Abstract
Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) is an example of highly regulated alternative splicing in which exons IIIb and IIIc are utilized in a mutually exclusive manner in different cell types. The importance of this splicing choice is highlighted by studies which indicate that deregulation of the FGF-R2 splicing is associated with progression of prostate cancer. Loss of expression of a IIIb exon-containing isoform of FGF-R2 [FGF-R2 (IIIb)] accompanies the transition of a well-differentiated, androgen-dependent rat prostate cancer cell line, DT3, to the more aggressive, androgen-independent AT3 cell line. We have used transfection of rat FGF-R2 minigenes into DT3 and AT3 cancer cell lines to study the mechanisms that control alternative splicing of rat FGF-R2. Our results support a model in which an important cis-acting element located in the intron between these alternative exons mediates activation of splicing using the upstream IIIb exon and repression of the downstream IIIc exon in DT3 cells. This element consists of 57 nucleotides (nt) beginning 917 nt downstream of the IIIb exon. Analysis of mutants further demonstrates that an 18-nt “core sequence” within this element is most crucial for its function. Based on our observations, we have termed this sequence element ISAR (for intronic splicing activator and repressor), and we suggest that factors which bind this sequence are required for maintenance of expression of the FGF-R2 (IIIb) isoform.
Alternative splicing, the process whereby a single pre-mRNA is spliced differentially, represents a means by which gene expression can be modulated posttranscriptionally. An example of alternative splicing in which tight regulation results in a very defined cell type discrepancy in the differential expression of isoforms occurs during the splicing of the second half of the third immunoglobulin-like domain of the fibroblast growth factor receptor 2 (FGF-R2). Mutually exclusive splicing results in a mRNA containing either the 148-nucleotide IIIb exon or the 145-nucleotide IIIc exon, a choice which is specific for a given cell type (Fig. 1A). These mRNAs encode receptors with physiologically relevant differences. FGF-R2 (IIIb) is the predominant isoform expressed in epithelial cells and displays high affinity for FGF-7 (or keratinocyte growth factor [KGF]) and low affinity for FGF-2 (or basic FGF), whereas FGF-R2 (IIIc) is expressed in certain cells of mesenchymal origin and exhibits high affinity for FGF-2 but not for FGF-7 (7, 32–34, 46, 47, 53, 66). Thus, the specificity of this regulated splicing event is crucial for the maintenance of proper pathways of cellular communication and control of cell proliferation. Loss of appropriate regulation of the splicing of FGF-R2 has been proposed to be one step at which dysregulated growth pathways may lead to progression of prostate cancer (10, 63). A well-differentiated, androgen-dependent rat prostate cancer cell line, DT3 (or DT-E), expresses the IIIb isoform of FGF-R2 exclusively, as do normal rat prostatic epithelial cells. On the other hand, a poorly differentiated, androgen-independent AT3 rat prostate cancer cell line expresses only the isoform containing IIIc. FGF-7, secreted by prostatic stromal cells in response to androgens, has been proposed to mediate controlled proliferation and differentiation of epithelial cells through its interaction with FGF-R2 (IIIb) (63, 64). Thus, it has been proposed that the loss of appropriate regulation of FGF-R2 splicing may result in the loss of a normal pathway of growth control in this model of cancer progression (63).
FIG. 1.
Structural organization of the FGF-R2 gene and demonstration of IIIb and IIIc mutually exclusive splicing. (A) Organization of the FGF-R2 protein domains (top) and genomic gene arrangement of the region in which alternative splicing yields transcripts containing either the IIIb or IIIc exon and encoding the second half of the third immunoglobulin (Ig)-like domain. TM, transmembrane domain, TK, tyrosine kinase domains. The solid box represents a highly acidic domain, and the thick line indicates the IIIb- or IIIc-encoded portion of the protein. Shaded boxes represent exons, and solid lines represent introns, with intron sizes indicated. U and D indicate the exons upstream and downstream of these alternative exons, respectively. (B) Scale representation of the exons (solid boxes) and introns (solid lines) with regions of high (at least 90%) rat-human intron sequence similarity (shaded boxes). Also shown are regions FS and FL and their sizes. nt, nucleotide.
It is implicit in models of alternative splicing that the pattern of splicing is affected by cell-specific factors via interactions with the constitutive splicing apparatus. Consensus sequences located at the 5′ splice site, the 3′ splice site and associated polypyrimidine tract, and the branch point are known to determine splicing efficiency. The degree to which these signals match the consensus sequences is in fact a determinant of the ability of the constitutive splicing apparatus to recognize the sequences and carry out the splicing reaction. The process of splicing is performed by the spliceosome, which is comprised of the small nuclear ribonucleoproteins U1, U2, U4/U6, and U5, and associated splicing factors (reviewed in reference 50). A mechanistic understanding of the means by which alternative splicing can be regulated has been acquired largely through study of the pathway of sex determination in Drosophila melanogaster (reviewed in references 1, 31, 42, 43, and 45). Here, examples of both activation and repression of specific splicing pathways have been demonstrated to be affected by sex-specific factors, which interact with components of the constitutive splicing apparatus. Although these studies have provided elegant models for regulated alternative splicing, there has not yet been an example of alternative splicing in mammalian cells for which the mechanism has been conclusively demonstrated or for which cell type-specific splicing factors have been identified. Several studies have shown that the pattern of splicing of a pre-mRNA can be modulated in vitro and in vivo by changing the levels of certain constitutive splicing factors known as SR proteins, as well as that of heterogeneous nuclear ribonucleoprotein particle A1 (hnRNP A1) (8, 18, 20, 42, 44). However, it is not clear that differences in the levels of these factors will suffice to explain the high degree of specificity observed in numerous examples of mammalian alternative splicing. In addition to the strength of splice sites, exon size, intron size, and the presence of specific exonic sequences have been shown to have positive or negative effects upon splicing (5, 13–15, 17, 23, 30, 37, 55, 62). Recently, intronic sequences neighboring alternative exons have also been described which appear to be targets of factors involved in regulating alternative splicing (2, 3, 9, 11, 14, 16, 22, 24, 26, 28, 29, 38, 40, 41, 48, 49, 52, 54, 58, 59, 67). Although several proteins have been shown to play roles in interactions with these sequences, the precise means through which these proteins mediate alternative splicing are still unclear. A common observation in alternative splicing is that the consensus splicing signals associated with one of the splicing choices are “weak” matches to the consensus, and thus a “default” splicing pathway, which may only require the cooperation of members of the constitutive splicing pathways, is regulated by a specific factor(s) that blocks the use of such a default pathway and/or activates the weaker pathway. Examples have been well described for Drosophila, and it would stand to reason that the existence of tissue- or developmental-stage-specific factors which regulate alternative splicing will eventually be demonstrated in mammalian systems.
The study of alternative splicing of human FGF-R2 has demonstrated several exonic and intronic sequences which play a role in the regulation of FGF-R2 splicing (14–16, 21). However, given the high degree of fidelity with which regulation is maintained during splicing of the IIIb and IIIc exons, the mechanism which would most easily explain the mutually exclusive use of these exons and the roles of required cis elements has not been completely delineated. Using FGF-R2 minigenes in the DT3 and AT3 rat prostate cancer cell lines, we have investigated sequences and mechanisms which regulate FGF-R2 splicing. Based on these studies, we have identified an intronic sequence between the IIIb and IIIc exons which is necessary for appropriate regulation of FGF-R2 splicing. This sequence has a high degree of homology with one of the human sequences which has been shown by other investigators to be involved in activation of splicing of the upstream IIIb exon in a human cell line which uses the IIIb exon (16). However, while we observed a similar, and in fact more dramatic, effect on activation of IIIb usage in our rat minigenes and cells, we have also demonstrated that this sequence mediates repression of use of the downstream IIIc exon in the same cell line in which activation occurs. Therefore, we propose that this sequence is required in order to achieve regulation through coordinated activation of the weaker IIIb exon and repression of the stronger IIIc exon in cells (DT3) which select IIIb over IIIc.
MATERIALS AND METHODS
Plasmid construction.
