Abstract
The ionizing-radiation-resistant fractions of two soil bacterial communities were investigated by exposing an arid soil from the Sonoran Desert and a nonarid soil from a Louisiana forest to various doses of ionizing radiation using a 60Co source. The numbers of surviving bacteria decreased as the dose of gamma radiation to which the soils were exposed increased. Bacterial isolates surviving doses of 30 kGy were recovered from the Sonoran Desert soil, while no isolates were recovered from the nonarid forest soil after exposure to doses greater than 13 kGy. The phylogenetic diversities of the surviving culturable bacteria were compared for the two soils using 16S rRNA gene sequence analysis. In addition to a bacterial population that was more resistant to higher doses of ionizing radiation, the diversity of the isolates was greater in the arid soil. The taxonomic diversity of the isolates recovered was found to decrease as the level of ionizing-radiation exposure increased. Bacterial isolates of the genera Deinococcus, Geodermatophilus, and Hymenobacter were still recovered from the arid soil after exposure to doses of 17 to 30 kGy. The recovery of large numbers of extremely ionizing-radiation-resistant bacteria from an arid soil and not from a nonarid soil provides further ecological support for the hypothesis that the ionizing-radiation resistance phenotype is a consequence of the evolution of other DNA repair systems that protect cells against commonly encountered environmental stressors, such as desiccation. The diverse group of bacterial strains isolated from the arid soil sample included 60 Deinococcus strains, the characterization of which revealed nine novel species of this genus.
Extreme ionizing-radiation resistance has been observed in several members of the domains Bacteria and Archaea. Of the genera containing ionizing-radiation-resistant organisms, Deinococcus and Rubrobacter show the highest levels of resistance, and all species of these genera have been shown to be either gamma radiation resistant or UV radiation resistant or both (2, 6, 19, 20, 22, 47, 61, 62, 73). The genus Deinococcus, which represents a deeply branching lineage within the Bacteria, comprises 11 validly described species, D. frigens, D. geothermalis, D. grandis, D. indicus, D. marmoris, D. murrayi, D. proteolyticus, D. radiodurans, D. radiophilus, D. radiopugnans, and D. saxicola (5, 19, 26, 52, 61). Other ionizing-radiation-resistant bacteria have been isolated and described; these include some species of the genera Acinetobacter, Chroococcidiopsis, Hymenobacter, Kineococcus, Kocuria, and Methylobacterium (4, 5, 11, 23, 24, 30, 49, 50, 53). Hyperthermophilic euryarchaeote species of the genera Thermococcus and Pyrococcus also contain ionizing-radiation-resistant strains (15, 32, 33). Species of the genera Deinococcus and Rubrobacter have been shown to survive exposure to doses greater than 25 kGy (3, 6, 19, 20, 47, 73), while species of the genus Chroococcidiopsis survive exposure to 15 kGy (4). Strains of the species Acinetobacter radioresistens, Hymenobacter actinosclerus, Kineococcus radiotolerans, Methylobacterium radiotolerans, Pyrococcus furiosus, Pyrococcus abyssi, Thermococcus gammatolerans, Thermococcus marinus, and Thermococcus radiotolerans are less resistant and have been shown to survive after exposure to much lower levels of radiation (15, 23, 30, 32, 33, 53).
The origin of ionizing-radiation resistance in these prokaryotes is obscure, and this resistance cannot be explained as an adaptation to environmental radiation. Natural sources of ionizing radiation on Earth emit at very low levels (35, 64), making it impossible to generate the acute doses to which these organisms show resistance. It has been suggested that DNA repair mechanisms may have evolved not to counter the damage of ionizing radiation but rather to compensate for desiccation, another naturally occurring stress that generates a pattern of DNA damage similar to that produced by ionizing radiation (41). The process of desiccation is inherently DNA damaging and results in DNA double-strand breaks (17, 41), the primary lethal lesions resulting from exposure to ionizing radiation, and it is assumed that desiccation-tolerant species, as well as ionizing-radiation-resistant species, can avoid or effectively repair these lesions.
Ionizing-radiation-resistant organisms have been isolated from a wide range of environments, including sawdust (29), sewage (31), paper mill machinery (36, 66), animal feeds (31), processed meat (69), dried food (39, 42), feather pillows (67), room dust (8), textiles (37), irradiated meat and fish (13, 23, 69), high-level nuclear waste sites at Savannah River in South Carolina (53) and at Hanford in Washington (22), thermally polluted water (6), and irradiated rice (28). Other environments from which radiation-resistant isolates have been obtained include soil (5, 47), feces (5, 52), warm freshwater geothermal springs (19, 20), and shallow and abyssal marine thermal springs (15, 21, 32, 33). A number of A. radioresistens strains have also been isolated from clinical sources (9, 50). Many of the environments from which ionizing-radiation-resistant organisms have been isolated can be considered to be dry or desiccated, and it has been shown that many of these strains are also desiccation resistant (4, 17, 41, 42, 47, 58). Arid lands of various degrees cover more than 30% of the Earth's land surface and can be considered to represent natural environments that are desiccated (43). Little is known about the microbial diversity of arid or hyperarid environments, and there are no data on the abundance or diversity of ionizing-radiation-resistant organisms in these habitats. In an attempt to correlate the possible link between ionizing-radiation resistance and desiccation resistance at the ecological level, we exposed a soil sample from an arid environment in the Sonoran Desert in Arizona to various levels of gamma radiation and determined the numbers and diversity of the surviving population, the ionizing-radiation-resistant organisms. A soil sample from a nonarid region was used for comparison. This study provided further insight into the extensive diversity of ionizing-radiation-resistant organisms in an arid soil and resulted in description of nine additional species of the genus Deinococcus.
MATERIALS AND METHODS
Sampling and selective enrichment of ionizing-radiation-resistant bacteria.
A 100-g surface (upper 2 cm) soil sample designated S97-3 was collected using a sterile scoop in a sparsely vegetated area along Route 79 between Phoenix and Tucson, Ariz. A soil sample from a nonarid region of the southern United States, a Louisiana forest (designated LRB98-2), was also collected and studied for comparative purposes. The soil samples were stored at the ambient temperature until they were processed. One-gram aliquots of soil were exposed to levels of radiation between 0 and 30 kGy at a dose of 2.57 kGy h−1 at room temperature using a JL Sheppard model 484 60Co irradiator. After exposure, the samples were serial dilution plated on rich medium (RM) (72), plate count agar (PCA) (Difco), 0.1× PCA, and nutrient agar (NA) (Difco). The dilutions were prepared with liquid media that had the same composition as the agar plate media. Plates were incubated at 28°C for 20 days. The number of CFU was determined after 20 days of incubation. Selected colonies were purified and maintained in the appropriate medium containing 15% (vol/vol) glycerol at −80°C.
Bacterial strains.
The type strains used for taxonomic comparison were D. geothermalis DSM 11300, D. grandis DSM 3963, D. indicus DSM 15307, D. murrayi DSM 11303, D. proteolyticus DSM 20540, D. radiodurans DSM 20539, and D. radiophilus DSM 20551 obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany, and D. radiopugnans ATCC 19172 obtained from the American Type Culture Collection, Manassas, Va.
16S rRNA gene sequence determination and determination of G+C content of DNA.
Extraction of genomic DNA for 16S rRNA gene sequence determination, PCR amplification of the 16S rRNA gene, and sequencing of the purified PCR products were carried out as described previously (55, 56). Purified reaction mixtures were electrophoresed using a model 3100 Genetic Analyzer (Applied Biosystems). The identities of the 16S rRNA gene sequences examined in this study were determined using the BLAST (blastn) facility at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/BLAST/). Sequences were then aligned with representative reference sequences of members of the lineage to which the BLAST search data assigned them using the ae2 editor (10). The method of Jukes and Cantor (34) was used to calculate evolutionary distances. Phylogenetic dendrograms were generated and bootstrap analyses were performed using various algorithms contained in the PHYLIP package (18). The DNA used for determination of the G+C content of the DNA was isolated as described by Cashion et al. (7). The G+C content of DNA was determined by high-performance liquid chromatography as described by Mesbah et al. (44).
Morphological, biochemical, and physiological characteristics.
