Abstract
The food-borne pathogen Listeria monocytogenes is a ubiquitous soil bacterium with the potential to contaminate fresh produce during cultivation and postharvest processing. In order to identify potential mechanisms by which L. monocytogenes may successfully attach to and colonize fresh produce, gene expression in L. monocytogenes cells inoculated onto fresh-cut cabbage was compared to gene expression in cells grown under control conditions. Differential display of reverse transcriptase PCR fragments amplified with a set of 81 arbitrary primers allowed the isolation and identification of 32 L. monocytogenes gene fragments that were observed to be more highly expressed under cabbage-associated conditions. Genes involved in carbohydrate, amino acid, and nucleic acid metabolism, motility and cell division, and transport were identified, as were a number of open reading frames (ORFs) encoding putative proteins with no known functions. Site-directed mutations in two ORFs encoding potential cell surface-associated proteins and a third ORF encoding a putative regulatory protein had no effect on the mutants' capacity to attach to fresh-cut cabbage. Although this study did not show clearly the impact of the differentially expressed genes on growth on cabbage, it is a first step in identifying some of the genetic factors that are potentially involved.
Contamination of fresh-cut produce by Listeria monocytogenes is a great concern for both consumers and food processors. In the United States, there are approximately 2,500 cases of listeriosis per year, which result in about 500 deaths (21). The disease is systemic and can result in severe complications, such as septicemia, liver failure, meningitis, and abortion, in susceptible individuals (29). This gram-positive, facultatively intracellular pathogen is a saprophyte in its natural soil and agricultural niche, from which it can contaminate both plants and animals (5, 33). L. monocytogenes also grows well in food processing environments due to its ability to grow at refrigeration temperatures and its ability to survive within biofilms that can resist sanitation procedures. These abilities allow postharvest contamination of food (14).
The first recognized instance of human listeriosis due to food contamination was a 1981 outbreak caused by contaminated coleslaw, which resulted in 41 cases and 18 deaths. The contamination was traced to a cabbage field on a farm known to have had cases of ovine listeriosis (30). An understanding of the physiology of L. monocytogenes when it is in association with plants may lead to better methods of eliminating it from the food supply. Toward that end, we set out to study the gene expression of L. monocytogenes in a fresh-cut cabbage system. We compared cDNA from L. monocytogenes grown in medium to cDNA from L. monocytogenes grown with cut cabbage, using a differential display method following reverse transcriptase PCR (RT-PCR), to identify genes that were upregulated when L. monocytogenes was grown in association with cabbage and in cabbage-associated bacteria. In an attempt to characterize L. monocytogenes growth and attachment to cabbage, several of the genes identified were selectively mutated in an attempt to determine their effects on growth and/or attachment in the produce environment.
MATERIALS AND METHODS
Bacterial strains and media.
L. monocytogenes strain 10403 (11) and derivatives of this strain were grown in brain heart infusion medium (BHI) (Difco, Becton Dickinson, Franklin Lakes, NJ) or Listeria minimal medium (LMM) (26). RM2980 is a strain 10403 derivative that has an insertion of transposon Tn917-LTV3 into an unknown gene within a flagellar biosynthetic operon, which results in a mutant that has no flagella and is nonmotile (12). L. monocytogenes colonies were isolated and enumerated on modified Oxford agar (MOX) (Difco), which inhibits gram-negative bacteria and many gram-positive bacteria. These media were supplemented with erythromycin (1 μg ml−1) and lincomycin (25 μg ml−1), with chloramphenicol (15 μg ml−1), or with kanamycin (40 μg ml−1) when necessary. Escherichia coli strain DH10B (13) was used as a host for general cloning, and E. coli strain S17-1 (31) was used for conjugative transfer of plasmids into L. monocytogenes. E. coli strains were grown on LB medium (Difco) at 37°C, and this medium was supplemented with ampicillin (100 μg ml−1), chloramphenicol (15 μg ml−1), kanamycin (40 μg ml−1), or erythromycin (300 μg ml−1) when necessary.
Preparation of L. monocytogenes total RNA.
L. monocytogenes strain 10403 was grown overnight at 25°C in BHI. To isolate total RNA from cells expressing plant-associated genes, cultures were pelleted by centrifugation and washed once by resuspending them in an equal volume of sterile phosphate-buffered saline (PBS) (150 mM NaCl, 10 mM sodium phosphate; pH 7.2). Cells were diluted 1:1,000 in sterile PBS, and 20 ml of the cell suspension was added to 50 g of shredded green cabbage in polyethylene 1-qt bags (Ziploc). The cabbage was purchased at a local grocery store and used within 2 days of purchase. The outer four leaves were removed, and the cabbage was shredded into strips that were approximately 3 to 6 mm wide. The liquid was dispersed so that all the cabbage was wetted by the cell suspension, and the bags were incubated for 16 h at 25°C. The cabbage was washed three times with 50 ml of PBS, and attached L. monocytogenes cells were removed with 50 ml of fresh PBS by sonication for 15 min in an ice-water bath (Aquasonic model 75T; VWR Scientific Products, West Chester, PA). Bacteria were recovered by centrifugation, and the cell pellets were stored at −70°C until they were used.
The L. monocytogenes cultures used for isolation of control RNA consisted of PBS-washed cells diluted 1:1,000 in LMM and incubated for 16 h at 25°C without shaking. Twenty-milliliter cultures were then pelleted by centrifugation, resuspended in 50 ml PBS, and added to shredded green cabbage that had been incubated for 16 h at 25°C without inoculation. Samples were immediately sonicated on ice for 15 min to mimic the experimental conditions described above. The cell suspension was removed from the cabbage and pelleted by centrifugation. The cell pellets were stored at −70°C until they were used.