The pPIP11 adenoviral splicing construct is based on the L1 and L2 exons and is similar to the previously described pPIP7A (36), except that the sequence of the 3′ exon between the PstI and HindIII sites has been replaced by the sequence 5′-CTGCAGGACAAACTCTTCGCGGTCTCTATGCATCCTCCGAACGGTAAGACCCTAAGCTT-3′. The sequences of pPIP11 were PCR amplified with primers PIP10-F (5′-CCCGGGGGTACCGGGCGAATTCGAATTCGAGCTCACTC-3′) and PIP11-R (5′-CCCGGGACTAGTAAGCTTAGGCTCTTGGCGTT-3′). The PCR product was digested with EcoRI and SpeI and inserted into the EcoRI and XbaI sites of the eukaryotic expression vector PCDNA3 (Invitrogen). All cloning was done by standard methodologies. PCR amplification of genomic DNA from AT3 cells with the FGF-R2-specific primer pairs Int 3BF2 (5′-CCGGACTAGTCACTACCGTTCTCCACCACT-3′) and Int 3CR (5′-CCGGCTCGAGGGTCGGAAATCATTCGAAAC-3′) and Intron 1F (5′-CCGGACTAGTAAGCCCAAGGGGCCAGCAGT-3′) and Intron 3R (5′-CCGGCTCGAGACGAAGAGCCAAGGGCGCCT-3′) yielded fragments FL and FS, respectively (Fig. 1B). These products were digested with SpeI and XhoI and inserted into the XbaI and XhoI sites of the pI-11 intron to yield pI-11-FL and pI-11-FS. Constructs derived from intron deletions in pI-11-FS (see Fig. 4A) (for example, pI-11-FS-ΔBcl I/Nde I) were obtained by first cloning the FS sequences into the SpeI and XhoI sites of pBluescript (Stratagene) to generate pBlue-FS, since these enzymes cut within PCDNA3 but not pBluescript. Deletions were performed by sequential digestion of pBlue-FS with the indicated restriction endonucleases. The digested ends were blunted with Pfu polymerase (Stratagene), and the resulting plasmids were gel purified and religated with T4 DNA ligase. These plasmids were digested with SpeI and XhoI, and the minigenes were cloned back into the XbaI and XhoI sites of pI-11. Plasmids pI-11-IIIb-plus and pI-11-IIIb-minus were obtained with primers Int 3BF2 and Intron 2R2 (5′-CCGGCTCGAGGGCTAGACATAGGAATGATT-3′) and PCR amplification of pI-11-FS-ΔBcl I/Nde I and pI-11-FS-ΔBcl I/Nsi I, respectively. After SpeI and XhoI digestion, the PCR products were cloned into the XbaI and XhoI sites of pI-11. Plasmids pI-11-IIIc-plus and pI-11-IIIc-minus were similarly obtained by PCR amplification with primers Intron 2F (5′-CCGGACTAGTCAACGTTTTTGTGTTTGTGT-3′) and Int 3CR to amplify pI-11-FS-ΔBcl I/Nde I and pI-11-FS-ΔBcl I/Nsi I, respectively, followed by digestion with SpeI and XhoI and insertion into the XbaI and XhoI sites of pI-11. pI-11-FS-Not/Cla-ISAR resulted from PCR of pI-11-FS with primers Nde/Not-F (5′-CCGGCATATGGCGGCCGCCAAACAAATTCAAAGAGAAC-3′) and Nsi/Cla-R (5′-CCGGATGCATATCGATGCGATTGAACACATGGAAAA-3′), digestion of these products with NdeI and NsiI, and cloning into the NdeI and NsiI sites in pBlue-FS. The minigene sequence was removed with SpeI and XhoI and cloned into the XbaI and XhoI sites of pI-11 to generate pI-11-FS-Not/Cla-ISAR. The ISAR mutant constructs pI-11-Not/Cla: Blue1, SAR 5′, SAR 3′, SAR-20, Mut1, Mut2, Mut3, Blue2, Rep1, and Rep3 were obtained by deleting ISAR from pI-11-FS-Not/Cla-ISAR with NotI and ClaI and then inserting annealed oligonucleotides with complementary NotI and ClaI sites, as represented by the following oligonucleotide pairs: Blue1-F, 5′-GGAA GCGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCA TCTGGCCCCAGTGAT-3′; Blue1-R, 5′-CGATCACTGGGGCCAGATGGT AAGCCCTCCCGTATCGTAGTTATCTACACGACGGGGAGTCGC-3′; SAR 5′-F, 5′-GGCCGCAAACAAATTCAAAGAGAACGGACTCTGTAT-3′; SAR 5′-R, 5′-CGATACAGAGTCCGTTCTCTTTGAATTTGTTTGGC-3′; SAR 3′-F, 5′-GGCCGCGGGCTGATTTTTCCATGTGTTCAATCGCAT-3′; SAR 3′-R, 5′-CGATGCGATTGAACACATGGAAAAATCAGCCCGC-3′; SAR-20-F, 5′-GGCCGCCAAAGAGAACGGACTCTGTGGGCTGATTTTTC CATGTAT-3′; SAR-20-R, 5′-CGATACATGGAAAAATCAGCCCACAGAGTCCGTTCTCTTTGGC-3′; Mut1-F, 5′-GGCCGCCAAACTCTACGGACTCTGTGGGCTGATTTTTCCATGTAT-3′; Mut1-R, 5′-CGATACATGGAAAAATCAGCCCACAGAGTCCGTAGAGTTTGGC-3′; Mut2-F, 5′-GGCCGCCAAAGAGAACGGACTCTGTGGGCTGAAAGATCCATGTAT-3′; Mut2-R, 5′- CGATACATGGATCTTTCAGCCCACAGAGTCCGTTCTCTTTGGC-3′; Mut3-F, 5′-GGCCGCCAAAGAGAACGGACTCTGTGGGCTGATTTTTCACGCTAT-3′; Mut3-R, 5′-CGATAGCGTGAAAAATCAGCCCACAGAGTCCGTTCTCTTTGGC-3′; Blue2-F, 5′-GGCCGCAAGTGGTGGCCTAACTACGGCTACACTAGAAGGACACAT-3′; Blue2-R, 5′-CGATGTGTCCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTGC-3′; Rep1-F, 5′-GGCCGCGGGCTGATTTTTCCATGTAT-3′; Rep1-R, 5′-CGATACATGGAAAAATCAGCCCGC-3′; Rep3-F, 5′-GGCCGCGGGCTGATTTTTCCATGTGGGCTGATTTTTCCATGTGGGCTGATTTTTCCATGTAT-3′; and Rep3-R, 5′-CGATACATG GAAAAATCAGCCCACATGGAAAAATCAGCCCACATGGAAAAATCA GCCCGC-3′. All plasmid minigenes were prepared with plasmid maxi kits from Qiagen. The identities of all minigenes were confirmed by automated DNA sequencing with an ABI sequencer.
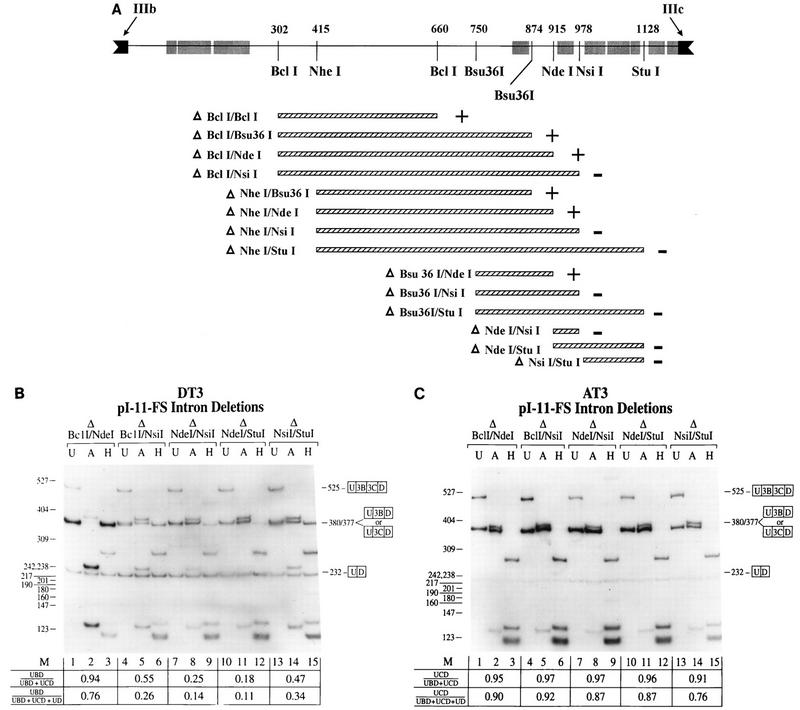
FIG. 4.