Cell morphology and motility were examined by phase-contrast microscopy and differential interference contrast microscopy after cultivation on agar plates. Photomicrographs were taken after cells were placed on coverslips that had a thin film of 1% (wt/vol) agarose on the surface. The temperature range for growth was determined on nutrient and plate count agar plates incubated for 10 days at temperatures between 5 and 50°C. The pH range for growth was determined at 28°C on agar plates with the media buffered between pH 5.5 and pH 9.0 as described previously (19). Control media containing each buffer adjusted to pH 6.7 were used to assess possible inhibitory effects of the buffering agents. Catalase activity, cytochrome oxidase activity, and the hydrolysis of starch, casein, and gelatin were determined as described by Smibert and Krieg (60).
Assimilation tests.
Single-carbon-source assimilation studies were performed in a defined medium solidified with deionized water-washed agar (approximately 2%; Oxoid) containing Degryse basal salts medium 162 (14) to which filter-sterilized yeast extract (0.05 g liter−1), a carbon source (2.0 g liter−1), ammonium sulfate (0.5 g liter−1), and a vitamin and nucleotide solution (59) were added at a final pH of 7.5. Strains KR-235T and KR-242 did not grow well on this medium, and for these organisms it was necessary to assess single-carbon-source assimilation in a minimal medium in which the Degryse macroelements and salts were replaced by K2HPO4 (0.5 g liter−1) and MgSO4 · 7H2O (0.1 g liter−1). The inocula were grown on nutrient agar (Difco) at 30°C for 48 to 72 h. Cells were scraped off the agar plates and resuspended in the Degryse basal salts medium to a turbidity equal to the McFarland no. 1 standard. Plates containing each single carbon source were spotted (diameter, about 1 cm) with the cell suspension. Growth was examined visually on plates incubated at 30°C for up to 7 days. Negative control plates did not contain the carbon source. Positive control cultures were grown in nutrient agar and solidified Degryse medium 162 (14). We also attempted to determine carbon source assimilation profiles in liquid media having the same compositions as the media mentioned above, but this was unsuccessful due to a lack of growth or clumping of cells.
Hydrolysis of chromogenic substrates.
The chromogenic substrates (2 mM) were dissolved in 5 ml of the appropriate medium, the pH was adjusted to 7.0, and the solutions were sterilized by filtration. Aliquots (50 μl) were placed in microtiter plates. Most strains were grown in the appropriate liquid medium. The exceptions were strains KR-114 and KR-245; these strains were grown on agar plates, and cell suspensions were prepared. After growth in the liquid medium was visible, 50 μl of each of the cultures was added to the wells of the prepared microtiter plates. The plates were incubated at 28°C for 24 h or 5 days. Development of yellow color indicated positive results.
Polar lipid, lipoquinone, and fatty acid composition.
The cultures used for polar lipid analysis were grown on NA (Difco). Harvesting of the cultures and extraction of lipids were performed as described previously (16, 54). Lipoquinones were extracted from freeze-dried cells, purified by thin-layer chromatography, and separated with a Gilson high-performance liquid chromatograph (63). Cultures for fatty acid analysis were grown on agar plates incubated in sealed plastic bags submerged in a water bath at the optimum growth temperature for 72 h. Fatty acid methyl esters were obtained from fresh wet biomass by saponification, methylation, and extraction as described previously by Kuykendall et al. (38), and they were separated, identified, and quantified as described previously (46).
Determination of levels of ionizing-radiation and desiccation resistance.
In order to determine the survival of isolates after exposure to various doses of gamma radiation, strains were grown in the appropriate liquid medium to the exponential phase. The cells were recovered by centrifugation, washed with 0.067 M potassium phosphate buffer at pH 7.0, and resuspended in the same buffer. Aliquots were exposed at room temperature to gamma radiation from a cobalt 60 source at a dose of 2.57 kGy h−1. After exposure to 5, 10, 15, and 20 kGy, suspensions were dilution plated in triplicate on the appropriate solid medium. Growth after 15 days was scored as positive or negative in comparison to an unirradiated control. To determine the desiccation resistance of the isolates, 100-μl aliquots of cultures of strain LB-34T and D. radiodurans R1 were dried over CaSO4 (Dri-Rite) and kept at 5% relative humidity until rehydration. Titers were determined before desiccation and immediately after rehydration, and the values were used to calculate the surviving fraction.
16S rRNA gene sequence accession numbers.
The 16S rRNA gene sequences of the Deinococcus strains described in this study have been deposited under accession numbers AY743256 to AY743285. The 16S rRNA gene sequences recovered from the Central Arizona Phoenix Long-Term Ecological Research (CAP-LTER) site have been deposited under accession numbers AY905380 to AY905384.
RESULTS AND DISCUSSION
Survival and isolation of bacteria after exposure to different doses of gamma radiation.
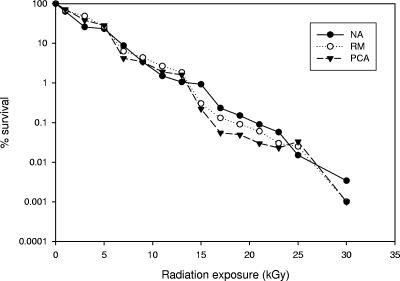
The unirradiated soil sample, designated S97-3, contained bacterial concentrations ranging from 5.5 × 106 to 1.3 × 107 CFU/g depending on the culture medium used for dilution plating. The colonies on the plates containing the unirradiated samples exhibited extensive diversity in terms of colony morphology, in contrast to the plates containing the irradiated samples, on which the majority of the colonies were yellow, orange, pink, or red. There was a decrease in the number of CFU/g recovered from the soil samples with increasing doses of gamma irradiation, and this was observed on a number of different culture media (Fig. 1). When samples were dilution plated on PCA and RM, the concentrations decreased from 6.6 × 106 and 5.5 × 106 CFU/g, respectively, for the unirradiated sample to undetectable after exposure to 30 kGy. In the case of NA, radiation exposure resulted in a decrease in the concentration from 1.3 × 107 to 4.4 × 102 CFU/g. It should be noted that the values below 3.0 × 103 CFU/g derived from less than 30 colonies on an agar surface which were obtained for samples exposed to more than 23 kGy are outside the statistical limits of the dilution plating technique used but still provide an indication of the decrease in the number of organisms with increasing radiation dose. The colonies on these plates were, however, a source of isolates for further characterization. As shown in Fig. 1, there was an exponential decrease in the percentage of recoverable CFU/g as the radiation dose increased. Approximately 30% of the culturable population (on all media tested) was lost after exposure of the soil sample to 1.0 kGy. Increasing proportions of the original population were eliminated as the radiation dose was increased. After exposure to 15 kGy, less than 1% of the original culturable population could be recovered by dilution plating. The data demonstrated that there are organisms in natural environmental samples, such as the arid soil examined here, that comprise populations of ionizing-radiation-resistant bacteria. These organisms are resistant to levels of gamma radiation that far exceed the background levels in the natural environment, which for southern Arizona are about 30 × 10−5 Gy per year (48, 65). The unirradiated soil sample from the nonarid Louisiana forest, designated LRB98-2, contained bacterial concentrations ranging from 4.4 × 106 to 1.1 × 107 CFU/g depending on the culture medium used for dilution plating, which are comparable to the concentrations found in the unirradiated arid soil. In contrast to the arid soil, from which ionizing-radiation-resistant organisms were recovered after exposure to 30 kGy, no survivors were recovered from the nonarid soil after exposure to doses above 13 kGy. Exposure of the nonarid soil to 11 kGy resulted in concentrations below 103 CFU/g, in contrast to the results obtained for the arid soil, in which the concentration dropped to 103 CFU/g only after exposure to 23 kGy.
FIG. 1.
Effect of radiation dose on the survival of culturable heterotrophic bacteria in the arid soil sample (S97-3).
Diversity of ionizing-radiation-resistant bacteria based on 16S rRNA gene sequence data.