Cell pellets were resuspended in 100 μl of Tris-EDTA buffer containing 3 mg/ml lysozyme and 20 U each of HPL118 and HPL511 recombinant bacteriophage endolysins (18) and incubated at room temperature for 10 min. RNA was isolated using an RNeasy Mini kit (QIAGEN, Inc., Valencia, CA). Residual DNA contamination in RNA samples was removed by heating the samples to 95°C for 3 min and cooling them on ice to denature any DNA-RNA heteroduplexes, followed by treatment with DNase (DNA-free; Ambion, Austin, TX). RNA samples from three independent preparations were combined to minimize the variability, diluted to a concentration of 0.1 μg/μl, and stored at −70°C until they were used.
RT-PCR.
A set of 81 oligonucleotide primers was used for RT-PCRs, as described by Brzostowicz et al. (7). The primer sequences were 5′-GGTACGGGCATTCnnnn-3′, where n is A, G, or C. The 5′ 13-mer portion of each primer was designed to minimize annealing to rRNA sequences in L. monocytogenes. RT-PCRs were performed using a ProSTAR HF single-tube RT-PCR kit (Stratagene, La Jolla, CA). Briefly, each 25-μl reaction mixture contained 15 pmol of a single primer, 0.1 μg of total RNA, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 2.5 μl of 10× HF RT-PCR buffer, 1.25 U of StrataScript reverse transcriptase, and 1.25 U of TaqPlus Precision DNA polymerase mixture. Each primer was used in combination with total RNA from control and induced L. monocytogenes cells. The RT-PCR program was as follows: 37°C for 30 min for reverse transcription; 95°C for 1 min, 37°C for 5 min, and 68°C for 5 min for second-strand synthesis; and 40 cycles of 95°C for 30 s, 52 to 59°C (depending on the calculated melting temperature of the primer) for 30 s, and 68°C for 2 min.
Identification of differentially expressed genes.
The results of RT-PCRs with control and induced RNAs were analyzed side by side following electrophoresis on 5% polyacrylamide Tris-borate-EDTA gels and staining with SYBR Gold nucleic acid stain (Molecular Probes, Eugene, OR). Amplified DNA fragments that were present in reactions with induced RNA but absent in reactions with control RNA were reamplified from gels using the “bandstab” method (36). Briefly, each of these fragments was stabbed from the differential display gel with a gel-loading pipette tip and swirled in a 25-μl PCR mixture containing the same primer used for RT-PCR of the fragment. Reamplified fragments were cloned into pDRIVE (PCR cloning kit; QIAGEN). Cloned fragments were sequenced with the M13(-20) and M13R sequencing primers. Sequences were identified based on alignment with the L. monocytogenes strain EGD-e genomic sequence using the ListiList database tool (http://genolist.pasteur.fr/ListiList/index.html) (23). In cases where amplified fragments resulted from RNA from non-Listeria cabbage-associated bacteria, sequences were identified by similarity to sequences in the GenBank database using the BLASTN and BLASTX programs (1) at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST).
Real-time RT-PCR.
For real-time RT-PCR RNA was prepared as described above. The primers used are shown in Table 1. Each real-time RT-PCR mixture (total volume, 50 μl) consisted of 25 μl of 2× SYBR Green PCR master mixture (Applied Biosystems, Foster City, CA), 0.1 μg of total RNA, 25 pmol of the forward primer, 25 pmol of the reverse primer, and 10 U of StrataScript reverse transcriptase (Stratagene). The cycling program was as follows: 48°C for 30 min, 95°C for 10 min, and then 40 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 45 s. Incorporation of SYBR Green into RT-PCR products was monitored with an iCycler-iQ real-time PCR detection system (Bio-Rad, Hercules, CA).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a | Restriction site |
|---|---|---|
| Real-time RT-PCR | ||
| flgKf | TCGCGTGAACCAAAATGAAAT | |
| flgKr | CCGCCGCGATAACACCTA | |
| tcsAf | GCATGGGAAGGCTTACAAAAAT | |
| tcsAr | TGCAAAGACACCGTTACCAGT | |
| lmo0160f | GATGGCAACACGGTAGGTCA | |
| lmo0160r | TCGCCGTGGAATGCTTTTAT | |
| lmo0449f | TGAAGGACAAAACAAAGAAAAGAA | |
| lmo0449r | CGGACTCATCGAAAGGGTAAT | |
| lmo0528f | GGCGGAAATCCACCAACAT | |
| lmo0528r | GGCCCTGCACTCACGATAAA | |
| lmo0770f | TTGGCGATACAGAGCGAGAAG | |
| lmo0770r | AACAACGGATAAATCAGCAGGAA | |
| lmo0998f | TGTTTTATGCGCCCTACTCGT | |
| lmo0998r | ACTCGGGAACATCATTACCATACA | |
| lmo1726f | GCACGCCACATTCAAGAGC | |
| lmo1726r | GGGCCAGCGAGTTCAGTTT | |
| lmo2381f | TTCCGGTGCTTGCTTTGTTT | |
| lmo2381r | CCCGCCGACCATTTGAC | |
| lmo2678f | GAAAAGAAGCGCTAGAACAGACG | |
| lmo2678r | GTGGCGAGCTAACAGGAGTAAAA | |
| Mutant construction | ||
| lmo0160 5′fB | GCCGGATCCAACTGTGCCACCAACAAA | BamHI |
| lmo0160 5′rG | CCCAGATCTTTTCAAAAAATCGGACCCATAAT | BglII |
| lmo0160 3′fG | GCCAGATCTACGGTTGCTGATTTACTTCC | BglII |
| lmo0160 3′rB | GGGGGATCCGGCCGTGTAGATCGTGAAA | BamHI |
| lmo2678 5′fB | GGCGGATCCTTGTTGGTGAGGCGGTTG | BamHI |
| lmo2678 5′rG | GCCAGATCTGTCTGTTCTAGCGCTTCTTTTC | BglII |
| lmo2678 3′fG | TCCAGATCTAAATCAAGCGCTTCGAGTCAAT | BglII |
| lmo2678 3′rB | GGGGGATCCGTTCGGGTAGCGTTCTTC | BamHI |
| tcsA 5′fB | CCCGGATCCGCAATTAACGAAGCCATAC | BamHI |
| tcsA 5′rG2 | GTCAGATCTACGGTCATCAACGCCACCAG | BglII |
| tcsA 3′fG2 | TGAAGATCTTGTTTGGGTAATCGGTGTTGAC | BglII |
| tcsA 3′rB | CCCGGATCCTCCCCTTGTCGTCTGGTTC | BamHI |
Restriction sites are indicated by boldface type.