Deletions which result in loss of sequences between the NdeI and NsiI sites in intron 2 result in loss of regulation in DT3 cells. (A) The IIIb and IIIc exons (solid boxes) and the intron (intron 2) between them (solid line) are shown. Also indicated are the restriction enzymes used to generate these deletions and the regions of high rat-human sequence homology (shaded boxes). The locations of these restriction sites are represented as the position (in nucleotides) from the start of the intron and are measured to the center position of each recognition sequence. The minigenes tested consisted of deletions (hatched boxes) from the parent construct, pI-11-FS, and the results of these deletions in DT3 cells are summarized. Delta, construct in which the sequence between the indicated restriction enzymes was deleted from pI-11-FS; plus, deletion constructs which still demonstrated >80% IIIb inclusion in DT3 cells; minus, deletion constructs with ≤55% IIIb inclusion in DT3 cells. (B) Results of the most representative intron 2 deletions in DT3 cells. Deletion of over half of the intron from BclI to NdeI did not affect regulation, whereas deletions spanning NdeI to NsiI caused loss of regulation. A deletion of NsiI-to-StuI sequences also caused some loss of regulation, but less than a NdeI-to-NsiI deletion. (C) The same deletions had no effect upon splicing in AT3 cells. Efficient IIIc usage was seen in these deletions, as well as all deletions summarized in panel A. Abbreviations are defined in the legends to Fig. 2 and 3.
PCR amplification of DNA templates.
PCR from DNA templates was performed with 1 to 2 ng of plasmid DNA or 1 to 5 μg of genomic DNA and Taq DNA polymerase (Boehringer Mannheim) according to the supplier’s recommendations. Amplifications were performed with a Perkin-Elmer 2400 thermal cycler. A typical cycle consisted of initial denaturation at 94°C for 4 min, followed by 30 to 40 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 1 min, and extension at 72°C for 1 to 2 min. After completion of the final cycle, a final extension was done at 72°C for an additional 7 min. For amplification of templates longer than 2 kb, we used TaqPlus (Stratagene) according to the manufacturer’s recommendations and cycles similar to those described above except that extension times were generally 5 to 8 min.
Cell culture and transfections.
AT3 and DT3 cells were maintained in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (Hyclone). All transfections were performed in 35-mm-diameter wells with 5 μl of Lipofectamine (Gibco) according to the supplier’s recommendations. Each well was seeded with 3 × 105 cells 16 to 24 h prior to transfection. Pilot experiments with chloramphenicol acetyltransferase reporter plasmids demonstrated that DT3 and AT3 cells were transfected with equivalent efficiency. Transient transfections were done for 2 h with 50 ng to 2 μg of minigene DNA, and RNA was harvested 24 h later. Stable transfections were performed with 2 μg of minigene DNA, and after 24 to 48 h the cells were trypsinized and reseeded in 75-cm2 flasks containing Geneticin (Gibco) at an active concentration of 400 μg/ml. Selection was performed until isolated colonies were obtained and no cells remained from a control transfection with a plasmid lacking neomycin resistance genes (usually 12 to 16 days). Pooled colonies were then harvested for RNA preparation.
RNA purification and RT-PCR analysis.
Total cellular RNA was isolated from transfected cells by the method of Chomczynski and Sacchi (12). Two micrograms of total RNA was heated to 100°C, chilled on ice, and reverse transcribed in a reaction volume of 20 μl containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 1 mM deoxynucleoside triphosphates, 100 ng of random hexamers, 2 U of RNasin (Promega), and 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco) at 37°C for 1 h. Samples were then heated to 90°C for 5 min and then chilled on ice. Two microliters of each reverse transcription (RT) reaction mixture was amplified in a 100-μl PCR reaction mixture containing 50 mM KCl; 10 mM Tris-HCl (pH 8.8); 1.5 mM MgCl2; 0.1% gelatin; 200 μM (each) dATP, dGTP, and TTP; 50 μM dCTP; 100 nM each primer; 10 μCi of [α-32P]dCTP; and 2.5 U of Taq DNA polymerase (Boehringer Mannheim). The primers used were FGF-FB (5′-CCCGGGTCTAGATTTATAGTGATGCCCAGCCC-3′) and FGF-RB (5′-CCCGGGGAATTCACCACCATGCAGGCGATTAA-3′) for analysis of the endogenous gene and standard T7 and SP6 promoter primers for analysis of transfected minigenes. Amplification conditions consisted of an initial denaturation at 94°C for 4 min followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, and extension at 72°C for 1 min and a final extension at 72°C for 7 min. In all amplification reactions, a water control and a mock RT control were included, which resulted in no PCR product in all experiments. PCR products were added directly to restriction endonuclease digestions with either AvaI or HincII (New England Biolabs). We always observed complete digestion when using this protocol. Aliquots representing equal amounts of each PCR reaction mixture with undigested and digested PCR products were loaded directly on nondenaturing 5% polyacrylamide gels at 100 V for 3 to 4 h, followed by drying and exposure to Amersham Hyperfilm-MP. Analysis was performed with a Molecular Dynamics PhosphorImager. Because equal amounts of AvaI- and HincII-digested PCR products were loaded onto each gel, quantification of cDNAs containing exon IIIb or IIIc (UBD or UCD, respectively, where U and D are the 5′ and 3′ exons of pI-11) was obtained by using the quantification of the band at 380 or 377 bp which remained following HincII or AvaI digestion, respectively, as the numerator. The denominator consisted of the sum of the bands remaining at 380 or 377 bp from both digests (UBD + UCD). When these results were also expressed with the contribution of products with IIIb and IIIc skipped, the average value of the 232-bp band was also used in the sum of the denominator (UBD + UCD + UD), corrected for molar equivalents. Quantification of experiments with minigenes with only one (IIIb or IIIc) internal exon was determined as the quantification of the 380- or 377-bp band divided by the sum of the same band and the 232-bp band, corrected for molar equivalents.
RESULTS
Nucleotide sequences of introns flanking alternative exons IIIb and IIIc are highly conserved between rat and human FGF-R2 genes.
Because phylogenetic sequence comparisons often help to identify sequences with important functions, we chose to sequence the genomic DNA from the regions of both rat and human FGF-R2 genes containing the alternative IIIb and IIIc exons, the constitutive exons located upstream and downstream of them, and the three introns between the exons. As seen in Fig. 1 the intron sizes flanking the alternative exons are roughly 1.1, 1.2, and 1.9 nucleotides in both rat and human genomic sequences. Henceforth, these exons will be referred to as intron 1, intron 2, and intron 3, respectively. We used the University of Wisconsin Sequence Analysis Package GAP program to align the rat and human sequences for direct sequence comparison. As expected, the exon sequences were highly similar and corresponded to previously reported cDNA sequences for rat and human FGF-R2 (25, 46, 65). Interestingly, the introns contained a number of regions with very high levels of sequence similarity, and these regions were clustered around the IIIb and IIIc exons, whereas the intron sequences adjacent to the constitutive exons did not show appreciable levels of similarity. We screened the rat sequence and highlighted all intronic regions in which 90% of the nucleotides were identical to the corresponding human sequence for a stretch of at least 20 consecutive nucleotides. These data are presented graphically in Fig. 1B. While we suspected that some of these regions represented evolutionary vestiges, we also expected that regulatory sequences involved in mediating alternative splicing of these exons, which have been conserved between rat and human genes, would also be likely to be represented by such conserved sequences. Thus, this information was used to direct the construction of a variety of minigenes, which will be described below.
Minigenes pI-11-FL and pI-11-FS recapitulate the splicing pattern of the endogenous gene in AT3 and DT3 cells.
We used an RT-PCR-based assay similar to one used by other researchers investigating the splicing of human FGF-R2 to assay for the splicing pattern of exons IIIb and IIIc (14, 21). For analysis of the endogenous FGF-R2 transcript, we performed RT-PCR with primers (FGF-FB and FGF-RB) specific for the constitutive exons located upstream of IIIb and downstream of IIIc, as shown in Fig. 2A. These products were separately digested with AvaI and HincII and analyzed by gel electrophoresis, as shown in Fig. 2B. Because exon IIIb contains an AvaI site not present in IIIc, and exon IIIc contains two HincII sites not present in IIIb, we expected the inclusion of IIIb to result in a 367-bp product which is cut with AvaI but not HincII, and we expected the inclusion of IIIc to result in a 364-bp product which is cut only by HincII. As expected, DT3 cells produce an FGF-R2 transcript which contains only exon IIIb and AT3 cells consist entirely of transcripts containing IIIc, results which are consistent with the original report describing alternative splicing of FGF-R2 in these cells (63). We also tested the validity of this assay by performing the same assay with titrations in which we mixed RNAs that contained only exon IIIb with RNAs containing only exon IIIc, and we observed that the proportion of each isoform seen by RT-PCR directly correlated with the fraction of the same isoform in the mixture (data not shown). Thus, mRNAs containing exon IIIb or IIIc were amplified with equivalent efficiencies in this assay. Furthermore, sequencing of these RT-PCR products verified that they were correctly identified by this approach (data not shown).