A collection of 210 ionizing-radiation-resistant bacteria was recovered from soil sample S97-3. Partial 16S rRNA gene sequences were determined for 133 of the isolates recovered from this arid soil sample. Comparison of these 16S rRNA gene sequences to the public databases using the BLAST (blastn) facility (www.ncbi.nlm.nih.gov/BLAST/) enabled us to assign each isolate to a taxonomic group at the family or genus level and in some cases at the species level. Complete sequences were determined for representative isolates, so phylogenetic dendrograms could be constructed. Since the organisms that survived high doses of gamma radiation were of special interest, a large number of the isolates sequenced (108 strains) were isolates recovered from the soil sample after irradiation at doses greater than 17 kGy. The 133 isolates whose taxonomic identities were determined based on partial 16S rRNA gene sequence comparisons (∼800 nucleotide positions) fell into 14 different taxonomic groups based on their closest relatives (Table 1). Seventy-three isolates were assigned to taxonomic groups that have previously been shown to contain ionizing-radiation-resistant bacteria, namely, the genera Deinococcus, Hymenobacter, Kineococcus, Kocuria, and Methylobacterium. With increasing doses of radiation the diversity of the isolates recovered was found to decrease. After exposure to 17 kGy and higher doses, only species of the genera Deinococcus, Geodermatopilus, and Hymenobacter were recovered. Seven of the eight isolates from the unirradiated sample and a sample exposed to 3 kGy were from the unirradiated sample and were selected based on their pigmentation in order to determine if these pigmented isolates did in fact represent the same taxa that survived higher doses of radiation. These isolates fell into five different taxonomic groups, including two genera with known ionizing-radiation-resistant species, Hymenobacter and Methylobacterium. Seventeen isolates from soil samples exposed to 5 to 9 kGy fell into 11 taxonomic groups, five of which contain known ionizing-radiation-resistant species; these isolates included eight isolates belonging to the genera Deinococcus, Hymenobacter, Kineococcus, Kocuria, and Methylobacterium. The remaining nine isolates fell into taxonomic groups not previously shown to contain ionizing-radiation-resistant organisms, including the genera Bosea, Chelatococcus, Corbulabacter, Planococcus, and Spirosoma and the family Sphingomonadaceae. The 32 isolates recovered from the soil samples exposed to between 11 and 15 kGy fell into seven taxonomic groups. All of these groups except the genus Geodermatophilus were recovered from samples exposed to lower doses. After exposure to 11 to 15 kGy the numbers of Deinococcus strains recovered increased compared to the numbers obtained with lower doses, and 16 strains were recovered at these levels. This predominance of Deinococcus species, along with members of the Geodermatophilus group, was seen at all higher doses. Species of these two genera made up 82% of the isolates recovered from the soil sample after exposure to doses of ionizing radiation of 17 kGy or more. At the highest doses (23, 25, and 30 kGy), Deinococcus species accounted for more than 72% of the isolates recovered, and Geodermatophilus and Hymenobacter species accounted for 23 and 5%, respectively. Of the 126 isolates recovered and identified from the soil sample exposed to various levels of ionizing radiation in this study, 106 belonged to these three genera (Deinococcus, 60 strains; Geodermatophilus, 40 strains; and Hymenobacter, 6 strains).
TABLE 1.
Summary of the taxonomic affiliations of bacterial strains recovered from an arid soil sample after it was exposed to various doses of gamma ionizing radiation
| Dose (kGy) | No. of isolates identified | Taxonomic groups (no. of isolates) |
|---|---|---|
| 0-3.0 | 8 | Arthrobacter sp. (3), Friedmanniella sp., Geodermatophilus sp., Hymenobacter sp., Methylobacterium sp., Nocardioides sp. |
| 5.0-9.0 | 17 | Bosea sp., Chelatococcus sp. (2), Corbulabacter sp., Deinococcus sp. (2), Hymenobacter sp. (2), Kineococcus sp., Kocuria sp., Methylobacterium sp. (2), Planococcus sp. (2), Sphingomonadaceae (2), Spirosoma sp. |
| 11.0-15.0 | 32 | Chelatococcus sp. (3), Corbulabacter sp., Deinococcus sp. (16), Geodermatophilus sp. (7), Hymenobacter sp., Methylobacterium sp. (2), Sphingomonadaceae (2) |
| 17.0-21.0 | 38 | Deinococcus sp. (15), Geodermatophilus sp. (23) |
| 23.0-25.0 | 24 | Deinococcus sp. (17), Geodermatophilus sp. (5), Hymenobacter sp. (2) |
| 30.0 | 14 | Deinococcus sp. (10), Geodermatophilus sp. (4) |
The diversity of the isolates recovered from the forest soil from the nonarid region after irradiation doses greater than 5 kGy was limited. Morphologically, the majority of these isolates were very similar; they had white pigmentation and producted aerial mycelia. 16S rRNA gene sequence analysis of these isolates showed that they are members of the genus Streptacidiphilus. The other isolates recovered from the nonarid soil after exposure to ionizing-radiation doses between 3 and 13 kGy were identified as members of the genera Bacillus, Nocardia, and Micrococcus and a group related to Dehalococcoides for which only environmental 16S rRNA gene sequences exist. Among the 110 isolates obtained from the irradiated nonarid soil, no members of the genus Deinococcus were identified. The nonarid soil from a Louisiana forest examined in this study did not contain as large a fraction of ionizing-radiation-resistant bacteria as the arid soil sample contained. The identities of the species which showed resistance to ionizing radiation were different in the two samples. The ionizing-radiation-resistant population of the nonarid soil was dominated by a single genus, while the arid soil contained a diverse group of ionizing-radiation-resistant species. Interestingly, no isolate belonging to the genus Deinococcus, Geodermatophilus, or Hymenobacter was isolated from the nonarid soil even after exposure to doses greater than 11 kGy.
This study expanded our knowledge of the diversity of ionizing-radiation-resistant bacteria. This is the first report of strains related to the taxonomic groups Bosea, Chelatococcus, Corbulabacter, Geodermatophilus, Planococcus, species of the family Sphingomonadaceae, and Spirosoma that are resistant to gamma radiation. The high number of Deinococcus isolates recovered from the soil samples exposed to the high ionizing-radiation levels further establishes that the species of this genus are extremely ionizing radiation resistant. The isolation of high numbers of ionizing-radiation-resistant members of the genus Geodermatophilus, a member of a group of organisms previously isolated from desert soils and arid environments (40, 45), was a novel finding and provided strains for comparison with Deinococcus species.
Maxcy and Rowley (42) were the first workers to obtain experimental evidence that there could be a link between a prokaryote's ability to survive dehydration and ionizing-radiation resistance. They demonstrated that by selecting for desiccation tolerance in natural microflora, it was possible to simultaneously isolate radioresistant species. Subsequently, detailed evaluations of members of the genera Deinococcus and Chroococcidiopsis supported the notion that these phenotypes could be interrelated by demonstrating that ionizing radiation and desiccation introduced similar types of DNA damage (4, 41) and that the loss of DNA repair capacity in a radioresistant species can result in a strain that is no longer capable of surviving dehydration (41). This study demonstrated that ionizing-radiation-resistant species are present at higher numbers in arid soil of the Sonoran Desert than in a nonarid Louisiana forest soil and that the ionizing-radiation-resistant organisms in the arid soil are recovered after exposure to higher doses than the doses which allow recovery of organisms from the nonarid soil. This is consistent with the hypothesis that organisms with heightened DNA repair capacity are among the species that have a selective advantage in an arid environment. We assume that the ionizing-radiation resistance of species isolated from these environments are an incidental consequence of their ability to survive desiccation-induced DNA damage. The isolation of a large number of deinococcal strains from an arid soil sample and the extensive diversity of these strains were surprising considering that a culture-independent study of >10,000 environmental 16S rRNA gene sequences from 14 desert or desert remnant soils at the CAP-LTER site in Arizona recovered only five sequences that fell in the Deinococcus lineage (57). Further study at the taxonomic level of the predominant group among the ionizing-radiation-resistant isolates recovered in this study revealed that they comprise nine novel species of the genus Deinococcus.
16S rRNA gene-based phylogenetic analysis of the Deinococcus isolates.
