% rpoA, recA, and pyrH sequence similarity, respectively.
FIG.1.
Phylogenetic trees based on neighbor-joining method using 16S rRNA (1,300 nt), rpoA (928 nt), pyrH (443 nt), and recA (613 nt) gene sequences of vibrios. Distance estimations were obtained by the model of Jukes and Cantor (16). Bootstrap percentages (≥50) after 1,000 simulations are shown. Bars, 1% (16S rRNA) and 10% (rpoA, pyrH, and recA) estimated sequence divergence. The Campylobacter NCTC 11168 sequence was used as an outgroup.
Within the Vibrionaceae, some pairs of highly related species, i.e., V. aerogenes-V. gazogenes, V. fluvialis-V. furnissii, V. cholerae-V. mimicus, V. diazotrophicus-V. hispanicus, V. ichthyoenteri-V. scophthalmi, and V. anguillarum-V. ordalii, appeared in the different phylogenetic trees. These pairs are indeed known to have highly related genomes, with about 70% DNA-DNA similarity. V. halioticoli-, V. harveyi-, V. splendidus-, and V. tubiashii-related species formed groups in each tree. Roughly, the grouping of Vibrio species obtained with different genes is in agreement with that in previous polyphasic taxonomy studies (31-38, 40). V. harveyi-related species had at least 96.5%, 97%, and 95% rpoA, recA, and pyrH gene sequence similarity, respectively. V. harveyi was closely related to V. parahaemolyticus (98.8 to 99%), but it had only 96.5%, 97.4 to 97.7%, and 91 to 95% rpoA, recA, and pyrH gene sequence similarity, respectively, to its sister species V. campbellii and V. rotiferianus (9). V. harveyi LMG 11659, LMG 20370, and R-14913 clustered apart from the other cospecific strains, having 98.3% similarity to the other V. harveyi strains. These three strains were found in the former AFLP clusters A30 and A31 (31) and had about 70% DNA-DNA similarity with V. harveyi LMG 4044T, suggesting that they may belong to a new species (10). The gut abalone vibrios were all grouped together with at least 91.2, 97, and 93% rpoA, recA, and pyrH similarity, respectively. V. halioticoli, V. ezurae, and V. neonatus grouped together (93 to 99.5%) in all trees. The gut abalone vibrios have similar 16S rRNA sequences (≥98%) (12, 28), indicating that the three loci studied here are alternatives for the identification of these organisms. It is also evident that the three genetic loci are useful for the identification of the species V. nigripulchritudo, V. penaeicida, V. tapetis, V. mediterranei, V. rumoiensis, V. proteolyticus, V. metschnikovii, and V. cincinnatiensis, as they had <96% gene sequence similarity with their closest neighbors.
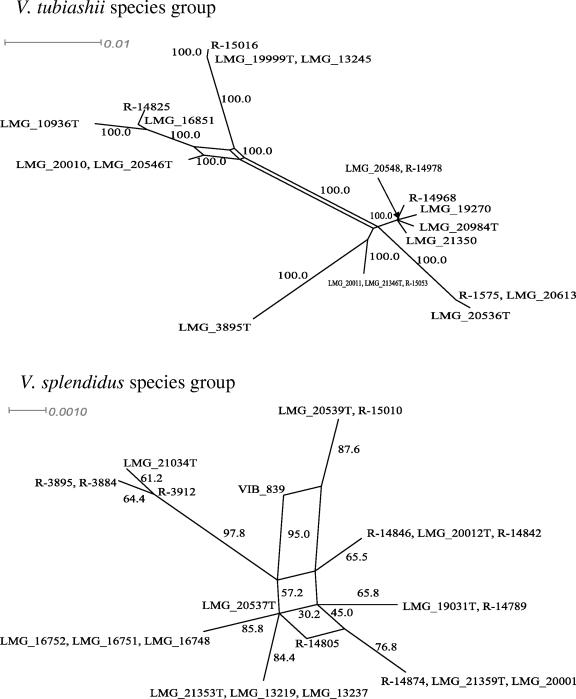
The high similarity of genomes found among different species of vibrios, e.g., in the V. splendidus- and V. tubiashii-related groups, may be explained by niche adaptation (4). V. splendidus-related species probably coexist in the same environment. These organisms have been associated with bivalve mollusks and with the so-called summer mortality syndrome (19). According to Cohan (4), natural selection is the main driving force in the evolution of bacterial species. He suggests that sexual isolation is not a milestone in the origin and maintenance of bacterial species. Indeed, candidate gene transfers, including paralog acquisition or the displacement and acquisition of new genes, have been detected in 5.6% (215 genes) of the genome of V. cholerae N16961 (17). Horizontal gene transfer (HGT) may indeed be an important force in the evolution of vibrios. The positions of various pairs of species, e.g., V. hispanicus and V. diazotrophicus, V. mytili and V. diabolicus, V. coralliilyticus and V. neptunius, and V. brasiliensis and V. tubiashii, changed in the trees constructed from the different genetic loci, suggesting that recombination might have occurred among these vibrios. Although rpoA and recA are thought to belong to the bacterial core genome and for this reason may be refractory to HGT (11), a recent study reported the HGT of rpoA in Aquifex, Thermotoga, and Fusobacterium (14). Our study did provide clear evidence of gene