FIG. 2.
Splicing of the endogenous gene transcript in DT3 and AT3 cells. (A) Map illustrating PCR products containing exon IIIb or IIIc amplified with primers FGF-FB and FGF-RB and sizes (in nucleotides) of fragments which result from AvaI or HincII digestion. U, upstream exon; D, downstream exon. (B) Gel showing the RT-PCR products following digestion with AvaI and HincII. DT3 cells express only products containing IIIb, and AT3 cells express products containing IIIc. U, uncut products; A, AvaI-digested products; H, HincII-digested products; M, pBR322/Msp I DNA size markers.
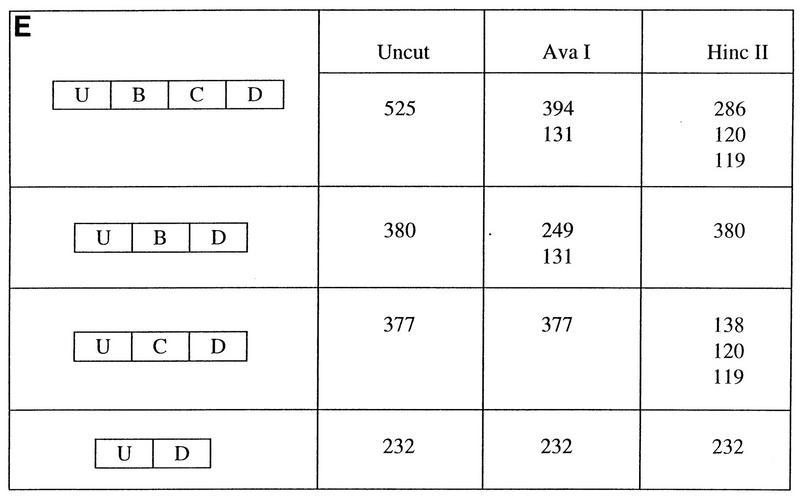
In order to further characterize the cis elements required for maintenance of the cell type-specific splicing patterns in DT3 and AT3 cells, we used PCR to amplify genomic regions FL and FS from the rat FGF-R2 gene (Fig. 1B and 3A). These sequences were cloned into the intron of splicing construct pI-11, which is a two-exon, one-intron adenovirus-derived splicing construct adapted for eukaryotic expression. As seen in Fig. 1B and 3A, the resulting minigenes, pI-11-FL and pI-11-FS, consisted of 4,378 and 1,804 nucleotides of genomic FGF-R2 sequence, which represented exons IIIb and IIIc, the entire intron 2 between IIIb and IIIc, and variable amounts of introns 1 and 3. In the case of pI-11-FL, nearly all of introns 1 and 3 are included except intron sequences of the normal 5′ splice site of the upstream exon-intron boundary and the polypyrimidine tract and the 3′ splice site in the intron downstream of exon IIIc. In constructing pI-11-FS we chose to limit the sequences from introns 1 and 3 to those regions closest to exons IIIb and IIIc which contained greater-than-90% intron homology, as described previously. It should be noted that in constructing these and all subsequent rat FGF-R2 minigenes we substituted the adenoviral pPIP11 exons for the constitutive exons normally used upstream and downstream of exons IIIb and IIIc. We performed stable and transient transfections of these minigenes in DT3 and AT3 cells with Lipofectamine and harvested RNA 24 h later for transient transfections or after colony formation 12 to 16 days later for stable transfection analysis. We then performed labeled RT-PCR with primers (the T7 and SP6 promoter primers) specific for PCDNA3. As we will detail later, results from transient transfections were highly dependent on the amount of minigene DNA transfected, and thus we predominantly used stable transfections due to their easier reproducibility. Initial experiments with pI-11 in both DT3 and AT3 cells showed that the adenoviral strong consensus splice sites directed highly efficient splicing in both cell lines, with the pre-mRNA transcript being spliced essentially to completion (Fig. 3B). In order to analyze the splicing products of pI-11-FL and pI-11-FS, we digested the RT-PCR products with AvaI and HincII and performed polyacrylamide gel electrophoresis. When stable transfections with pI-11-FL and pI-11-FS were analyzed as shown in Fig. 3C and D, the primary product was 380 or 377 bp, and this product contained almost exclusively exon IIIb in DT3 cells and IIIc in AT3 cells. In addition, we observed a minor 525-bp band, which corresponded to a product which contained both IIIb and IIIc, as well as a 232-bp product, which results when the pI-11 exons are directly spliced together. The origins of the products obtained when these products are digested with AvaI and HincII are shown in Fig. 3E. Of note, digestion of the 525-bp product, which contains both exons IIIb and IIIc, results in a band at 394 bp, just above the location of the predominant product of 380 bp, which is nearly completely digested by AvaI (Fig. 3C and D, lanes 2). Thus, this product is not to be confused with undigested 380-bp products. Although the splicing product containing both exons was seen reproducibly in all experiments, it was a minor product on a molar basis, and thus, for simplicity we did not include it in our further analysis. We did note in these and subsequent experiments that the amount of product seen in which both IIIb and IIIc were skipped and the pI-11 exons were directly spliced was slightly higher in DT3 cells than in AT3 cells, which may in part reflect the fact that the IIIb exon contains a weaker polypyrimidine tract than that associated with exon IIIc. In order to evaluate the efficiency of usage of IIIb and IIIc in our experiments, we used PhosphorImager analysis to quantify the levels of products containing IIIb (in DT3 cells) or IIIc (in AT3 cells). Because loss of a pathway selecting either IIIb or IIIc (but not both) has two consequences (i.e., a switch to use the other exon or to skip both and directly splice the pI-11 exons), we quantified our results both including and excluding the contribution of this skipping pathway. These experiments with stable transfections demonstrated that we could reproduce the endogenous gene’s pattern of regulated splicing of the IIIb and IIIc exons with a high degree of precision. We were also able to demonstrate that the sequences in the exons normally located upstream of exon IIIb and downstream of IIIc, as well as most of the sequences of introns 1 and 3, are dispensable for maintenance of regulated splicing of these exons of FGF-R2.
FIG. 3.
Rat FGF-R2 minigenes transfected into DT3 and AT3 cells reproduce the splicing pattern of the endogenous gene. (A) Representation of the two-exon, one-intron splicing construct pI-11 and insertion of FGF-R2 genomic sequences FL and FS (which were generated with the primer sets indicated at bottom) to create minigenes pI-11-FL and pI-11-FS, respectively. CMV indicates the efficient immediate early CMV promoter, and pA indicates the bovine growth hormone polyadenylation sequence. The XbaI and XhoI sites used for cloning and the T7 and SP6 vector-specific primers are also indicated. U, the 5′ exon of pI-11; D, the 3′ exon of pI-11. (B) pI-11 pre-mRNA is spliced almost completely and with equal efficiency in DT3 and AT3 cells, indicating no differences in the abilities of these cells to splice the exons. RT-PCR products for this and subsequent minigenes were obtained with the T7 and SP6 promoter primers. (C and D) Minigenes pI-11-FL and pI-11-FS reproduce the endogenous gene splicing pattern. The major PCR product containing either IIIb (3B or B) or IIIc (3C or C) is 380 or 377 bp, respectively. Products containing exons U, IIIb, IIIc, and D are indicated to the right. The sizes of products of AvaI and HincII digestion are also indicated. Quantification was performed to yield values for the fraction of the expected IIIb (in DT3) or IIIc (in AT3) exon as a fraction of products containing IIIb and IIIc and also as a fraction of products skipping IIIb and IIIc (see Results and Materials and Methods). (E) Representation of the origins (in nucleotides) of the products obtained when UBD, UCD, and UBCD products are cut with AvaI and HincII. Sizes are indicated in base pairs. Lanes are labeled as in Fig. 2.
Deletion of nucleotide sequences between the NdeI and NsiI sites located in intron 2 causes loss of splicing regulation.