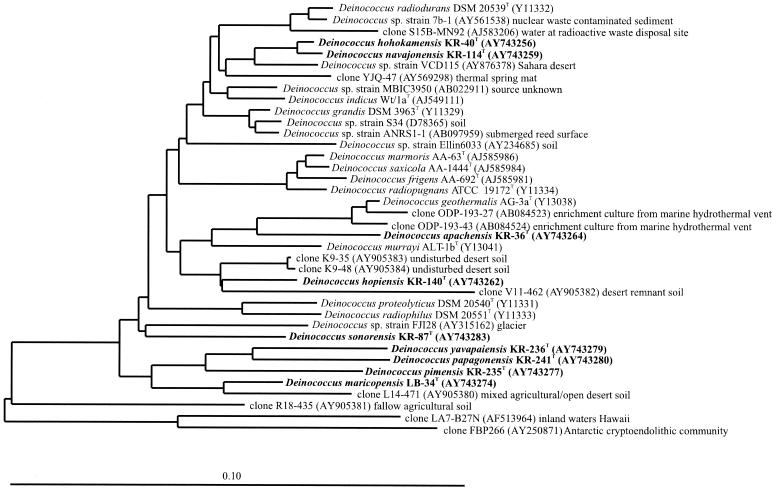
Almost complete 16S rRNA gene sequences comprising between 1,453 and 1,464 nucleotides were determined for 30 of the Deinococcus strains that were assigned to this genus on the basis of partial 16S rRNA gene sequences. The phylogenetic analysis, represented by a neighbor-joining tree in Fig. 2, demonstrated the relationship of the new isolates to the 11 previously described species of the genus Deinococcus, as well as undescribed deinococcal strains and environmental 16S rRNA gene sequences deposited in the public databases. None of the new isolates showed a close relationship to the previously described species; the values for 16S rRNA gene sequence similarity between the new isolates and the previously described species were in the range from 88 to 95%. The 30 Deinococcus isolates for which almost complete 16S rRNA gene sequences were determined were found to represent nine new lineages in this genus cluster (Fig. 2). Each of these lineages except species 7 (strain KR-236T) is represented by more than one strain (data not shown); species 1 is represented by KR-40T, KR-88, and KR-245, species 2 is represented by KR-33, KR-39, and KR-114T, species 3 is represented by KR-125 and KR-140T, species 4 is represented by KR-30, KR-31, KR-32, KR-35, KR-36 T, KR-37, KR-51, KR-53, KR-54, and KR-55, species 5 is represented by LB-34T, KR-1, and KR-23, species 6 is represented by KR-235T and KR-242, species 8 is represented by KR-119, KR-241T, and KR-237, and species 9 is represented by KR-87T, KR-90, and KR-136. In the public databases there are a number of full and partial sequences for as-yet-undescribed strains or environmental 16S rRNA gene sequences, including a strain isolated from the Sahara Desert (accession no. AY876378) (Fig. 2), that fall within the radiation of the genus Deinococcus. The 16S rRNA gene sequences of the new species isolated and described in this study are not closely related to any of the database sequences of strains or environmental 16S rRNA gene sequences, further demonstrating the diversity of this genus yet to be described from the strains already isolated or from isolates still to be obtained from environments in which 16S rRNA gene sequences that fall within the radiation of the genus Deinococcus have been detected (Fig. 2).
FIG. 2.
16S rRNA gene sequence-based phylogeny indicating the relationship of the type strains of the nine new species of the genus Deinococcus to previously described species, strains, and environmental sequences falling in this lineage. Scale bar = 10 inferred nucleotide substitutions per 100 nucleotides.
Morphological, physiological, biochemical, desiccation resistance, and radioresistance characteristics of the deinococci.
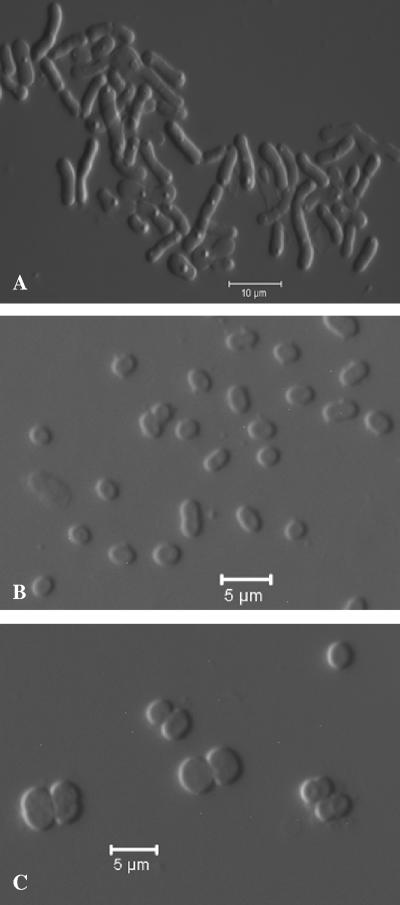
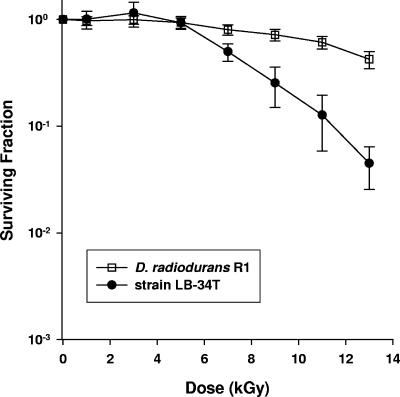
The colonies of the Sonoran Desert deinococcal isolates varied from light pink to orange-red. The cells were either spherical or rod shaped (Fig. 3). One species, represented by strains KR-87T and KR-136, produced very short rod-shaped or spherical cells (Fig. 3). The new isolates, unlike the type strains of all other species of the genus Deinococcus previously described, grew very poorly or not at all in liquid media. Even solid media that generally support the growth of the previously validly described species did not support the growth of many of these isolates. For example, most of the strains from the Sonoran Desert did not grow or grew poorly on nutrient agar from Oxoid, which contains (per liter) 1.0 g Lab-Lemco powder, 2.0 g yeast extract, 5.0 g peptone, 5.0 g NaCl, and 15.0 g agar and has a pH of 7.4, but they grew very well on nutrient agar from Difco, which contains (per liter) 3.0 g beef extract, 5 g peptone, and 15 g agar and has a pH 6.8. At first, we thought that the NaCl in the Oxoid formulation inhibited growth, but this did not seem to be the case. The salt tolerance of the new strains was very low, but the NaCl level in the Oxoid NA did not inhibit growth when it was added to the Difco NA. Many of the strains did not grow on NA (Difco) containing 1.0% (wt/vol) NaCl, and none grew in this medium with 1.5% (wt/vol) NaCl. All organisms were cytochrome oxidase positive, but catalase could not be detected in the strains of the species represented by strains KR-40T and KR-245 and the species represented by strains KR-114T and KR-33. The new isolates assimilated a large variety of carbon sources, including carbohydrates, organic acids, and amino acids, although none of the strains utilized l-sorbose, raffinose, acetate, citrate, cysteine, lysine, or methionine (see Table A in the supplemental material). Strains KR-235T and KR-242, which represent one of the new species, grew poorly in the medium used to examine carbon source assimilation for the other species, and we used a slightly different medium for these organisms. The strains of all species could be distinguished from each other by carbon source assimilation tests (see Table A in the supplemental material). The new isolates hydrolyzed a range of chromogenic substrates (see Table B in the supplemental material). These data differentiate organisms at the strain level but are not useful at the species level. The growth temperatures for the new species fall in the range from 10°C to 45°C, and the majority of the strains grow at temperatures ranging from 10°C to 37°C. The three strains of species 5 are capable of growth at 45°C but not at 47°C, while the strains of species 6 do not grow at 15°C or 45°C. It is interesting that there were a number of thermotolerant strains in a sample from which temperature-sensitive strains were obtained. Since the novel species described here were isolated from a desert environment, we attempted to establish if they, like D. radiodurans R1, exhibited unusual tolerance to desiccation. This proved to be difficult because the majority of the new species clumped extensively in liquid culture, which made it impossible to accurately determine cell numbers before and after desiccation. The only exception was strain LB-34T, which remained evenly distributed in liquid. As indicated in Table 2, this species showed levels of desiccation tolerance comparable to that of R1 cultures. As described previously, the viability of R1 cultures decreases to between 80 and 85% after 6 weeks of desiccation (41). Strain LB-34T cultures exhibited similar kinetics but showed a greater loss of viability after 5 and 6 weeks compared to R1. This increased sensitivity to DNA damage was also observed when the ionizing-radiation resistance of these strains was compared. Figure 4 shows the survival curves generated following irradiation of stationary-phase cultures of strain LB-34T and strain R1 of the type species D. radiodurans. At doses of 7,000 Gy or less, these strains exhibit identical radioresistance, but at higher doses the R1 strain is more resilient. At 13,000 Gy strain LB-34T cultures are approximately 10-fold more sensitive to ionizing radiation than an R1 culture, which reinforced the differences in viability observed following irradiation. The type strains of the new Deinococcus species were tested for survival after exposure to gamma radiation, and all of these strains were shown to survive after exposure to at least 10 kGy, which is comparable to the results obtained for the previously described species of this genus.