conversion events in the rpoA, recA, and pyrH (P < 0.05) genes of V. splendidus- and V. tubiashii-related species using Sawyer's test. In addition, our splits-tree decomposition analysis revealed a network-like tree for these groups (Fig. 2). The presence of parallelograms in splits trees is a hallmark of recombination (6). Thus, we may conclude that recombination is a rather common evolutionary process within different species of vibrios. Due to such recombinational events, different vibrios, e.g., V. tubiashii- and V. splendidus-related species, may group close to one another, hampering their identification. In order to overcome the effects of gene conversion and recombination in species identification, several loci should be indexed simultaneously and used to construct multigene phylogenetic trees. The four families of vibrios were clearly apart from one another in our multigene tree (Fig. 3). The groups obtained with this tree corresponded to those in the single-locus trees, except for the position of V. mediterranei along with those of V. tubiashii-related species. All species had <97% concatenated sequence similarity, with the exception of the pairs V. coralliilyticus-V. neptunius and V. anguilarum-V. ordalii.
FIG. 2.
Splits tree showing networks in V. tubiashii (A)- and V. splendidus (B)-related groups on the basis of rpoA gene sequences (931 bp). Bootstrap percentages (≥50) after 500 simulations are shown. Bar, 10% estimated sequence divergence.
FIG. 3.
Phylogenetic tree based on neighbor-joining method using the concatenated sequences (3,284 nt) of 16S rRNA, rpoA, pyrH, and recA of type strains. Distance estimations were obtained by the model of Jukes and Cantor (16). Bootstrap percentages (≥50) after 1,000 simulations are shown. Bar, 2% estimated sequence divergence.
So far, there is not a single gene that can differentiate well all species of vibrios. Different genes show different degrees of discrimination according to the group of vibrios under analysis. For instance, V. cyclitrophicus, V. splendidus, V. pomeroyi, and V. chagasii had about 99% rpoA sequence similarity. V. splendidus-related species are indeed highly related, having little variation (<2%) in gyrB and 16S rRNA gene sequences, phenotypic features, and DNA-DNA hybridization (19, 36). On the other hand, these species have <97% recA and pyrH gene sequence similarity (except for V. lentus and V. splendidus, which have 97.2% gene sequence similarity) (30). Nevertheless, the different V. splendidus-related species clearly formed separated clusters on the basis of the three loci, suggesting that these genes are indeed useful for the differentiation of highly related species.
Another interesting example of high gene sequence similarity in different loci is found within the V. tubiashii-related species. These organisms had at least 92.9% rpoA gene sequence similarity. V. coralliilyticus had 98.2, 97.9, 97.1, and 95.1% rpoA sequence similarity with V. xuii, V. neptunius, V. nereis, and V. tubiashii, respectively. The corresponding 16S rRNA similarity values were 96.9, 98.2, 96.7, and 97.1%, respectively (35), while those for pyrH (except those for V. neptunius and V. coralliillyticus, which had a 96% sequence similarity) and recA were below 87 and 97%, respectively. V. tubiashii and V. brasiliensis had a 98.2% gene sequence similarity for both rpoA and 16S rRNA (35). Overall, rpoA and 16S rRNA gene sequences have similar discriminatory powers for V. tubiashii-related species that are inferior to those of recA and pyrH (40). V. pacinii, V. pectenicida, and the pair V. scophthalmi-V. ichthyoenteri had <96% rpoA gene sequence similarity to V. tubiashii-related species, suggesting that this gene is useful for the identification of these species. Overall, rpoA gene sequences were more discriminatory than 16S rRNA sequences. 16S rRNA and recA similarities above 97 and 94% corresponded to rpoA similarities above 88 and 97%, respectively. This fact highlights the need for future studies aiming at additional loci.