To further characterize intron sequences required for splicing regulation, we constructed a series of deletions in intron 2 of pI-11-FS. The deletions tested are shown in Fig. 4A along with a map in which the regions of high rat-human homology are superimposed on the locations of the restriction enzymes used to create these deletions. The resulting minigenes were stably transfected into DT3 and AT3 cells. The results of the entire series of deletions are summarized in Fig. 4A, and selected gels representing the most informative deletions are shown in Fig. 4B and C. It should first be noted that a deletion from the BclI site at position 302 to the NdeI site at 915 which removed more than half of intron 2 did not appreciably affect appropriate splicing regulation in either DT3 or AT3 cells; DT3 cells used IIIb and AT3 cells used IIIc (Fig. 4B and C). However, any deletion which included the sequences between the NdeI and NsiI sites at positions 915 and 978 resulted in a dramatic loss of exon IIIb usage by DT3 cells. This loss of regulation in DT3 cells was reflected by increased usage of the IIIc exon as well as by increased skipping of both exons, as seen in Fig. 4B. In addition, we noted that a deletion from NsiI to StuI also caused a decrease in IIIb usage, but not nearly as much as a much smaller deletion from the NdeI to NsiI sites. As demonstrated in Fig. 4C, when any of these minigenes with deletions were stably transfected into AT3 cells, appropriate use of the IIIc exon was not affected nor was any increased level of skipping observed. Because nearly identical results were obtained in AT3 cells with all manipulations of intron 2 sequences, we will subsequently mainly show results obtained in DT3 cells.
Sequences between the NdeI and NsiI sites mediate regulation in DT3 cells by activating use of the upstream IIIb exon as well as by repressing use of the downstream IIIc exon.
The results obtained with deletions in intron 2 suggested that the requirements for exon IIIc inclusion in AT3 cells are less stringent than those for IIIb inclusion in DT3 cells. In fact, with our sets of minigenes we did not observe any intronic sequences outside of the conserved splice junctions or polypyrimidine tract which impeded splicing of IIIc in AT3 cells. As we have discussed, this is not surprising given the stronger polypyrimidine tract associated with the IIIc exon compared to that of IIIb. Therefore, while we cannot completely rule out the possibility that there are other untested intron sequences or exon IIIc sequences which interact with AT3 cell-specific factors to mediate IIIc inclusion in these cells, we hypothesized that IIIc exon inclusion is a default splicing pathway which may only require the cooperation of factors involved in the constitutive splicing process. Thus, regulation may be achieved by proteins in DT3 cells which are able to switch the splicing pattern from exon IIIb to IIIc. Consistent with this view are the observations that several of our deletions caused not only skipping of both exons but also a switch towards some IIIc inclusion. Therefore, if FGF-R2 mutually exclusive alternative splicing is predominantly regulated only in DT3 cells, the sequences which are involved in this regulation could be acting by activating IIIb splicing or repressing IIIc splicing or by performing both of these functions. To investigate these alternatives, we constructed a series of minigenes in which we inserted either IIIb or IIIc (but not both) into pI-11 and we used our previous deletions in such a manner that IIIb was inserted either with (pI-11-IIIb-plus) or without (pI-11-IIIb-minus) the NdeI-to-NsiI sequences located downstream and IIIc was inserted with (pI-11-IIIc-plus) or without (pI-11-IIIc-minus) these same sequences upstream (Fig. 5A). Because these minigenes only offered a choice of including an internal exon or skipping, we quantified use of the internal IIIb or IIIc exon versus that of the skipped product. As shown in Fig. 5B, when these minigenes were transfected into AT3 cells, the IIIc exon was included highly efficiently, and this inclusion was not affected by the presence of the NdeI-to-NsiI sequences located upstream. In addition, AT3 cells did not include exon IIIb efficiently and this effect was essentially unchanged whether or not these sequences were located downstream. In DT3 cells, on the other hand, IIIb inclusion was seen to occur with fairly high efficiency, but this inclusion was largely dependent on the presence of the NdeI-to-NsiI sequence located downstream; when this sequence was deleted, IIIb inclusion was dramatically reduced from 68 to only 13%. In addition, we noted that when the NdeI-to-NsiI sequences were present upstream of IIIc, DT3 cells rarely included IIIc, but when these sequences were deleted, the proportion of IIIc included approximately tripled, from 11 to 35%. These data were consistent with a model in which regulation of FGF-R2 alternative splicing in DT3 cells is achieved by the interaction of a cell-specific factor or complex of factors with intronic sequences in intron 2 and the coordinated activation of exon IIIb splicing and repression of the stronger IIIc exon. The fact that the sequences between NdeI and NsiI were necessary for both of these effects to occur prompted us to call this sequence ISAR (for intronic splicing activator and repressor), and it suggests that this element is required for the formation of a regulatory complex which acts in DT3 cells to force the use of IIIb instead of IIIc.
FIG. 5.
Sequences contained between the NdeI and NsiI sites of intron 2 normally function to activate upstream IIIb splicing and repress downstream IIIc splicing. (A) Method used to generate minigene constructs containing either the IIIb or IIIc exon with NdeI-to-NsiI sequences (crosshatched boxes) present or deleted. All constructs had sequences BclI to NdeI, which were previously shown to be dispensable for regulation, deleted. The primers used to generate these regions in relation to the sequences of pI-11-FS are shown. The hatched box represents polylinker sequences present in PCDNA 3. (B) Transfection of the minigenes into DT3 and AT3 cells reveals that AT3 cells use exon IIIc highly efficiently and do not use exon IIIb efficiently regardless of the presence of NdeI-to-NsiI sequences. DT3 cells use exon IIIb efficiently only when these NdeI-to-NsiI sequences are present downstream. DT3 cells do not use exon IIIc efficiently, but when these sequences are deleted, IIIc usage triples. Quantifications were performed as described in Materials and Methods. Abbreviations are defined in the legend to Fig. 3.
A core sequence of 18 nucleotides within the ISAR sequence mediates splicing regulation in DT3 cells.
In order to further characterize the sequences in this NdeI-to-NsiI ISAR element which are required for regulation, we prepared a series of deletion mutants to see which regions appeared most crucial. In order to facilitate easier manipulation of the ISAR sequence, we created a new minigene, pI-11-FS/Not/Cla-ISAR, in which we used PCR to engineer a NotI site directly 3′ of the NdeI site and also placed a ClaI site directly 5′ of the NsiI site in intron 2. Thus, this minigene was identical to pI-11-FS except that the NdeI and NsiI restriction sequences were separated from the 57 nucleotides normally located between them by the NotI and ClaI sites, respectively. We stably transfected pI-11-FS/Not/Cla-ISAR into DT3 and AT3 cells and observed essentially no differences between the splicing patterns with this construct and that of pI-11-FS; splicing regulation was preserved (Fig. 3D, lanes 1 to 3, and 6B, lanes 1 to 3). We then replaced the 57-nucleotide ISAR element with sequences consisting of parts of the ISAR sequence or containing mutations within the ISAR sequence (Fig. 6A). In addition we also replaced the ISAR element with random sequences chosen from pBluescript as controls, to verify that the effects of our deletions were a result of loss of this sequence and not due to changes in the size of the intron and thus possibly in the proximity of other cis-acting elements.
FIG. 6.
A critical 18-nucleotide sequence within the 57-nucleotide ISAR sequence between NdeI and NsiI nearly restores splicing regulation in DT3 cells. (A) The 57-nucleotide ISAR sequence is indicated at the top, and deletions and mutants of this sequence are shown below, as are control pBluescript sequences. The 18-nucleotide core sequence (Rep1) is boxed, and mutant sequences are underlined and in boldface. All sequences were tested by deleting ISAR sequences from pI-11-FS/Not/Cla-ISAR and inserting the indicated sequences. (B) SAR-20 and SAR 3′ sequences restore regulation, whereas SAR 5′ does not. (C) Mutations in the 18-nucleotide sequence shared by SAR-20 and SAR 3′ (Mut2 and Mut3) cause loss of regulation, whereas a mutation outside this region (Mut1) preserves regulation. (D) One or three copies of the 18-nucleotide core sequence restore splicing regulation, with three repeats of the sequence being slightly more efficient than one repeat. Abbreviations are defined in the legends to Fig. 2 and 3.