FIG. 3.
Differential interference contrast photomicrographs showing the range of morphological types within the genus Deinococcus. (A) Strain KR-241T. (B) Strain KR-87T. (C) Strain KR-140T.
TABLE 2.
Survival of strain LB-34T and D. radiodurans R1 following prolonged desiccation
| Week | Surviving fraction
|
|
|---|---|---|
| D. radiodurans R1 | Strain LB-34T | |
| 1 | 0.98 ± 0.02a | 0.99 ± 0.07 |
| 2 | 1.01 ± 0.08 | 1.05 ± 0.14 |
| 3 | 0.97 ± 0.06 | 0.94 ± 0.08 |
| 4 | 0.85 ± 0.02 | 0.87 ± 0.02 |
| 5 | 0.85 ± 0.02 | 0.75 ± 0.15 |
| 6 | 0.82 ± 0.03 | 0.55 ± 0.18 |
The values are the means ± standard deviations for the surviving fraction obtained from three independent trials (n = 9).
FIG. 4.
Representative survival curve for D. radiodurans R1 and strain LB-34T. The values are the means ± standard deviations of three independent experiments (n = 9 for R1 cultures and n = 12 for strain LB-34T cultures).
Polar lipid, lipoquinone, and fatty acid composition of the deinococci.
The type strains of the Deinococcus species described previously and the strains of the new species had fairly similar polar lipid patterns consisting of phosphoglycolipids and glycolipids (1, 27); however, strain KR-236T (species 7) does not contain the major phosphoglycolipid found in all other strains examined (results not shown). Menaquinone 8 was the major respiratory quinone of all the strains examined.
The strains of the new species of Deinococcus possess a large variety of fatty acids, including saturated straight-chain, hydroxy, iso- and anteiso-branched-chain, and monounsaturated fatty acids. Unexpectedly, all species could be easily distinguished from each other and from each of the type strains of the previously described species on the basis of the relative proportions of the fatty acids (see Table C in the supplemental material). Strains KR-119, KR-237 and KR-241T (species 8) and strains KR-235T and KR-242 (species 6), unlike all other species of this genus (19), possessed 3OH fatty acids (namely, iso 16:0 3OH and iso 17:0 3OH). All strains of the new species except strains KR-140T and KR-125 (species 3) possess branched-chain iso and anteiso fatty acids. In this respect the strains of species 3 resemble those of D. proteolyticus, D. radiodurans, and D. radiophilus, which possess straight-chain saturated and unsaturated fatty acids.
Taxonomic conclusions and description of new taxa.
The species of the genus Deinococcus have been shown to represent a distinct phylogenetic lineage that branches with the species of the family Thermaceae to form a phylum-level lineage within the domain Bacteria (12, 25, 26, 56, 68, 70, 71). 16S rRNA gene sequence data have been used to differentiate the species of the genus Deinococcus both from other taxa and from each other (19, 56, 61). The low levels of 16S rRNA gene sequence similarity (<95%) between the new isolates and the previously described species indicate that the new isolates represent a number of novel Deinococcus species. The phylogenetic analysis showed that the strains fall into nine new lineages in the radiation of the genus Deinococcus, each of which can be considered to represent a new species of this genus (Fig. 2). The strains representing the nine new species of the genus Deinococcus were easily distinguished from each other and from the previously validly described species of this genus by 16S rRNA gene sequence data, single-carbon-source assimilation patterns, and fatty acid profiles. With the exception of D. grandis and D. indicus, which form rod-shaped cells, all strains of the previously validly described species of the genus Deinococcus form spherical cells (2, 3, 19, 47, 56, 61). The description of the desert soil strains adds four additional species with rod-shaped morphologies to the the genus Deinococcus. It should also be noted that strains KR-40T and KR-245 and strains KR-114T and KR-33, belonging to two separate species, were catalase negative, like several of the species of the genus Meiothermus (51).
On the basis of the results presented in this study we describe the following nine new species of the genus Deinococcus: Deinococcus hohokamensis is proposed for the species represented by strains KR-40T, KR-88, and KR-245; Deinococcus navajonensis is proposed for strains KR-114T, KR-33, and KR-39; Deinococcus hopiensis is proposed for strains KR-140T and KR-125; Deinococcus apachensis is proposed for strains KR-36T, KR-30, KR-31, KR-32, KR-35, KR-37, KR-51, KR-53, KR-54, and KR-55; Deinococcus maricopensis is proposed for strains LB-34T, KR-1, and KR-23; Deinococcus pimensis is proposed for strains KR-235T and KR-242; Deinococcus yavapaiensis is proposed for strain KR-236T; Deinococcus papagonensis is proposed for strains KR-241T, KR-119, and KR-237; and Deinococcus sonorensis is proposed for strains KR-87T, KR-90, and KR-136.
Description of Deinococcus hohokamensis sp. nov. Rainey and da Costa.
Deinococcus hohokamensis (ho.ho.kam′en.sis. N. L. masc. adj. hohokamensis, named after the ancient people that inhabited central Arizona). Deinococcus hohokamensis forms spherical cells that are 2.0 to 3.0 μm in diameter. Gram staining is positive. Cells are nonmotile; spores are not observed. Colonies on RM are light pink. The strains were isolated from an arid soil sample after exposure to 15 to 30 kGy, and strain KR-40T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase negative. The major fatty acids are 16:1 ω7c, 17:1 ω8c, and iso 17:1 ω7c. Strains utilize galactose, l-rhamnose, l-arabinose, maltose, sucrose, cellobiose, malate, ornithine, glutamate, glutamine, and proline. Mannose and asparagine are used by strain KR-40T but not by strain KR-245. Starch, casein, and gelatin are degraded. The DNA of strain KR-40T has a G+C content of 67.9 mol%. Source: desert soil from Arizona. Strain KR-40 (LMG 22129 = NRRL B-23949) is the type strain. Strain KR-33 (LMG 22130 = NRRL B-23944) is a reference strain.
Description of Deinococcus navajonensis sp. nov. Rainey and da Costa.
Deinococcus navajonensis (na.va.jo′nen.sis. N. L. masc. adj. navajonensis, named after the Navajo Nation). Deinococcus navajonensis forms rod-shaped cells that are 3.0 to 5.0 μm long and 1.0 to 2.0 μm in diameter; short filaments are present. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on RM are pink. The strains were isolated from an arid soil sample after exposure to 15 to 25 kGy, and strain KR-114T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase negative. The major fatty acids are 16:1 ω7c, 17:1 ω8c, and iso 17:1 ω7c. Strains utilize galactose, fructose, mannose, l-rhamnose, l-arabinose, maltose, sucrose, trehalose, cellobiose, malate, ornithine, glutamate, glycine, and proline. Starch and gelatin are degraded. The DNA of strain KR-114T has a G+C content of 66.4 mol%. Source: desert soil from Arizona. Strain KR-114 (LMG 22131 = NRRL B-23951) is the type strain. Strain KR-33 (LMG 22132 = NRRL B-23947) is a reference strain.
Description of Deinococcus hopiensis sp. nov. Rainey and da Costa.