Construction of mutant L. monocytogenes strains.
Primers used for construction of mutant L. monocytogenes strains are shown in Table 1. A BamHI restriction fragment containing the erythromycin resistance cassette (Emr) from pAUL-A (28) was constructed by digesting pAUL-A with ClaI and KpnI and cloning the resulting 2-kb fragment into pBluescript SK(+) (Stratagene). To construct an insertion mutant with a mutation in open reading frame (ORF) lmo0160, the full-length ORF was PCR amplified with primers lmo0160 5′fB and lmo0160 3′rB, both of which contained a BamHI restriction site, and cloned into pDRIVE (QIAGEN), resulting in pDR0160. The central portion of the lmo0160 ORF was deleted by performing inverse PCR (Expand Long Template PCR kit; Roche, Indianapolis, IN) using primers lmo0160 5′rG and lmo0160 3′fG, both of which contained a BglII restriction site, followed by digestion with BglII and self-ligation, resulting in pDR0160Δ. The Emr BamHI cassette was then cloned into the BglII site, resulting in pDR0160Δ::Emr. The lmo0160Δ::Emr fragment from pDR0160Δ::Emr was excised as a BamHI fragment and cloned into pCON1, which carried a chloramphenicol resistance marker (3). The resulting plasmid, pCON0160Δ::Emr, was transformed into E. coli strain S17-1 for conjugation. Similar plasmid constructs for mutagenesis of the ORFs lmo2678 and tcsA were made by using the same methods that were used for lmo0160.
For conjugation, cultures of L. monocytogenes 10403 and E. coli S17-1 harboring each mutagenic construct were grown to the mid-exponential phase (optical density at 600 nm [OD600], 0.5). Five hundred microliters of each culture was mixed, pelleted by centrifugation, and resuspended in 50 μl of BHI. Cells were spotted onto BHI agar, allowed to dry, and incubated at 30°C overnight. Cells were then scraped off the agar and resuspended in 1 ml of PBS. Aliquots (100 μl) were spread on MOX (since E. coli does not grow on this medium) containing erythromycin, lincomycin, and chloramphenicol and incubated at 30°C to select for maintenance of the temperature-sensitive pCON1-derived plasmid. L. monocytogenes transconjugants were plated on the same medium and shifted to 42°C to select for integration of the plasmid by a single recombination event. Colonies arising on these plates were subcultured on MOX containing erythromycin and lincomycin and incubated at 30°C to allow a second recombination event, which excised the vector portion of the plasmid. Recombinant L. monocytogenes strains were confirmed by PCR using, for example, primers lmo0160 5′fB and lmo0160 3′rB to demonstrate replacement of the parental ORF with the engineered fragment containing an internal deletion and insertion of the 2-kb Emr fragment.
Cabbage colonization assays.
Cultures of L. monocytogenes parental and mutant strains were grown in 10 ml of LMM for 16 h at 28°C. Cells were pelleted by centrifugation and resuspended in 35 ml of sterile PBS. For each experiment, the OD600s of the parental and mutant cell suspensions were adjusted to 0.1, and 10 ml of each cell suspension was added to 25 g of shredded green cabbage in a 1-qt polyethylene bag (Ziploc). The inoculating cell suspensions were enumerated by dilution plating on MOX. Inoculated cabbage samples were incubated for 16 h at 25°C. Cabbage samples were washed three times with 50 ml of PBS, and colonizing bacteria were removed with 50 ml of fresh PBS by sonication for 15 min in an ice-water bath. Recovered L. monocytogenes cells were enumerated by dilution plating on MOX. For single-strain tests, colonization of cabbage by mutant strains and colonization of cabbage by the parental L. monocytogenes strain were compared following recovery of bacteria from cabbage samples inoculated with individual cell suspensions. For competitive colonization assays, cell suspensions of the parental and mutant L. monocytogenes strains were adjusted so that the OD600s were equal, and equal volumes were mixed before the OD600 of the inoculum was adjusted to 0.1. Inoculated and recovered cell suspensions were dilution plated on MOX and MOX containing erythromycin and lincomycin in order to enumerate total and mutant L. monocytogenes cells, respectively. In these experiments, the number of wild-type cells recovered was calculated by subtracting the number of CFU recovered on MOX with erythromycin and lincomycin from the number of CFU recovered on plain MOX. Each experiment consisted of three replicate samples for each strain, and each experiment was performed at least twice.