We first tested minigenes in which we replaced the full ISAR sequence with only the most 5′ 29 nucleotides (SAR 5′), the 3′ 28 nucleotides (SAR 3′), or the central 37 nucleotides (SAR-20) as outlined in Fig. 6A. We also introduced a 57-nucleotide sequence from pBluescript (Blue1) as a control for the size of ISAR. When these constructs were stably transfected into AT3 cells, we again observed highly efficient, nearly exclusive use of exon IIIc (data not shown). In DT3 cells we observed a dramatic reduction in IIIb inclusion with the control Blue1 sequences as well as with SAR 5′ (Fig. 6B). SAR 3′ and SAR-20, however, nearly restored splicing regulation, although IIIb inclusion with SAR 3′ was slightly less efficient than with SAR-20. Because SAR-20 restored splicing regulation almost to the levels seen with full-length ISAR, we also tested several mutations (Mut1, Mut2, and Mut3) within this sequence in which we mutated 4 consecutive nucleotides within SAR-20 by replacing purines with pyrimidines or vice versa. In addition, because both SAR-20 and SAR 3′ nearly restored regulation we tested the effect of the 18 nucleotides these sequences shared (Rep1 [Fig. 6A]) as well as three repeats of this sequence (Rep3). We also included a second negative control from a different part of pBluescript (Blue2), which was 37 nucleotides long in order to correspond to the size of SAR-20. We noted loss of regulation in DT3 cells when either of the Bluescript sequences was used, as well as when either Mut2 or Mut3, which were located within the 18 nucleotides which appeared to be most critical, were used (Fig. 6C). However, Mut1, Rep1, and Rep3 sequences were all capable of at least partially restoring splicing regulation (Fig. 6C and D). In fact, three repeats of this 18-nucleotide region appeared to be slightly more efficient than one repeat, although not better than full-length ISAR (Fig. 6D). Thus, this 18-nucleotide region contained most of the sequences required for regulation by the ISAR element, although given the slightly decreased level of restoration of regulation by SAR 3′ and Rep1 compared to that by ISAR or SAR-20, other sequences within ISAR may have minor contributing roles.
Regulated splicing in DT3 cells is dependent on a titratable factor or factors.
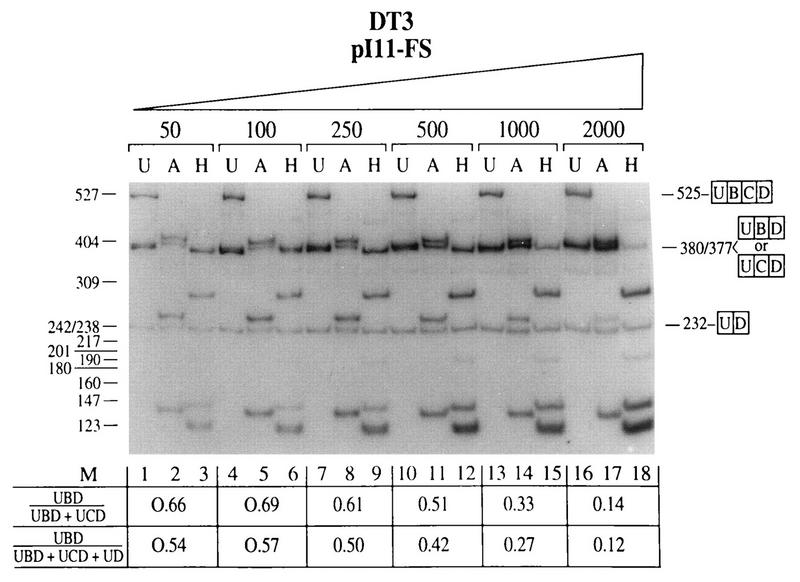
In our initial experiment using the FGF-R2 minigenes pI-11-FS and pI-11-FL in transient transfections with 2 μg of plasmid DNA, we noted that AT3 cells utilized the IIIc exon exclusively, as we observed in stable transfections; however, the DT3 cells also predominantly used the IIIc exon, with only a minor component of IIIb usage. Because transfection with Lipofectamine is highly efficient, we hypothesized that splicing regulation was not being seen in these experiments because the amount of RNA produced in the transient transfections may have been overwhelming the regulatory factors present in DT3 cells. Therefore, using pI-11-FS, we titrated the amount of plasmid DNA used in these transfections of DT3 cells as shown in Fig. 7. It can be seen that when we used amounts of DNA in the range of 50 to 100 ng we were able to achieve nearly (but not quite) the same degree of IIIb inclusion as that obtained with stable transfections and the endogenous gene. As the amount of DNA used in the transfection is increased, a stepwise reduction in IIIb inclusion is seen. In fact a similar effect was previously observed with human FGF-R2 minigenes, and in that case, curiously, the effect was abrogated through maintenance of an open reading frame in the minigene (21). Transient transfection of a minigene which does not contain ISAR (pI-11-FS Δ Nde I/Nsi I) resulted in nearly exclusive use of the IIIc exon even with only 50 ng of DNA (data not shown). Thus, ISAR is likewise required for maintenance of splicing regulation in transient transfections. However, this regulation is compromised when larger amounts of a minigene are transfected, most likely due to overtitration of a factor or factors required for IIIb inclusion.
FIG. 7.
DT3 cells contain a titratable factor or factors required for appropriate splicing regulation which can be overcome in transient transfections. Transient transfection of DT3 cells with increasing numbers of pI-11-FS minigenes resulted in stepwise loss of IIIb inclusion and increased IIIc inclusion, suggesting that a factor or factors required for regulation (i.e., IIIb inclusion and/or IIIc exclusion) is overwhelmed when large numbers of these minigenes are transfected. Abbreviations are defined in the legends to Fig. 2 and 3.
The core sequences of ISAR are highly homologous with a similar sequence in human FGF-R2 shown to mediate IIIb activation in human cells which express FGF-R2 (IIIb).
Recently Del Gatto and colleagues (16) described several sequences, located in the same region of the human gene between exons IIIb and IIIc as ISAR occupies in the rat gene, which were required for upstream IIIb activation, although the effects of these sequences on IIIc repression were not characterized. In addition, several other intronic sequences, as well as exon sequences in IIIb, have also been shown to be required for proper splicing regulation (14, 15). These investigators proposed that a sequence element similar to the 18-nucleotide core sequence in our rat ISAR could form an RNA secondary structure with another important intronic sequence, IAS2, approximately 800 nucleotides upstream from this sequence. It was further proposed that this secondary structure was a necessary element involved in exon IIIb activation. Interestingly, as shown in Fig. 8, the rat sequences corresponding to both of these elements are highly similar to the human sequences. While we did not directly test the validity of this model with rat FGF-R2 minigenes, it is interesting to note that in all of the rat minigenes tested here, including those with only one of the two exons, the corresponding upstream sequence was always included. Thus, possible synergistic effects of these separate sequences on FGF-R2 splicing regulation, whether or not they function via formation of a secondary structure, may be involved in the repression of IIIc as well as in IIIb activation.
FIG. 8.
Intron sequences important for regulation of rat and human FGF-R2 splicing are highly similar. (A) Rat intron sequences corresponding to a previously reported 21-nucleotide human sequence, IAS2 (see Results), which also mediates IIIb activation, contain only 1 nucleotide difference. (B) The 57-nucleotide rat ISAR sequence is highly similar to human sequences in this same region, including the 18 nucleotides shown to be most important for regulation (boxed sequences).
DISCUSSION
We have described the identification of an important cis-acting element in rat FGF-R2 which is required for proper splicing regulation of the mutually exclusive IIIb and IIIc exons. Because this 57-nucleotide element was shown to cause both activation of the upstream IIIb exon and repression of the downstream IIIc exon, effects exclusive to DT3 cells, we have termed this sequence ISAR. Within this sequence we have further identified 18 nucleotides which appear to be the most critical region for the regulation of splicing, and we have shown that this sequence is highly similar to a similarly situated sequence in the human FGF-R2 gene which has been shown to mediate IIIb activation. Based on our data, we suggest a model of regulation in this system, which our data support in several ways, and furthermore show that this ISAR sequence is a necessary, although perhaps not sufficient, element involved in this model. We suggest that in AT3 cells splicing of the IIIc exon occurs by a default pathway in which members of the constitutive splicing apparatus are capable of recognizing the splice sites associated with this exon and splicing occurs with high efficiency. DT3 cells, on the other hand, are a regulated cell line with respect to IIIb exon usage, requiring a factor(s) not present in AT3 cells or not present at a level sufficient to mediate IIIb inclusion and IIIc exclusion. We further propose that this DT3-specific regulation mediates both activation of splicing to the upstream, weak IIIb exon and repression of splicing to the downstream, strong IIIc exon (Fig. 9). Several lines of evidence support this model. First, the polypyrimidine tract associated with the IIIc exon is a significantly stronger match to the high-pyrimidine-content consensus sequence than that of the IIIb exon. Thus, in AT3 cells, the IIIb exon is recognized poorly by constitutive splicing factors in the absence of specialized factors. Second, although we cannot rule out regulation in AT3 cells by untested sequences, we performed a large series of intron deletions and never observed any decrease in the efficiency of IIIc usage in AT3 cells. Third, IIIc inclusion approaches 100% even when our minigenes are transfected at very high levels in these cells, whereas in DT3 cells IIIc inclusion increases proportionally as higher numbers of minigenes are transfected. These observations suggest the presence of a factor or factors in DT3 cells which are needed to achieve cell-appropriate IIIb-exclusive splicing. The fact that such a factor(s) can be titrated is further evidence for the presence of a specific protein(s) in DT3 cells which is distinct from factors normally associated with constitutive splicing processes and which regulates specific splicing events. The specificity of such a factor(s) is further supported by the demonstration of a cis element, ISAR, which is required for appropriate exon IIIb splicing in DT3 cells.