Deinococcus hopiensis (ho.pi′en.sis. N. L. masc. adj. hopiensis, named after the Hopi Nation). Deinococcus hopiensis forms spherical cells that are 2.0 to 3.0 μm in diameter. Cells occur in clusters. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on PCA are pink. The strains were isolated from an arid soil sample after exposure to 15 to 25 kGy, and strain KR-140T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase positive. The major fatty acids are 16:1 ω7c, 16:0, and 15:1 ω6c. Strains utilize l-arabinose, cellobiose, fructose, galactose, glucose, mannose, maltose, l-rhamnose, ribose, sucrose, trehalose, xylose, glucosamine, l-glutamine, proline, and ornithine. Lactose and glycerol are used by strain KR-125 but not by strain KR-140T. Starch, casein, and gelatin are degraded. The DNA of strain KR-140T has a G+C content of 66.2 mol%. Source: desert soil from Arizona. Strain KR-140 (LMG 22133 = NRRL B-23843) is the type strain. Strain KR-125 (LMG 22134 = NRRL B-23952) is a reference strain.
Description of Deinococcus apachensis sp. nov. Rainey and da Costa.
Deinococcus apachensis (a.pa′chen.sis. N. L. masc. adj. apachensis, named after the Apache Nation). Deinococcus apachensis forms spherical cells that are 1.5 to 3.0 μm in diameter. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on RM are pink. The strains were isolated from an arid soil sample after exposure to 15 to 17 kGy, and strain KR-36T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase positive. The major fatty acids are iso 15:0, 16:1 ω7c, and iso 17:0. Strains utilize l-arabinose, cellobiose, fructose, galactose, glucose, mannose, maltose, l-rhamnose, sucrose, trehalose, and proline. Lactose is used by strain KR-55 but not by strain KR-36T. Starch is degraded. Casein and gelatin are not degraded. The DNA of strain KR-36T has a G+C content of 68.5 mol%. Source: desert soil from Arizona. Strain KR-36 (LMG 22135 = NRRL B-23948) is the type strain. Strain KR-55 (LMG 22136 = NRRL B-23950) is a reference strain.
Description of Deinococcus maricopensis sp. nov. Rainey and da Costa.
Deinococcus maricopensis (ma.ri.co′pen.sis. N. L. masc. adj. maricopensis, named after the Maricopa Nation). Deinococcus maricopensis forms rod-shaped cells that are 3.0 to 6.0 μm long and 2.0 μm wide. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on RM are pink. The strains were isolated from an arid soil sample without exposure to irradiation or after exposure to 13 kGy, and strain LB-34T is resistant to >10 kGy. The optimum growth temperature is about 40°C, and growth occurs at 45°C. Oxidase positive and catalase positive. The major fatty acids are iso 15:0, iso 17:0, and 16:0. Strains utilize l-arabinose, cellobiose, galactose, glucose, mannose, maltose, sucrose, trehalose, glucosamine, glycerol, malate, asparagine, aspartate, glutamate, l-glutamine, ornithine, and proline. Fructose is used by strain KR-23 but not by strain LB-34T. Starch, casein, and gelatin are degraded. The DNA of strain LB-34T has a G+C content of 71.1 mol%. Source: desert soil from Arizona. Strain LB-34 (LMG 22137 = NRRL B-23946) is the type strain. Strain KR-23 (LMG 22138 = NRRL B-23945) is a reference strain.
Description of Deinococcus pimensis sp. nov. Rainey and da Costa.
Deinococcus pimensis (pi′men.sis. N. L. masc. adj. pimensis, named after the Pima Nation). Deinococcus pimensis forms rod-shaped cells that are 3.0 to 6.0 μm long and 2.0 μm wide. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on NA are pink. The strains were isolated from an arid soil sample after exposure to 30 kGy, and strain KR-235T is resistant to >10 kGy. The optimum growth temperature is 30°C. Strain KR-235T is cytochrome oxidase negative (strain KR-242 is positive) and catalase positive. The major fatty acids are iso 15:0, iso 17:0, and iso 15:1; iso 16:0 3OH and iso 17:0 3OH are present. Strains utilize l-arabinose, cellobiose, fructose, galactose, d-glucose, maltose, l-rhamnose, sucrose, trehalose, lactose, ribose, xylose, glycerol, glucosamine ornithine, glutamate, alanine, asparagine, glutamine, and proline. d-Mannose is used by strain KR-235T but not by strain KR-242. Starch, casein, and gelatin are degraded. The DNA of strain KR-235T has a G+C content of 71.5 mol%. Source: desert soil from Arizona. Strain KR-242 (LMG 22244 = NRRL B-23994) is the type strain. Strain KR-33 (LMG 22245 = NRRL B-23995) is a reference strain.
Description of Deinococcus yavapaiensis sp. nov. Rainey and da Costa.
Deinococcus yavapaiensis (ya.va.pa.i′en.sis. N. L. masc. adj. yavapaiensis, named after the Yavapai Nation). Deinococcus yavapaiensis forms rod-shaped cells that are 3.0 to 6.0 mm long and 2.0 mm wide. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on NA are dark pink or red. The organism was isolated from an arid soil sample after exposure to 30 kGy, and strain KR-236T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase positive. The major fatty acids are iso 15:0, 16:0, and iso 17:0. Strains utilize d-cellobiose, d-fructose, d-galactose, d-glucose, d-mannose, maltose, l-rhamnose, sucrose, trehalose, glucosamine, lactate, l-glutamine, and proline. Starch, casein, and gelatin are degraded. The DNA of strain KR-236T has a G+C content of 66.1 mol%. Source: desert soil from Arizona. Strain KR-236 (LMG 22171 = NRRL B-2360) is the type strain.
Description of Deinococcus papagonensis sp. nov. Rainey and da Costa.
Deinococcus papagonensis (pa.pa.go′nen.sis. N. L. masc. adj. papagonensis, named after the Tohono O'odham Nation, also known as the Papago Nation). Deinococcus papagonensis forms rod-shaped cells that are 2.0 to 6.0 μm long and 1.5 to 2.0 μm wide. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on NA are light pink. The strains were isolated after exposure to 25 to 30 kGy, and strain KR-241T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase positive. The major fatty acids are iso 15:0, 16:0, and iso 17:0; iso 16:0 3OH and iso 17:0 3OH are present. Strains utilize l-arabinose, cellobiose, fructose, galactose, glucose, d-mannose, maltose, l-rhamnose, sucrose, trehalose, xylose, glucosamine, glutamate, alanine, l-glutamine, ornithine, and proline. Lactose and malate are used by strain KR-241T but not by strain KR-119. Starch and casein are not degraded. Gelatin is degraded. The DNA of strain KR-241T has a G+C content of 69.0 mol%. Source: desert soil from Arizona. Strain KR-241 (LMG 22139 = NRRL B-23961) is the type strain. Strain KR-119 (LMG 22140 = NRRL B-23942) is a reference strain.
Description of Deinococcus sonorensis sp. nov. Rainey and da Costa.
Deinococcus sonorensis (so.no′ren.sis. N.L. masc. adj. sonorensis, named after the Sonora Desert of Arizona). Deinococcus sonorensis forms spherical or short rod-shaped cells that are 2.0 μm long and 1.5 to 2.0 μm wide. Gram staining is positive. The cells are nonmotile; spores are not observed. Colonies on PCA are light pink. The strains were isolated after exposure to 15 to 25 kGy, and strain KR-87T is resistant to >10 kGy. The optimum growth temperature is 30°C. Oxidase positive and catalase positive. The major fatty acids are 15:1 ω6c, iso 16:0, and 16:1 ω7c. Strains utilize l-arabinose, d-cellobiose, d-fructose, d-galactose, d-glucose, lactose, d-mannose, maltose, d-melibiose, l-rhamnose, sucrose, trehalose, d-xylose, glycerol, α-ketoglutarate, glucosamine, ornithine, and proline. Gelatin is degraded. The DNA of strain KR-87T has a G+C content of 70.3 mol%. Source: desert soil from Arizona. Strain KR-114 (LMG 22172 = NRRL B-23941) is the type strain. Strain KR-136 (LMG 22173 = NRRL B-23953) is a reference strain.
Supplementary Material
Acknowledgments
F.A.R. was supported by National Science Foundation awards DEB-971427 and MCB 9977882. This work was supported in part by Fundação para a Ciência e Tecnologia, Portugal PRAXIS/PCNA/BIO/46/96 and POCTI 35029/99, Portugal.