To determine if the mutants had defects early in the attachment process, we measured short-term attachment to cut cabbage. Cultures were grown in LMM at 28°C, and the OD600 was adjusted to 0.1 with PBS. Forty microliters of cell suspension was added to 50-ml polypropylene, disposable centrifuge tubes that contained 10 g of shredded cabbage in 20 ml of PBS. The tubes were placed on their sides in a plastic tub that was placed on a Belly Dancer shaker (Stovall Life Science, Inc., Greensboro, NC) at the maximum setting on a bench top. The tubes were incubated for 4 h, after which the liquid was removed and the cabbage was washed three times with 50 ml of PBS. Attached bacteria were removed with 50 ml of fresh PBS by sonication for 15 min in an ice-water bath and were enumerated by dilution plating onto MOX.
RESULTS
Differential display of L. monocytogenes gene expression.
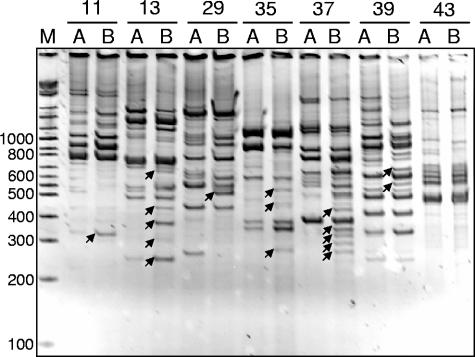
Each RNA sample was randomly amplified by RT-PCR, and reaction pairs were run in adjacent wells on polyacrylamide gels (Fig. 1). RT-PCR fragments that appeared to be unique or more abundant in lanes corresponding to reactions containing cabbage-associated RNA than in lanes corresponding to reactions containing control RNA were interpreted as a reflection of increased expression of genes encoded on the fragments. Of 145 RT-PCR fragments identified on differential display gels as fragments that were upregulated under cabbage-associated conditions, 81 were sufficiently resolved to allow reamplification using the same arbitrary primer that was used for the RT-PCR. Forty-six of these reamplified fragments were cloned and sequenced, and 32 of them were 97 to 100% identical to sequences present in the sequenced L. monocytogenes strain EGD-e genome (13). The remaining 14 fragments were homologous to genes from plant-associated bacteria, including Pantoea, Pseudomonas, and Erwinia species.
FIG. 1.
Differential display gels. RT-PCR mixtures containing template RNA from control (lanes A) and cabbage-associated (lanes B) L. monocytogenes cells and individual arbitrary primers (indicated by numbers above the lanes) were separated on 5% polyacrylamide gels. The positions of size markers (in base pairs) (lane M) are indicated on the left. Differentially amplified RT-PCR fragments (arrows) indicate differential gene expression in cabbage-associated L. monocytogenes cells.
Identification of differentially expressed genes.
Table 2 shows the L. monocytogenes genes identified by sequence analysis of cloned RT-PCR fragments, organized by putative operon function. Fragments of several genes related to nucleic acid and amino acid biosynthesis and metabolism were identified, including 23S rRNA genes, which were identified from three independent RT-PCR fragments. Genes for pyrimidine biosynthesis (pyrAB and pyrC) were cloned on different RT-PCR fragments. Other genes identified in this category encode DNA polymerases, endonuclease IV, threonyl-tRNA synthase, and ribosomal proteins.
TABLE 2.
L. monocytogenes ORFs identified by RT-PCR differential display
| Cloned fragmenta | Putative gene function | Putative operon function |
|---|---|---|
| Nucleic acid and amino acid metabolism | ||
| dnaE | DNA polymerase III alpha subunit | DNA metabolism |
| polA | DNA polymerase I | DNA metabolism |
| thrS | Threonyl-tRNA synthetase | DNA metabolism |
| pyrAB | Carbamoyl phosphate synthetase | Pyrimidine biosynthesis |
| pyrC | Dihydroorotase | Pyrimidine biosynthesis |
| lmo2565 | Unknown | RNA metabolism |
| lmo1449 | Endonuclease IV | |
| lmo1437 | Aspartate semialdehyde dehydrogenase | Amino acid biosynthesis |
| pheA | Prephenate dehydratase | Phenylalanine biosynthesis |
| rplX, rplE | Ribosomal protein L24, ribosomal protein L5 | Ribosomal proteins |
| 23S rRNA | 23S ribosomal RNA | rRNA |
| Carbohydrate metabolism | ||
| pgi | Glucose-6-phosphate isomerase | |
| ssrA | Small stable RNA A | Glycosidases |
| lmo1726 | Unknown | β-Glucosidase; cellobiose phosphorylase |
| Motility and cell division | ||
| flgK, flgL | Flagellar hook-associated proteins | Flagellar assembly |
| fliR | Flagellar biosynthetic protein | Flagellar assembly and motility |
| lmo1543 | RNase G | Cell shape determination, cell division |
| lmo2689, ftsW | Mg2+ transport ATPase, cell division protein | Cell division |
| Transporters | ||
| lmo2678 | Two-component response regulator KdpE | ATPase-type K+ transporter |
| lmo2677, lmo2678 | Similar to hydrolase (esterase); two-component response regulator KdpE | ATPase-type K+ transporter |
| tcsA | T-cell-stimulating antigen | Sugar ABC transporter |
| lmo1999, lmo1998 | Glucosamine-fructose-6-phosphate aminotransferase, opine catabolism | Mannose-specific phosphotransferase transport system |
| lmo2194 | Oligopeptide ABC transporter (permease) | Oligopeptide/pheromone ABC transporter |
| lmo2381 | Na+/pH homeostasis | Na+/H+ antiporter (pH homeostasis) |
| lmo2469 | Amino acid transporter | |
| Unknown function | ||
| lmo0160 | Peptidoglycan-bound protein | |
| lmo0449 | Unknown | |
| lmo0528 | Hypothetical secreted protein | |
| lmo0770 | Lacl family transcription regulator | |
| lmo0998 | Unknown |
The ORF designations correspond to those assigned for the L. monocytogenes strain EGD-e genome sequence, as shown in the ListiList database web server (http://genolist.pasteur.fr/ListiList/index.html).