FIG. 9.
Depiction of a model which can account for our results and the high fidelity of FGF-R2 splicing. AT3 cells use a default splicing pathway and choose the IIIc exon because of its stronger polypyrimidine tract (ppt); they splice IIIb inefficiently due to its weaker polypyrimidine tract. DT3 cells require a regulatory factor(s) which can activate (+) the weaker IIIb exon and at the same time repress (−) use of the IIIc exon. The ISAR element (indicated by a hatched box) is shown binding a factor or complex of factors (large shaded oval) which mediates both of these effects. The previously demonstrated contributions of other cis elements and associated factors (smaller shaded ovals) to IIIb activation are also shown, as well as the suggestion of possible cooperative interaction between proteins bound at several locations within the intron. Abbreviations are defined in the legend to Fig. 3.
Is ISAR alone capable of mediating the observed effects on FGF-R2 splicing? It appears that there are several cis elements involved in mediating appropriate regulation of FGF-R2 splicing. At present it is unclear whether these sequences are recognized in a coordinated manner or if each is recognized independently by both cell-specific and non-cell-specific factors which exert additive effects upon splicing. Investigation of alternative splicing in a number of systems has been complicated by the frequent observation that multiple intronic and exonic cis elements are involved in mediating the splicing efficiencies of alternative exons (2, 11, 14, 16, 28, 29, 57). An interesting observation has been that regulated exons often contain purine-rich enhancer sequences, which have been shown to enhance the activation of exons that contain them, effects which have been shown to involve the SR family of splicing factors (17, 30, 37, 55, 61, 62). However, so far these “exonic enhancers” have not been shown to exert tissue-specific effects but rather appear to influence the efficacy with which the exons containing them are spliced. Similarly, examples of repression by exon sequences as well as of both activation and repression of splicing by intronic sequences have also been described. Thus, in addition to the strength of splice site sequences, it may be that the relative strength of a given splicing pattern can be influenced by a number of cis elements whose effects may be general and not specific for a given cell type; rather they may set a balance between splicing choices, which can then be manipulated by cell-specific factors.
The challenge, then, is to identify regulatory sequences which function as the “switch” by binding a factor(s) which can exert cell-specific effects upon splicing. In the case of FGF-R2, several cis elements have been demonstrated to modulate the splicing efficiency of the IIIb exon (14–16). These elements include a specific sequence within the IIIb exon which represses its use; a polypyrimidine-rich sequence just downstream of exon IIIb (IAS1); and another sequence element, IAS2, just downstream from IAS1, as well as the ISAR sequence reported here, which is similar to a human sequence, IAS3, just recently described. It is worth noting that each of these sequences is highly homologous between rats and humans. Because we noted that deletion of sequences downstream of ISAR also causes some loss of regulation, it is likely that a cis element is present there as well. In the case of the first three elements, the original investigators, using various combinations of these sequences, showed that their effects upon splicing were not necessarily cell type specific, as each could exert these effects in both HeLa cells (which use IIIc) and SVK14 cells (which use IIIb), depending upon the context in which each was tested (14). Thus, one possibility is that some of these elements in both exons and introns exert general effects upon splicing efficiency. The fact that ISAR is required for both IIIb activation and IIIc repression suggests that this sequence may be more likely to be involved in the binding of cell-specific factors, although it may require additional sequences (with their associated factors) in order to exert tight regulation in this system. Del Gatto and colleagues (16) recently proposed that a human sequence with high similarity to ISAR, IAS3, may form a secondary structure with IAS2 which is a prerequisite for its function in the activation of IIIb splicing, although a role for IAS3 in IIIc repression was not described. As noted previously, sequences in the same region of the rat sequence as IAS2 occupies in the human sequence are also highly similar to their human homologs and would be predicted to form a similar secondary structure with ISAR. When we tested for the effects of ISAR on activation of IIIb and repression of IIIc we serendipitously included these sequences, and therefore we cannot rule out the possibility that they are required in a cooperative role in order for these effects to be exerted. Interestingly, however, even when only the effect of either IAS2 or IAS3 on the activation of human IIIb splicing was studied, the effect of deleting these sequences was less pronounced than that which we achieved with deletion of the rat ISAR, and complete loss of regulation in the experiments of Del Gatto et al. required mutation of IAS1. Studying the rat gene, however, we have demonstrated in this paper that we can obtain nearly complete loss of regulation by mutating ISAR alone, without altering a sequence homologous to IAS1. Thus, it is possible that there are some key differences in the degree to which these various cis elements function to mediate alternative splicing of rat and human FGF-R2.
Clarification of the mechanism by which ISAR and other intronic sequences change the pattern of splicing will require further biochemical study, but several other systems have provided clues as to how intron sequences can affect splicing. Intron sequences which are capable of activating splicing have been identified in gene transcripts from c-src (4, 48, 49), fibronectin (28, 29), calcitonin/CGRP (41), cardiac troponin T (9, 57), and β-tropomyosin (59). In the case of c-src, an intronic activating sequence has been shown to promote inclusion of an upstream neural-cell-specific exon via formation of a multiprotein complex containing heterogeneous nuclear ribonucleoprotein particle F (hnRNP F) and a novel protein, KH-type splicing regulatory protein (KSRP), although neither of these proteins is neural cell specific (48, 49). It has been proposed that one function of these activators, which are mostly located in the intron downstream of a regulated exon, is to promote binding of U1 to the upstream 5′ splice site (4, 49), which could then promote U2AF binding to the upstream polypyrimidine through cross-exon interactions which mediate exon definition (27, 56). Repression mediated by intronic sequences has also been described in several genes; examples include c-src (11) and the genes encoding α- and β-tropomyosin (22, 39, 52, 54, 58) and the γ2 subunit of the GABA receptor (2). In these cases an emerging theme has been the participation of polypyrimidine tract binding protein (PTB) (6, 19), which may block U2 binding to the branch point (39) and possibly interfere with U2AF binding as well (58), although given the ubiquitous expression of PTB it is still unclear how the specificity of repression can be achieved by PTB alone.
Mutually exclusive models of alternative splicing contain an additional level of complexity. In the case of mutually exclusive exons 2 and 3 of the α-tropomyosin gene, the presence of a branch point only 41 nucleotides downstream of exon 2 prevents inclusion of both exons, and exon 3 is the default exon in unregulated cells (22, 51, 60). When exon 3 is repressed, exon 2 usage can be achieved, even in the absence of specific factors promoting its use. In the chicken tropomyosin gene, a 33-nucleotide sequence, located in the intron between mutually exclusive exons 6A and 6B, was suggested to function in a manner similar to that which we propose for ISAR, as its deletion led to both loss of activation of the upstream 6A exon and derepression of the downstream 6B exon (3). However, this conclusion was weakened by the observation that replacing this sequence with its complementary sequence showed a similar effect, and unlike ISAR, any repositioning of this element abrogated the effect. Thus, our data are perhaps the most definitive to date which demonstrate a specific effect of a cis-acting intronic element which is required to facilitate activation of an upstream exon while also repressing a downstream exon. The ability of one cis-acting sequence to mediate both activation and repression is somewhat (although not directly) analogous to the experiments of Kanopka et al. (35), in which an exonic sequence which normally activates splicing at the upstream polypyrimidine tract causes repression of splicing when placed in the intron upstream of the same polypyrimidine tract. Although our ISAR sequence is positioned only in an intronic location, it may be that factors which bind this sequence are involved in mediating the opposite effects on splicing of an upstream versus a downstream exon. Identification of proteins which bind to ISAR and possibly other cis elements in the FGF-R2 gene transcript will hopefully yield a clearer picture of the process through which these multiple elements are used to modulate the activity of the constitutive splicing apparatus to yield distinct mRNAs in different cell types.