Vibrios have been implicated in the phenomenon of coral bleaching (2, 25). We used rpoA gene sequences to allocate taxonomically fresh isolates of vibrios associated with coral bleaching. These isolates were identified as V. fortis, V. campbellii, V. coralliilyticus, V. harveyi, V. mediterranei, and V. rotiferianus, suggesting that the process of coral bleaching may be carried out by different Vibrio species. V. harveyi has been implicated in diseases of a wide range of marine animals (1), including different coral species (10, 29). Although vibrios isolated in Hawaii were always associated with healthy corals, our data suggest that highly related strains of potentially pathogenic vibrios, e.g., R-21432 and R-23286, are present in both Kaneohe Bay (Hawaii) and the Great Coral Barrier (Australia). Environmental conditions in Australia may favor the prevalence of coral infection caused by vibrios, but this remains to be determined in future studies.
Considerable numbers of representative strains of the species V. campbellii (n = 9), V. cholerae (n = 10), V. coralliilyticus (n = 9), V. harveyi (n = 14), and V. mediterranei (n = 7) were examined in this study in order to unambiguously determine the intraspecies variation of rpoA, recA, and pyrH gene sequences. The intraspecies gene sequence heterogeneity for most species was well below 2, 6, and 6%, respectively (Table 1). Overall, the 192 type and reference strains represent well the currently known genomic diversity of most Vibrionaceae species. Strains within each of the examined species fulfill the criteria of a ≥60% mutual AFLP pattern similarity, ≥70% DNA-DNA similarity, and ≥97.5% 16S rRNA gene sequence similarity. Representative strains were selected in order to represent the currently known genomic diversity of these species. The V. cholerae strains, for instance, comprise the serogroups O1, O139, and non-O1/non-O139 and represent the known genomic diversity of this species, as revealed by FAFLP analysis (39, 40). We could therefore conclude that strains of the same species will have at least 98%, 94%, and 94% sequence similarity in the rpoA, recA, and pyrH genes, respectively.
The data generated in this study are well suited to be used for the rapid detection and identification of pathogenic vibrios in the environment through, e.g., real-time PCR (3, 7). The data could also be an alternative to 16S rRNA gene sequences in studies of the ecology and community dynamics of vibrios in coastal waters (42, 43). The advantages of the loci studied here are that they belong to the bacterial core genome (11), have a high phylogenetic signal, and are single-copy genes and that different species of vibrios have different gene sequences that thus enable the reliable identification of these organisms. Our multilocus sequence analysis data will be used as the basis for the creation of a free-access online identification system for vibrios (http://lmg.ugent.be/bnserver/MLSA/Vibrionaceae/).Work is under way on other genes, including the atpA, obg, uvrB, pheS, and serS genes encoding tRNA synthases and thd, which together will enhance the discrimination of all currently recognized species of vibrios.
Acknowledgments
F.L.T. acknowledges a postdoctoral fellowship from BCCM/LMG Bacteria Collection (Ghent, Belgium) and a young researcher fellowship from FAPESP, Brazil (no. 2004/00814-9). D.G. acknowledges financial support from BOF (project no. 01110803). J.S. acknowledges grants from the Fund for Scientific Research (FWO), Belgium. S.N. acknowledges a Ph.D. scholarship from the Palestinian Ministry of Higher Education.
We thank K. Vandemeulebroecke and R. Coopman for their technical assistance. We thank M. Maiden (University of Oxford) for suggestions in the early stages of this work.
REFERENCES
- 1.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: diseases of farmed and wild fish, 3rd ed. Springer, Berlin, Germany.
- 2.Ben-Haim, Y., F. L. Thompson, C. C. Thompson, M. C. Cnockaert, B. Hoste, J. Swings, and E. Rosenberg. 2003. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53:309-315. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, M. S., and A. C. Wright. 2003. Real-time PCR analysis of Vibrio vulnificus from oysters. Appl. Environ. Microbiol. 69:7137-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohan, F. M. 2004. Concepts of bacterial biodiversity for the age of genomics, 175-194. In C. M. Fraser, T. D. Read, and K. E. Nelson (ed.), Microbial genomes. Humana Press Inc., Totowa, N.J.