We thank Jean Euzéby, Laboratoire de Bactériologie, École Nationale Vétérinaire, Toulouse, France, for the etymology of the species epithets.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, R., and Y. Haung. 1992. Fatty acids are precursors of alkylamines in Deinococcus radiodurans. J. Bacteriol. 174:7168-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battista, J. R. 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51:203-224. [DOI] [PubMed] [Google Scholar]
- 3.Battista, J. R., and F. A. Rainey. 2001. Order 1. Deinococcales Rainey, Nobre, Schumann, Stackebrandt, and da Costa 1997, 513VP, p. 395-403. In D. Boone, R. Castenholz, and G. Garrity (ed.), Bergey's manual of systematic bacteriology, vol. 1. Springer, New York, N.Y. [Google Scholar]
- 4.Billi, D., E. I. Friedmann, K. G. Hofer, M. G. Caiola, and R. Ocampo-Friedmann. 2000. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl. Environ. Microbiol. 66:1489-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, B. W., and R. G. E. Murray. 1981. Nomenclature for “Micrococcus radiodurans” and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int. J. Syst. Bacteriol. 31:353-360. [Google Scholar]
- 6.Carreto, L., E. Moore, M. F. Nobre, R. Waite, P. W. Riley, R. J. Sharp, and M. S. da Costa. 1996. Rubrobacter xylanophilus sp. nov., a new thermophilic species isolated from a thermally polluted effluent. Int. J. Syst. Bacteriol. 46:460-465. [Google Scholar]
- 7.Cashion, P., M. A. Holder-Franklin, J. McCully, and M. Franklin. 1977. A rapid method for the base ratio determination of bacterial DNA. Anal. Biochem. 81:461-466. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, E. A., and H. Kristensen. 1981. Radiation resistance of microorganisms from air in clean premises. Acta. Pathol. Microbiol. Scand. Sect. B 89:293-301. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, E. A., P. Gerner-Smidt, and H. Kristensen. 1991. Radiation resistance of clinical Acinetobacter spp.: a need for concern? J. Hosp. Infect. 18:85-92. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, M. D., R. A. Hutson, I. R. Grant, and M. F. Patterson. 2000. Phylogenetic characterisation of a novel radiation resistant bacterium from irradiated pork: description of Hymenobacter actinosclerus sp. nov. Int. J. Syst. Evol. Microbiol. 50:731-734. [DOI] [PubMed] [Google Scholar]
- 12.da Costa, M. S., M. F. Nobre, and F. A. Rainey. 2001. The genus Thermus, p. 404-414. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 13.Davis, N. S., G. J. Silverman, and E. B. Masurovsky. 1963. Radiation resistant, pigmented coccus isolated from haddock tissue. J. Bacteriol. 86:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degryse, E., N. Glansdorff, and A. Pierard. 1978. A comparative analysis of extremely thermophilic bacteria belonging to the genus Thermus. Arch. Microbiol. 177:189-196. [DOI] [PubMed] [Google Scholar]
- 15.DiRuggiero, J., N. Santangelo, Z. Nackerdien, J. Ravel, and F. Robb. 1997. Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 179:4643-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donato, M. M., E. A. Seleiro, and M. S. da Costa. 1990. Polar lipid and fatty acid composition of strains of the genus Thermus. Syst. Appl. Microbiol. 13:234-239. [Google Scholar]
- 17.Dose, K., A. Bieger-Dose, M. Labusch, and M. Gill. 1992. Survival in extreme dryness and DNA-single-strand breaks. Adv. Space Res. 12:221-229. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP (phylogenetic inference package), version 3.5.1. Department of Genetics, University of Washington, Seattle.
- 19.Ferreira, A. C., M. F. Nobre, F. A. Rainey, M. T. Silva, R. Waite, J. Burghardt, A. P. Chung, and M. S. da Costa. 1997. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int. J. Syst. Bacteriol. 47:939-947. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira, A. C., M. F. Nobre, E. D. Moore, F. A. Rainey, J. R. Battista, and M. S. da Costa. 2000. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 3:235-238. [DOI] [PubMed] [Google Scholar]
- 21.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 22.Fredrickson, J. K., J. M. Zachara, D. L. Balkwill, D. Kennedy, S. M. Li, H. M. Kostandarithes, M. J. Daly, M. F. Romine, and F. J. Brockman. 2004. Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington state. Appl. Environ. Microbiol. 70:4230-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, I. R., and M. F. Patterson. 1989. A novel radiation-resistant Deinobacter sp. isolated from irradiated pork. Lett. Appl. Microbiol. 8:21-24. [Google Scholar]
- 24.Green, P., and I. J. Bousfield. 1983. Emendation of Methylobacterium Patt, Cole, and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito and Iizuka 1971) comb. nov., corrig.; and Methylobacterium mesophilicum (Austin and Goodfellow 1979) comb. nov. Int. J. Syst. Bacteriol. 33:875-877. [Google Scholar]
- 25.Hensel, R., W. Demharter, O. Kandler, R. M. Kroppenstedt, and E. Stackebrandt. 1986. Chemotaxonomic and molecular-genetic studies of the genus Thermus: evidence for a phylogenetic relationship of Thermus aquaticus and Thermus ruber to the genus Deinococcus. Int. J. Syst. Bacteriol. 36:444-453. [Google Scholar]
- 26.Hirsch, P., C. A. Gallikowski, J. Siebert, K. Peiss, R. Kroppenstedt, P. Schumann, E. Stackebrandt, and R. Anderson. 2004. Deinococcus frigens sp. nov., Deinococcus saxicola sp. nov., and Deinococcus marmoris sp. nov., low temperature and draught-tolerating, UV-resistant bacteria from continental Antarctica. Syst. Appl. Microbiol. 27:636-645. [DOI] [PubMed] [Google Scholar]
- 27.Huang, Y., and R. Anderson. 1995. Glucosyl diglyceride lipid structures in Deinococcus radiodurans. J. Bacteriol. 177:2567-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizuka, H., and H. Ito. 1968. Effect of gamma-irradiation on the microflora of rice. Cereal Chem. 45:503-511. [Google Scholar]
- 29.Ito, H. 1977. Isolation of Micrococcus radiodurans occurring in radurized sawdust culture media of mushroom. Agric. Biol. Chem. 41:35-41. [Google Scholar]
- 30.Ito, H., and H. Iizuka. 1971. Taxonomic studies on a radio-resistant Pseudomonas. Part XII. Studies on the microorganisms of cereal grain. Agric. Biol. Chem. 35:1566-1571. [Google Scholar]
- 31.Ito, H., H. Watanabe, M. Takehisa, and H. Iizuka. 1983. Isolation and identification of radiation-resistant cocci belonging to the genus Deinococcus from sewage sludges and animal feeds. Agric. Biol. Chem. 47:1239-1247. [Google Scholar]
- 32.Jolivet, E., S. L'Haridon, E. Corre, P. Forterre, and D. Prieur. 2003. Thermococcus gammatolerans sp. nov., a hyperthermophilic archaeon from a deep-sea hydrothermal vent that resists ionizing radiation. Int. J. Syst. Evol. Microbiol. 53:847-851. [DOI] [PubMed] [Google Scholar]
- 33.Jolivet, E., S. L'Haridon, E. Corre, S. L'Haridon, P. Forterre, and D. Prieur. 2004. Thermococcus marinus sp. nov., and Thermococcus radiotolerans sp. nov., two hyperthermophilic archaea from deep-sea hydrothermal vents that resist ionizing radiation. Extremophiles 8:219-227. [DOI] [PubMed] [Google Scholar]
- 34.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 35.Karam, P. A., and S. A. Leslie. 1999. Calculations of background beta-gamma radiation dose through geologic time. Health Phys. 77:662-667. [DOI] [PubMed] [Google Scholar]
- 36.Kolari, M., J. Nuutinen, F. A. Rainey, and M. S. Salkinoja-Salonen. 2003. Colored moderately thermophilic bacteria in paper-machine biofilms. J. Ind. Microbiol. Biotechnol. 30:225-238. [DOI] [PubMed] [Google Scholar]
- 37.Kristensen, H., and E. A. Christensen. 1981. Radiation-resistant microorganisms isolated from textiles. Acta Pathol. Microbiol. Scand. Sect. B 89:303-309. [DOI] [PubMed] [Google Scholar]
- 38.Kuykendall, L. D., M. A. Roy, J. J. O'Neill, and T. E. Devine. 1988. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Bacteriol. 38:358-361. [Google Scholar]
- 39.Lewis, N. F. 1973. Radio resistant Micrococcus radiophilus sp. nov. isolated from irradiated Bombay duck (Harpodon nehereus) J. Gen. Microbiol. 66:29-35. [DOI] [PubMed] [Google Scholar]
- 40.Luedemann, G. 1968. Geodermatophilus, a new genus of the Dermatophilaceae (Actinomycetales). J. Bacteriol. 96:1848-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxcy, R. B., and D. B. Rowley. 1978. Radiation-resistant vegetative bacteria in a proposed system of radappertization of meats, p. 347-359. In Food preservation by irradiation, vol. 1. International Atomic Energy Agency, Vienna, Austria. [Google Scholar]
- 43.McGinnies, W. G. 1988. Climatic and biological classifications of arid lands: a comparison, p. 61-68. In E. E. Whitehead, C. F. Hutchinson, B. N. Timmermann, and R. G. Varady (ed.), Arid lands today and tomorrow. Westview Press, Boulder, Colo.