Three RT-PCR fragment clones contained sequences identified as genes associated with carbohydrate metabolism. These included pgi, encoding glucose-6-phosphate isomerase, a glycolytic and gluconeogenic enzyme. Other RT-PCR fragments in this category showed sequence identity to ssrA, encoding a ribozyme involved in tagging improperly folded peptides for degradation, and to the ORF lmo1726, encoding a putative protein with an unknown function. These fragments were included in this category based on context. ssrA is located in an intergenic region downstream of two putative glycosidase genes and upstream of a putative transcription regulator with an unknown function, and lmo1726 occurs downstream of three ORFs with sequence similarities to β-glucosidase, cellobiose phosphorylase, and a LacI family transcription regulator. Both ssrA and lmo1726 are potentially cotranscribed with their surrounding ORFs, although the structure of each transcription unit is unknown.
Several RT-PCR fragments were related to motility and cell division, including fragments of operons encoding flagellar structural and assembly proteins. Also included in this category were two fragments, one containing lmo1543, encoding a putative RNase G homolog, which occurs downstream of (and may be cotranscribed with) several cell shape-determining protein homologs, and the other containing lmo2689, encoding a putative Mg2+ transport ATPase, upstream of two ORFs encoding proteins with similarity to cell division protein FtsW.
Several transport operons were identified in the cloned sequences, including an ATPase-type K+ transporter, which was identified in two independent RT-PCR fragments. The other transporters included a putative ABC-type sugar transporter, a putative ABC-type oligopeptide/pheromone transporter, a putative amino acid transporter, and a putative Na+/H+ antiporter system involved in pH homeostasis. Also included in this category was an RT-PCR fragment with sequence similarity to lmo1999 and lmo1998, encoding putative glucosamine-fructose-6-phosphate aminotransferase and opine catabolism proteins, respectively, which are located in an operon encoding a putative mannose-specific phosphotransferase transport system. Interestingly, tcsA, a putative lipoprotein gene, which is located upstream of the putative ABC-type sugar transporter gene, encodes a protein that functions as a T-cell-stimulating antigen, suggesting that it is cell surface bound.
A number of differentially displayed RT-PCR fragments contained sequences with homology to genes having unknown functions. ORFs lmo0770 and lmo0998 appear to be transcribed divergently from adjacent genes that also have no known functions, and lmo0160 appears to be transcribed as a monocistron, so no functional data for these genes can be discerned by their context. ORF lmo0528 occurs downstream of a gene encoding a putative transmembrane protein with an unknown function and upstream of a gene encoding a putative glucosaminyltransferase homolog. ORF lmo0449 occurs downstream of genes encoding putative proteins with homology to penicillin acylase and to conjugated bile acid hydrolase (lmo0446), a putative glutamate decarboxylase (lmo0447), and a putative amino acid antiporter (lmo0448).
Confirmation of differential expression.
Real-time RT-PCR was performed with a subset of the genes identified by differential display to measure the extent to which they were differentially expressed under the two growth conditions. The genes selected for real-time RT-PCR are shown in Table 3. PCR primers (Table 1) were designed using the genomic sequence of each identified ORF to amplify a <500-bp fragment, and relative fluorescence due to incorporation of SYBR Green into each PCR product was measured relative to the RNA sample type. As shown in Table 3, 9 of the 10 genes assayed by real-time RT-PCR showed a decrease in the number of cycles needed to reach a threshold fluorescence level in samples containing RNA from cabbage-associated L. monocytogenes cells relative to samples containing RNA from control cells. The difference in the number of cycles needed to pass the threshold level between sample RNAs for each gene ranged from −0.2 to −12.5, indicating that there were differences in the extent of differential expression of each gene under the two conditions tested. Thus, with the exception of lmo0528, increased expression of these genes as determined by random-primed RT-PCR and differential display was corroborated by real-time RT-PCR using gene-specific primers.
TABLE 3.
Evidence of differential gene expression obtained by quantitative RT-PCR
| ORF | Threshold cycle
|
Difference in expressionb | |
|---|---|---|---|
| Control RNA | “+ Cabbage” RNAa | ||
| flgK | 20.6 | 18.6 | −2.0 |
| tcsA | 31.3 | 18.8 | −12.5 |
| lmo0160 | 27.0 | 25.0 | −2.0 |
| lmo0449 | 27.4 | 23.1 | −4.3 |
| lmo0528 | 23.3 | 23.9 | 0.6 |
| lmo0770 | 23.5 | 21.7 | −1.8 |
| lmo0998 | 33.2 | 28.7 | −4.5 |
| lmo1726 | 18.9 | 18.7 | −0.2 |
| lmo2381 | 20.5 | 17.6 | −2.9 |
| lmo2678 | 22.7 | 18.5 | −4.2 |
“+ Cabbage” RNA, RNA from cells expressing plant-associated genes.
Calculated as follows: number of threshold cycles in “+ cabbage” samples − number of threshold cycles in control samples.
Cabbage colonization by site-directed mutants.