- 5.Farmer, J. J., III. 1992. The family Vibrionaceae, p. 2938-2951. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, and applications, 2nd ed. Springer-Verlag, Berlin, Germany.
- 6.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483-495. [DOI] [PubMed] [Google Scholar]
- 7.Fukushima, H., Y. Tsunomori, and R. Seki. 2003. Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J. Clin. Microbiol. 41:5134-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevers, D., K. Vandepoele, C. Simillion, and Y. Van de Peer. 2004. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 12:148-154. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Gil, B., F. L. Thompson, C. C. Thompson, and J. Swings. 2003. Vibrio rotiferianus sp. nov., isolated from cultures of the rotifer Brachionus plicatilis. Int. J. Syst. Evol. Microbiol. 53:239-245. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Gil, B., S. Soto-Rodríguez, A. García-Gasca, A. Roque, R. Vazquez-Juarez, F. L. Thompson, and J. Swings. 2004. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 150:1769-1777. [DOI] [PubMed] [Google Scholar]
- 11.Harris, J. K., S. T. Kelley, G. B. Spiegelman, and N. R. Pace. 2003. The genetic core of the universal ancestor. Genome Res. 13:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, K., J. Moriwaki, T. Sawabe, F. L. Thompson, J. Swings, N. Gudkovs, R. Christen, and Y. Ezura. 2003. Vibrio superstes sp. nov., isolated from the gut of Australian abalones Haliotis laevigata and Haliotis rubra. Int. J. Syst. Evol. Microbiol. 53:1813-1817. [DOI] [PubMed] [Google Scholar]
- 13.Huson, D. H. 1998. Splits tree: analysing and visualising evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 14.Iyer, L. M., E. V. Koonin,, and L. Aravind. 2004. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene 23:73-88. [DOI] [PubMed] [Google Scholar]
- 15.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. H. Munro (ed.), Mammalian protein metabolism. Academic Press, London, United Kingdom.
- 17.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok, A. Y., J. T. Wilson, M. Coulthart, L. K. Ng, L. Mutharia, and A. W. Chow. 2002. Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gene sequences. Can. J. Microbiol. 48:903-910. [DOI] [PubMed] [Google Scholar]
- 18a.Lerat, E., V. Daubin, and N. A. Moran. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-proteobacteria. PLOS Biol. 1:E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Roux, F., M. Gay, C. Lambert, M. Waechter, S. Poubalanne, B. Chollet, J. L. Nicolas, and F. Berthe. 2002. Comparative analysis of Vibrio splendidus-related strains isolated during Crassostrea gigas mortality events. Aquat. Liv. Res. 15:251-258. [Google Scholar]
- 20.Le Roux, F., M. Gay, C. Lambert, J. L. Nicolas, M. Gouy, and F. Berthe. 2004. Phylogenetic study and identification of Vibrio splendidus-related strains based on gyrB gene sequences. Dis. Aquat. Org. 58:143-150. [DOI] [PubMed] [Google Scholar]
- 21.Macian, M. C., W. Ludwig, R. Aznar, P. D. A. Grimont, K. H. Schleifer, E. Garay, and M. J. Pujalte. 2001. Vibrio lentus sp. nov., isolated from Mediterranean oysters. Int. J. Syst. Evol. Microbiol. 51:1449-1456. [DOI] [PubMed] [Google Scholar]
- 22.Macian, M. C., E. Garay, P. A. D. Grimont, and M. J. Pujalte. 2004. Vibrio ponticus sp. nov., a neighbour of V. fluvialis-V. furnissii clade, isolated from gilthead sea bream, mussels and seawater. Syst. Appl. Microbiol. 27:535-540. [DOI] [PubMed] [Google Scholar]
- 23.Nishiguchi, M. K., and V. S. Nair. 2003. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int. J. Syst. Evol. Microbiol. 53:2019-2026. [DOI] [PubMed] [Google Scholar]
- 24.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 25.Rosenberg, E., and Y. Loya. 2004. Coral health and disease. Springer, New York, N.Y.