- 44.Mesbash, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 45.Mevs, U., E. Stackebrandt, P. Schumann, C. A. Gallikowski, and P. Hirsch. 2000. Modestobacter multiseptatus gen. nov., sp. nov., a budding actinomycete from soil of the Asgard Range (Transantarctic Mountains). Int. J. Syst. Evol. Microbiol. 50:337-346. [DOI] [PubMed] [Google Scholar]
- 46.Moreira, C., F. A. Rainey, M. F. Nobre, M. T. da Silva, and M. S. da Costa. 2000. Tepidimonas ignava gen. nov., sp. nov., a new chemolithoheterotrophic and slightly thermophilic member of the β-Proteobacteria. Int. J. Syst. Evol. Microbiol. 50:735-742. [DOI] [PubMed] [Google Scholar]
- 47.Murray, R. G. E. 1992. The family Deinococcaceae, p. 3732-3744. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, vol. 4. Springer-Verlag, New York, N.Y. [Google Scholar]
- 48.National Research Council Committee on the Biological Effects of Ionizing Radiation (BEIR III). 1987. The effects on the populations of exposure to low levels of ionizing radiation. NCRP report no. 93. National Council on Radiation Protection and Measurements, Bethesda, Md.
- 49.Nishimura, Y., T. Ino, and H. Iizuka. 1988. Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int. J. Syst. Bacteriol. 38:209-211. [Google Scholar]
- 50.Nishimura, Y., K. Uchida, K. Tanaka, T. Ino, and H. Ito. 1994. Radiation sensitivities of Acinetobacter strains isolated from clinical sources. J. Basic Microbiol. 34:357-360. [DOI] [PubMed] [Google Scholar]
- 51.Nobre, M. F., and M. S. da Costa. 2001. The genus Meiothermus, p. 414-420. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 52.Oyaizu, H., E. Stackebrandt, K. H. Schleifer, W. Ludwig, H. Pohla, H. Ito, A. Hirata, Y. Oyaizu, and K. Komagata. 1987. A radiation resistant odd-shaped bacterium, Deinococcus grandis gen. nov., sp. nov., with peptidoglycan containing ornithine. Int. J. Syst. Bacteriol. 37:62-67. [Google Scholar]
- 53.Phillips, R. W., J. Wiegel, C. J. Berry, C. Fliermans, A. D. Peacock, D. C. White, and L. J. Shimkets. 2002. Kineococcus radiotolerans sp. nov., a radiation-resistant, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:933-938. [DOI] [PubMed] [Google Scholar]
- 54.Prado, A., M. S. da Costa, and V. M. C. Madeira. 1988. Effect of growth temperature on the lipid composition of two strains of Thermus sp. J. Gen. Microbiol. 134:1653-1660. [Google Scholar]
- 55.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 56.Rainey, F. A., M. F. Nobre, P. Schumann, E. Stackebrandt, and M. S. da Costa. 1997. Phylogenetic diversity of the deinococci as determined by 16S ribosomal DNA sequence comparison. Int. J. Syst. Bacteriol. 47:510-514. [DOI] [PubMed] [Google Scholar]
- 57.Rash, B. A. 2004. Analysis of bacterial diversity and biogeography at the Central Arizona-Phoenix Long Term Ecological Research (CAP LTER) site. Ph.D. dissertation. Louisiana State University, Baton Rouge.
- 58.Sanders, S. W., and R. B. Maxcy. 1979. Isolation of radiation-resistant bacteria without exposure to irradiation. Appl. Environ. Microbiol. 38:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharp, R. J., and R. A. D. Williams. 1988. Properties of Thermus ruber strains isolated from Icelandic hot springs and DNA:DNA homology of Thermus ruber and Thermus aquaticus. Appl. Environ. Microbiol. 54:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smibert, R. M., and N. R. Krieg. 1981. General characterization, p. 409-443. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. H. Phillips (ed.), Manual methods for general microbiology. American Society for Microbiology, Washington, D.C..
- 61.Suresh, K., G. S. N. Reddy, S. Sengupta, and S. Shivaji. 2004. Deinococcus indicus sp. nov., an arsenic-resistant bacterium from an aquifer in West Bengal, India. Int. J. Syst. Evol. Microbiol. 54:457-461. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki, K., M. D. Collins, E. Iijima, and K. Komagata. 1988. Chemotaxonomic characterization of a radiotolerant bacterium Arthrobacter radiotolerans: description of Rubrobacter radiotolerans gen. nov., comb. nov. FEMS Microbiol. Lett. 52:33-40. [Google Scholar]
- 63.Tindall, B. J. 1989. Fully saturated menaquinones in the archaebacterium Pyrobaculum islandicum. FEMS Microbiol. Lett. 60:251-254. [Google Scholar]
- 64.United Nations Scientific Committee on Effects of Atomic Radiation. 1982. Ionizing radiation: sources and biological effects. United Nations publication no. E. 82.IX. 8. United Nations, New York, N.Y.
- 65.U.S. Environmental Protection Agency. 1993. Radiation: risks and realities. U.S. Environmental Protection Agency, Washington, D.C.
- 66.Vaisanen, O. M., A. Weber, A. Bennasar, F. A. Rainey, H.-J. Busse, and M. S. Salkinoja-Salonen. 1998. Microbial communities of printing paper machines. J. Appl. Microbiol. 84:1069-1084. [DOI] [PubMed] [Google Scholar]
- 67.Weernink, A., W. P. Severin, I. Tjernberg, and L. Dijkshoorn. 1995. Pillows, an unexpected source of Acinetobacter. J. Hosp. Infect. 29:189-199. [DOI] [PubMed] [Google Scholar]
- 68.Weisberg, W. G., S. J. Giovannoni, and C. R. Woese. 1989. The Deinococcus-Thermus phylum and the effects of rRNA composition on phylogenetic tree construction. Syst. Appl. Microbiol. 11:128-134. [DOI] [PubMed] [Google Scholar]
- 69.Welch, A. B., and R. B. Maxcy. 1979. Characteristics of some radiation-resistant hemolytic micrococci isolated from chicken. J. Food Sci. 44:673-675. [Google Scholar]
- 70.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woese, C. R., E. Stackebrandt, T. J. Macke, and G. E. Fox. 1985. A phylogenetic definition of the major eubacterial taxa. Syst. Appl. Microbiol. 6:143-151. [DOI] [PubMed] [Google Scholar]
- 72.Yamada, K., and K. Komagata. 1972. Taxonomic studies on coryneform bacteria. IV. Morphology, cultural, biochemical, and physiological characteristics. J. Gen. Appl. Microbiol. 18:399-416. [Google Scholar]
- 73.Yoshinaka, T., K. Yano, and H. Yamaguchi. 1973. Isolation of highly radioresistant bacterium, Arthrobacter radiotolerans nov. sp. Agric. Biol. Chem. 37:2269-2275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.