The capacity of mutants with mutations in lmo0160, lmo2678, and tcsA to colonize cut cabbage was compared with the capacity of parental L. monocytogenes strain 10403, using both single-strain assays and competitive assays in which mutant and parental strains were coinoculated onto cabbage samples. In addition to these mutants, we also tested RM2980, a flagallar mutant found in a previous study to be 1-log reduced in attachment to cut radish tissue (12). The results of 16-h colonization assays (Table 4) indicated that none of the mutants were impaired for colonization of cabbage surfaces compared to the parental levels, either singly or in competition with wild-type L. monocytogenes. In single cultures, approximately 7 log CFU/g of each strain was recovered. In mixed cultures, 6.2 to 7.8 log CFU/g was the range recovered; however, the results were consistent since the ratio of mutant cells to wild-type cells recovered was approximately 1:1. Since no differences were discerned with 16 h of incubation, short-term assays were used to determine whether mutant cells were impaired in the initial stages of colonization compared to parental cells. Again, the results of these assays (Table 4) showed that the mutations in the genes examined did not alter the initial colonization capacity of the mutants compared to the parental strain, and approximately 3 log CFU/g was recovered for all strains.
TABLE 4.
Cabbage colonization by mutant and parental L. monocytogenes strains in long- and short-term colonization assays
| Strain | Recovered population (log CFU/g cabbage)a |
|---|---|
| 16-h single-strain assay | |
| Wild type | 7.2 ± 0.22 |
| lmo0160 | 7.4 ± 0.15 |
| lmo2678 | 7.0 ± 0.04 |
| tcsA | 7.3 ± 0.20 |
| fla− | 7.8 ± 0.10 |
| 16-h competition assay | |
| lmo0160 vs wild type | 6.75 ± 0.20, 6.2 ± 0.30b |
| lmo2678 vs wild type | 7.75 ± 0.25, 7.8 ± 0.35 |
| tcsA vs wild type | 7.20 ± 0.09, 7.3 = 0.08 |
| fla− vs wild type | 6.70 ± 0.07, 6.80 ± 0.13 |
| 4-h single-strain assay | |
| Wild type | 3.1 ± 0.05 |
| lmo0160 | 3.1 ± 0.10 |
| lmo2678 | 2.9 ± 0.20 |
| tcsA | 3.2 ± 0.04 |
| fla− | 3.3 ± 0.02 |
The data are means ± standard deviations for at least two replicate samples.
In competition assays, the first and second values are the values for the mutant and wild-type populations, respectively.
DISCUSSION
While L. monocytogenes is best known as a food-borne pathogen, in nature it lives in association with plants and decaying plant tissue, and little is known of its physiology in this environment. The capacity of this organism to grow in a fresh-cut produce environment, including cabbage, has been well documented (4, 8, 10, 16, 25). We used a differential display method that compared the transcription of L. monocytogenes in two different environments: growth on plant tissue and growth in minimal medium. To our knowledge, this is the first assessment of L. monocytogenes transcription in a complex environment interacting with nonsterile plant surfaces. Wilson et al. identified genes induced by L. monocytogenes in J774 tissue culture cells by use of fluorescence-activated cell sorting of a chromosomal library of L. monocytogenes DNA cloned in front of a promoterless green fluorescent protein gene (35). Other studies that showed a snapshot of transcription or global protein expression by L. monocytogenes, including studies of biofilm versus planktonic cells, studies of cells after acid exposure, at low temperature, and during salt stress, studies of wild-type and rpoN mutant cells, studies of virulence gene expression controlled by the major virulence gene regulator prfA, and studies of cells in transition from exponential growth to the stationary phase, have been done with pure cultures in medium (2, 9, 15, 17, 22, 32, 34). Some of these studies identified genes or operons similar to those found in our study, and they are mentioned below.
On a plant surface either in soil or in fresh-cut produce, L. monocytogenes must interact not only with the plant tissue but also with other bacteria. When control and cabbage-associated RT-PCRs with any given primer were compared (Fig. 1), the resulting band patterns were practically identical, indicating that the RNA preparations from the two different environments were of comparable quality. While RT-PCR fragments arising from contaminating bacteria were isolated, the majority of the fragments that were analyzed resulted from L. monocytogenes.
While the genes identified in this screening were not an exhaustive list of every gene induced under plant growth conditions, the method did provide a snapshot of genes induced in the cabbage growth and attachment milieu. Under the conditions examined, distinctions could not be made between genes upregulated as a result of cursory surface attachment and genes upregulated due to tight, specific interactions, although we attempted to minimize the contribution from the former via extensive washing and sonication. Many of the genes identified did make sense when we compared bacterial transcription for two different growth environments, minimal medium and cut cabbage. The nutrients available for growth in the cabbage system are in the form of organic acids, monosaccharides, peptides, and amino acids as a result of leakage from the damaged portion of the cabbage tissue, since these compounds are known to be in exudates from roots and cut produce (19, 24). L. monocytogenes can use sugars as carbon sources, and while it is unable to hydrolyze whole proteins for growth, studies have shown that it can scavenge peptides from proteins hydrolyzed by other bacteria, such as Pseudomonas spp., which were present in the system (6, 20). It would be expected that multiple transporter systems for various growth substrates would be induced to aid the cell in using the available nutrients. The transporters induced included a mannose-specific phosphotransferase system, a sugar ABC transporter, and potassium and peptide transporters. The sugar transport systems would be induced in response to the presence of new carbon sources resulting from degradation or scavenging of cell wall components (15). The source of the sugars could be either L. monocytogenes, the other bacteria present, or the cabbage cells themselves.