ACKNOWLEDGMENTS
We thank members of the Garcia-Blanco laboratory for helpful advice and suggestions. We also thank members of the McKeehan laboratory who developed and characterized the prostate cancer cell lines used in this study. In addition, we thank Dan Johnson for the generous gift of human FGF-R2 genomic clones, Laura Lindsey and Zhi-Ren Liu for critical review of the manuscript, and Ed Lobenhofer for assistance with cloning.
This work was supported by a grant from the NIH-NIGMS and by a Discovery Grant from the Duke Comprehensive Cancer Center to M.A.G.-B. M.A.G.-B. is an Established Investigator of the American Heart Association. R.P.C. was supported by an NIDDK training grant. M.A.G.-B. acknowledges the Keck foundation for support of the Leon Levine Science Research Center, where this research was performed.
REFERENCES
- 1.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 2.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 3.Balvay L, Libri D, Gallego M, Fiszman M Y. Intronic sequence with both negative and positive effects on the regulation of alternative transcripts of the chicken β tropomyosin transcripts. Nucleic Acids Res. 1992;20:3987–3992. doi: 10.1093/nar/20.15.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 5.Black D L. Finding splice sites in a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 6.Bothwell A L, Ballard D W, Philbrick W M, Lindwall G, Maher S F, Bridgett M M, Jamison S F, Garcia-Blanco M A. Murine polypyrimidine tract binding protein. Purification, cloning, and mapping of the RNA binding domain. J Biol Chem. 1991;266:24657–24663. [PubMed] [Google Scholar]
- 7.Bottaro D P, Rubin J S, Ron D, Finch P W, Florio C, Aaronson S A. Characterization of the receptor for keratinocyte growth factor. J Biol Chem. 1990;265:12767–12770. [PubMed] [Google Scholar]
- 8.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 9.Carlo T, Sterner D A, Berget S M. An intronic splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA. 1996;2:342–353. [PMC free article] [PubMed] [Google Scholar]
- 10.Carstens R P, Eaton J V, Krigman H R, Walther P J, Garcia-Blanco M A. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene. 1997;15:3059–3065. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 11.Chan R C, Black D L. Conserved intron elements repress splicing of a neuron-specific s-src exon in vitro. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Cote G J, Stolow D T, Peleg S, Berget S M, Gagel R F. Identification of exon sequences and an exon binding protein involved in alternative RNA splicing of calcitonin/CGRP. Nucleic Acids Res. 1992;20:2361–2366. doi: 10.1093/nar/20.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gatto F, Gesnel M, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gatto F, Plet A, Gesnel M, Fort F, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirksen W P, Hampson R K, Sun Q, Rottman F M. A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J Biol Chem. 1994;269:6431–6436. [PubMed] [Google Scholar]
- 18.Fu X D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Blanco M A, Jamison S F, Sharp P A. Identification and purification of a 62,000 dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 20.Ge H, Manley J L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert E, Del Gatto F, Champion-Arnaud P, Gesnel M, Breathnach R. Control of Bek and K-sam splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–5468. doi: 10.1128/mcb.13.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gooding C, Roberts G, Moreau G, Nadal-Ginard B, Smith C W J. Smooth muscle-specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham I R, Hamshere M, Eperon I C. Alternative splicing of a human α-tropomyosin muscle-specific exon: identification of determining sequences. Mol Cell Biol. 1992;12:3872–3882. doi: 10.1128/mcb.12.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo W, Mulligan G J, Wormsley S, Helfman D M. Alternative splicing of β-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991;5:2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- 25.Hattori Y, Odagiri H, Hakatani H, Miyagawa K, Naito K, Sakamoto H, Katoh O, Yoshida T, Sugimura T, Terada M. K-sam, an amplified gene in stomach cancer, is a member of the heparin-binding growth factor receptor genes. Proc Natl Acad Sci USA. 1990;87:5983–5987. doi: 10.1073/pnas.87.15.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helfman D M, Roscigno R F, Mulligan G J, Finn L A, Weber K S. Identification of two distinct intron elements involved in alternative splicing of β-tropomyosin pre-mRNA. Genes Dev. 1990;4:98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 28.Huh G S, Hynes R O. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh G S, Hynes R O. Regulation of alternative splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 30.Humphrey M B, Bryan J, Cooper T A, Berget S M. A 32 nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue K, Ohno M, Shimura Y. Aspects of splice site selection in constitutive and alternative pre-mRNA splicing. Gene Expr. 1995;4:177–182. [PMC free article] [PubMed] [Google Scholar]
- 32.Jaye M, Schlessinger J, Dionne C A. Fibroblast growth factor receptor tyrosine kinases: molecular analysis and signal transduction. Biochim Biophys Acta. 1992;1135:185–199. doi: 10.1016/0167-4889(92)90136-y. [DOI] [PubMed] [Google Scholar]
- 33.Johnson D E, Lu J, Chen H, Werner S, Williams L T. The human fibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–4634. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson D E, Williams L T. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 35.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 36.Kjems J, Frankel A D, Sharp P A. Specific regulation of mRNA splicing in vitro by a peptide from HIV Rev. Cell. 1991;67:178. doi: 10.1016/0092-8674(91)90580-r. [DOI] [PubMed] [Google Scholar]
- 37.Lavigueur A L, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 38.Libri D, Goux-Pelletan M, Brody E, Fiszman M Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the β tropomyosin gene. Mol Cell Biol. 1990;10:5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin C, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 40.Lou H, Gagel R F, Berget S M. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 41.Lou H, Yang Y, Cote G J, Berget S M, Gagel R F. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol Cell Biol. 1995;15:7135–7142. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991;251:33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- 43.Mattox W, Ryner L, Baker B S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992;267:19023–19026. [PubMed] [Google Scholar]
- 44.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 45.McKeown M. Sex differentiation: the role of alternative splicing. Curr Opin Genet Dev. 1992;2:299–303. doi: 10.1016/s0959-437x(05)80288-6. [DOI] [PubMed] [Google Scholar]
- 46.Miki T, Bottaro D P, Fleming T P, Smith C L, Burgess W H, Chan A M, Aaronson S A. Determination of ligand binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miki T, Fleming T P, Bottaro D P, Rubin J S, Ron D, Aaronson S A. Expression cDNA cloning of the KGF receptor by creation of a transforming autocrine loop. Science. 1991;251:72–75. doi: 10.1126/science.1846048. [DOI] [PubMed] [Google Scholar]
- 48.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 49.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 50.Moore M J, Query C C, Sharp P A. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 51.Mullen M P, Smith C W J, Patton J G, Nadal-Ginard B. α-Tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991;5:642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- 52.Mulligan G J, Guo W, Wormsley S, Helfman D M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 53.Orr-Urtreger A, Bedford M T, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 54.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 55.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan K J, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R, Valcarel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 59.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken β-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith C W J, Nadal-Ginard B. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Hoffman H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 62.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 63.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan W L. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol Cell Biol. 1993;13:4513–4522. doi: 10.1128/mcb.13.8.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan G, Fukabori Y, Nikolaropolous S, Wang F, McKeehan W L. Heparin-binding keratinocyte growth factor is a candidate stromal to epithelial cell andromedin. Mol Endocrinol. 1992;6:2123–2128. doi: 10.1210/mend.6.12.1491693. [DOI] [PubMed] [Google Scholar]
- 65.Yan G, McBride G, McKeehan W L. Exon skipping causes alteration of the COOH-terminus and deletion of the phospholipase C gamma 1 interaction site in the FGF receptor 2 kinase in normal prostate epithelial cells. Biochem Biophys Res Commun. 1993;194:512–518. doi: 10.1006/bbrc.1993.1849. [DOI] [PubMed] [Google Scholar]
- 66.Yayon A, Zimmer Y, Guo-Hong S, Avivi A, Yarden Y, Givol D. A confined variable region confers ligand specificity on fibroblast growth factor receptors: implications for the origin of the immunoglobulin fold. EMBO J. 1992;11:1885–1890. doi: 10.1002/j.1460-2075.1992.tb05240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Ashiya M, Sherman T G, Grabowski P J. Essential nucleotides direct neuron-specific splicing of γ2 pre-mRNA. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]