- 26.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sawabe, T., K. Hayashi, J. Moriwaki, F. L. Thompson, J. Swings, and R. Christen. 2004. Vibrio neonatus sp. nov. and Vibrio ezurae sp. nov. isolated from the gut of Japanese abalones. Syst. Appl. Microbiol. 27:527-535. [DOI] [PubMed] [Google Scholar]
- 28a.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland, K. P., J. W. Porter, and C. Torres. 2004. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266:273-302. [Google Scholar]
- 30.Thompson, C. C., F. L. Thompson, K. Vandemeulebroecke, B. Hoste, P. Dawyndt, and J. Swings. 2004. Use of recA as an alternative phylogenetic marker in the family Vibrionaceae. Int. J. Syst. Evol. Microbiol. 54:919-924. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, F. L., B. Hoste, K. Vandemeulebroecke, and J. Swings. 2001. Genomic diversity amongst Vibrio isolates from different sources determined by fluorescent amplified fragment length polymorphism. Syst. Appl. Microbiol. 24:520-538. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, F. L., B. Hoste, C. C. Thompson, G. Huys, and J. Swings. 2001. The coral bleaching Vibrio shilonii Kushmaro et al. 2001 is a later synonym of Vibrio mediterranei Pujalte and Garay 1986. Syst. Appl. Microbiol. 24:516-519. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, F. L., B. Hoste, C. C. Thompson, J. Goris, B. Gomez-Gil, L. Huys, P. De Vos, and J. Swings. 2002. Enterovibrio norvegicus gen. nov., sp. nov., isolated from the gut of turbot (Scophthalmus maximus) larvae: a new member of the family Vibrionaceae. Int. J. Syst. Evol. Microbiol. 52:2015-2022. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, F. L., B. Hoste, K. Vandemeulebroecke, and J. Swings. 2003. Reclassification of V. hollisae as Grimontia hollisae gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 53:1615-1617. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, F. L., Y. Li, B. Gomez-Gil, C. C. Thompson, B. Hoste, K. Vandemeulebroucke, G. S. Rupp, A. Pereira, M. M. De Bem, P. Sorgeloos, and J. Swings. 2003. Vibrio neptunius sp. nov., V. brasiliensis sp. nov. and V. xuii sp. nov., isolated from the marine aquaculture environment (bivalves, fish, rotifers and shrimps). Int. J. Syst. Evol. Microbiol. 53:245-252. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, F. L., C. C. Thompson, Y. Li, B. Gomez-Gil, J. Vandenberghe, B. Hoste, and J. Swings. 2003. Description of Vibrio kanaloae sp. nov, Vibrio pomeroyi sp. nov. and Vibrio chagasii sp. nov., from sea water and marine animals. Int. J. Syst. Evol. Microbiol. 53:753-759. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, F. L., C. C. Thompson, and J. Swings. 2003. Vibrio tasmaniensis sp. nov., isolated from Atlantic salmon (Salmo salar L.). Syst. Appl. Microbiol. 26:65-69. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, F. L., C. C. Thompson, B. Hoste, K. Vandemeulebroecke, M. Gullian, and J. Swings. 2003. Description of Vibrio fortis sp. nov. and V. hepatarius sp. nov., isolated from aquatic animals and the marine environment. Int. J. Syst. Microbiol. 53:1495-1501. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, F. L., C. C. Thompson, A. C. P. Vicente, G. N. D. Theophilo, E. Hofer, and J. Swings. 2003. Genomic diversity of clinical and environmental Vibrio cholerae strains isolated in Brazil between 1991 and 2001 as revealed by fluorescent amplified fragment length polymorphism analysis. J. Clin. Microbiol. 41:1946-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, F. L., T. Iida, and J. Swings. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, F. L., C. C. Thompson, S. Naser, B. Hoste, K. Vandemeulebroecke, C. Munn, D. Bourne, and J. Swings. 2005. Photobacterium rosenbergii sp. nov. and Enterovibrio coralii sp. nov., vibrios associated with coral bleaching. Int. J. Syst. Evol. Microbiol. 55:913-917. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. R., M. A. Randa, L. A. Marcelino, A. Tomita-Mitchell, E. Lim, and M. F. Polz. 2004. Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70:4103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. L. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 25:1311-1313. [DOI] [PubMed] [Google Scholar]
- 44.Verschuere, L., G. Rombaut, P. Sorgeloos, and W. Verstraete. 2000. Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 64:655-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wachsmuth, I. K., P. A. Blake, and O. Olsvik. 1994. Vibrio cholerae and cholera. Molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 46.Zeigler, D. R. 2003. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 53:1893-1900. [DOI] [PubMed] [Google Scholar]