Several biosynthetic genes were identified; these genes included genes for amino acid production, possibly indicating that the concentrations of the amino acids in the growth environment were low. pheA is needed for phenylalanine biosynthesis, and aspartate semialdehyde dehydrogenase is involved in the synthesis of aspartate, which is needed for the biosynthesis of isoleucine, leucine, threonine, and methionine. Additionally, two genes involved in pyrimidine biosynthesis, pyrAB and pyrC, were induced under cabbage growth conditions. In addition to pyrimidine biosynthesis, pyrAB, which catalyzes the formation of carbamyl phosphate, is also used in the formation of arginine from glutamic acid. In other studies, amino acid biosynthetic genes were also found to be induced in L. monocytogenes upon salt stress (9), indicating possible adaptation to either stress and/or a new environment.
One of the induced genes was lmo2381, which is part of an operon involved in pH homeostasis. The experiment as it was set up was only weakly buffered, and with the metabolic activity of normal cabbage-associated bacteria, the pH was likely reduced during the 16 h of incubation, meaning that this stress had to be dealt with. Another possible indication of cellular stress was the induction of the mannose-specific phosphotransferase system (as mentioned above). This operon, which was recently shown to be downregulated in an rpoN mutant (2), was also induced by L. monocytogenes during intracellular growth in J774 macrophages (35), in response to salt stress (9) and glucose starvation (15), indicating that some of the L. monocytogenes population may have been stressed in the cabbage growth environment. However, none of the classic stress response genes (such as groEL or dnaK) were identified, as in previous studies of L. monocytogenes during salt or cold stress (9, 17).
It is possible that the cultures in the two environments were in different stages of growth. The control culture was most likely in the late exponential or early stationary phase when it was harvested, but in the cabbage-grown cells it is possible that there were different cell populations (e.g., attached and planktonic cells or logarithmic and stationary-phase cells). There was an indication, however, that L. monocytogenes was actively growing because of the upregulated operons that are important in metabolically active and growing cells. Actively growing cells express higher levels of the machinery for DNA replication, protein synthesis, and cell division, which includes the genes identified, such as polA, dnaE, thrS, and ftsW, as well as genes involved in RNA degradation (RNase G) and the degradation of incomplete polypeptides (ssrA). The induction of the glycolytic/gluconeogenic enzyme pgi is also an indication that the cells were metabolically active. In other studies, glycolytic enzymes were induced in L. monocytogenes during salt stress (9) and in the presence of glucose when the cells were in a biofilm (15), both of which are conditions in which the cells would be adapting to new environments.
Interestingly, several of the classes of genes identified in this study have been shown to be essential for the colonization of plants by saprophytic Pseudomonas spp. and Rhizobium meliloti. Genes important for motility and for amino acid, vitamin, and nucleotide biosynthesis have been shown to be important for colonization of legumes by R. meliloti and colonization of tomato by Pseudomonas fluorescens (19). Genes involved in nutrient acquisition and a stress response were induced in P. fluorescens in the rhizosphere (27). While the growth environment that we provided for L. monocytogenes contained fresh-cut cabbage rather than cabbage plants in soil, it is interesting that a similar battery of genes (nutrient transport, amino acid biosynthesis, motility, pH homeostasis) was induced in L. monocytogenes upon entrance into a plant environment. Since L. monocytogenes is a natural soil organism and a natural saprophyte, it might be expected that it would interact with plants in a fashion similar to that of bacteria that are normally thought of as being plant associated.
We selected genes for mutation based on the extent of their differential expression, as shown by real-time RT-PCR (tcsA) (Table 3); the potential for the gene product to be surface associated and thereby be involved directly in attachment to the cabbage tissue (lmo0160 and tcsA); or the potential for the gene product to have a regulatory function, indicating a possible regulatory cascade in response to the plant environment (lmo2678). None of the strains with mutations in these genes, however, were defective in the cabbage growth and attachment system, nor were they defective in a 4-h assay for cabbage attachment at the beginning of the growth process. In a previous study of L. monocytogenes defective in attachment to fresh-cut radish tissue, three mutants were obtained from a Tn917-LTV3 transposon library, and the greatest defect found was a 1-log reduction after 4 h for a mutant affected in flagellar biosynthesis (12). Since genes important for flagellar assembly were identified in this study, we tested this motility mutant in the cabbage system and found that it was not affected in cabbage attachment or colonization. Flagella may not be a factor in cabbage attachment, at least not to the extent seen in radish attachment. Differences in the matrices may play a role, since the radish is a tuber and the cabbage consisted of leaf surfaces, and more cut surface was available for binding in the radish system described previously than in the cabbage system. It is also possible that the cabbage assay was not sensitive enough to detect small differences in attachment and colonization levels between strains. It is possible that a combination of mutations of the genes that we have identified may result in a defect in attachment or growth on cabbage, and this possibility is being explored. In addition, it is possible that genes cotranscribed with the genes selected for mutagenesis are more directly involved in the plant-microbe interaction.
During adaptation to growth in a plant environment, bacteria must be capable of growth on available nutrients, biosynthesis, and/or transport of necessary building blocks, such as amino acids and nucleotides, and of dealing with potential stresses, such as fluctuations in pH and osmolarity. The genes induced by L. monocytogenes in this growth environment seem to reflect the capacity to deal with all of these things. While the differential display method did allow us to identify several genes that are upregulated by the growth and attachment of L. monocytogenes to cabbage, the protocol would most likely not allow us to discern genes expressed at very low levels, especially if the genes were not well resolved or the levels were below the staining detection limit of the polyacrylamide gels. Other methods for providing a global snapshot of gene expression, such as DNA microarrays, would be more useful in this regard. However, the benefit of the differential display method is that the contribution to the RNA from other bacteria in the mixed bacterial population can be directly assessed and the contaminating genes can be removed from the analysis.
Acknowledgments
We thank J. Barak, M. Cooley, J. McGarvey, R. Mandrell, W. Miller, and C. Parker for helpful discussions and advice. We thank N. Freitag for strains, plasmids, and advice. We also thank J. Duhe and D. Flaherty for technical assistance.
This research was funded by the U.S. Department of Agriculture (Agricultural Research Service CRIS project number 5325-42000-040-00D).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arous, S., C. Buchrieser, P. Folio, P. Glaser, A. Namane, M. Hébraud, and Y. Héchard. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581-1590. [DOI] [PubMed] [Google Scholar]
- 3.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrang, M. E., R. E. Brackett, and L. R. Beuchat. 1989. Growth of Listeria monocytogenes on fresh vegetables stored under controlled atmosphere. J. Food Prot. 52:702-705. [DOI] [PubMed] [Google Scholar]
- 5.Beuchat, L. R. 1996. Listeria monocytogenes: incidence on vegetables. Food Control 7:223-228. [Google Scholar]
- 6.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brzostowicz, P. C., K. L. Gibson, S. M. Thomas, M. S. Blasko, and P. E. Rouviere. 2000. Simultaneous identification of two cyclohexanone oxidation genes from an environmental Brevibacterium isolate using mRNA differential display. J. Bacteriol. 182:4241-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conner, D. E., R. E. Brackett, and L. R. Beuchat. 1986. Effect of temperature, sodium chloride, and pH on growth of Listeria monocytogenes in cabbage juice. Appl. Environ. Microbiol. 52:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duché, O., F. Trémoulet, P. Glaser, and J. Labadie. 2002. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farber, J. M., S. L. Wang, Y. Cai, and S. Zhang. 1998. Changes in populations of. Listeria monocytogenes inoculated on packaged fresh-cut vegetables. J. Food Prot. 61:192-195. [DOI] [PubMed] [Google Scholar]
- 11.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski, L., J. D. Palumbo, and R. E. Mandrell. 2003. Attachment of Listeria monocytogenes to radish tissue is dependent upon temperature and flagellar motility. Appl. Environ. Microbiol. 69:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravani, R. 1999. Incidence and control of Listeria in food-processing facilities, p. 657-709. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 15.Helloin, E., L. Jänsch, and L. Phan-Thanh. 2003. Carbon starvation survival of Listeria monocytogenes in planktonic state and in biofilm: a proteomic study. Proteomics 3:2052-2064. [DOI] [PubMed] [Google Scholar]
- 16.Jacxsens, L., F. Devlieghere, P. Falcato, and J. Debevere. 1999. Behavior of Listeria monocytogenes and Aeromonas spp. on fresh-cut produce packaged under equilibrium-modified atmosphere. J. Food Prot. 62:1128-1135. [DOI] [PubMed] [Google Scholar]
- 17.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loessner, M. J., A. Schneider, and S. Scherer. 1996. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl. Environ. Microbiol. 62:3057-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by pseudomonads. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 20.Marshall, D. L., and R. H. Schmidt. 1991. Physiological evaluation of stimulated growth of Listeria monocytogenes by Pseudomonas species in milk. Can. J. Microbiol. 37:594-599. [DOI] [PubMed] [Google Scholar]
- 21.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milohanic, E., P. Glaser, J.-Y. Coppée, L. Frangeul, Y. Vega, J.-A. Vázquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptosome analysis of Listeria monocytogenes identifies three groups of genes differentially regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 23.Moszer, I., P. Glaser, and A. Danchin. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen-The, C., and F. Carlin. 1994. The microbiology of minimally processed fresh fruits and vegetables. Crit. Rev. Food Sci. Nutr. 34:371-401. [DOI] [PubMed] [Google Scholar]
- 25.Prazak, A. M., E. A. Murano, I. Mercado, and G. R. Acuff. 2002. Prevalence of Listeria monocytogenes during production and postharvest processing of cabbage. J. Food Prot. 65:1728-1734. [DOI] [PubMed] [Google Scholar]
- 26.Premaratne, R. J., W.-J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 28.Schaferkordt, S., and T. Chakraborty. 1995. Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. BioTechniques 19:720-725. [PubMed] [Google Scholar]
- 29.Schlech, W. F., III. 2000. Epidemiology and clinical manifestations of Listeria monocytogenes infection, p. 473-479. In V. A. Fishetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 30.Schlech, W. F. I., P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 32.Weeks, M. E., D. C. James, G. K. Robinson, and C. M. Smales. 2004. Global changes in gene expression observed at the transition from growth to stationary phase in Listeria monocytogenes ScottA batch culture. Proteomics 4:123-135. [DOI] [PubMed] [Google Scholar]
- 33.Weis, J., and H. P. R. Seeliger. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30:29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wemekamp-Kamphuis, H. H., J. A. Wouters, P. L. A. de Leeuw, T. Hain, T. Chakraborty, and T. Abee. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, R. L., A. R. Tvinnereim, B. D. Jones, and J. T. Harty. 2001. Identification of Listeria monocytogenes in vivo-induced genes by fluorescence-activated cell sorting. Infect. Immun. 69:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilton, S. D., L. Lim, D. Dye, and N. Laing. 1997. Bandstab: a PCR-based alternative to cloning PCR products. BioTechniques 22:642-645. [DOI] [PubMed] [Google Scholar]