Abstract
In Pseudomonas fluorescens CHA0, mutation of the GacA-controlled aprA gene (encoding the major extracellular protease) or the gacA regulatory gene resulted in reduced biocontrol activity against the root-knot nematode Meloidogyne incognita during tomato and soybean infection. Culture supernatants of strain CHA0 inhibited egg hatching and induced mortality of M. incognita juveniles more strongly than did supernatants of aprA and gacA mutants, suggesting that AprA protease contributes to biocontrol.
Plant diseases caused by soilborne root pathogens account for major crop losses worldwide. Yet in a small number of environments, i.e., in suppressive soils, little or no disease is observed, despite the presence of pathogens. Disease suppression depends, in part, on microorganisms that are able to antagonize pathogens (5, 10, 14, 28). The root-colonizing bacterium Pseudomonas fluorescens CHA0, which was isolated from a suppressive soil, has been studied in detail as a model strain for the biological control of several fungal plant diseases, such as black root rot of tobacco and take-all disease of wheat (5, 27). In this strain, as well as in other biocontrol pseudomonads, antifungal secondary metabolites, e.g., 2,4-diacetylphloroglucinol, hydrogen cyanide, and pyoluteorin, are important for biocontrol activity. These biocontrol factors are synthesized in response to environmental conditions and to population densities of the producer strain, whereby the GacS/GacA two-component system exerts a crucial role as a positive control element (6, 8, 9, 11, 26). Some rhizosphere microorganisms, including P. fluorescens CHA0, can also act as antagonists of plant-pathogenic nematodes (23). For antagonistic fungi, this biological control has been shown to involve extracellular proteases (2, 21). In strain CHA0, the production of the major extracellular EDTA-sensitive protease, AprA, is controlled by the GacS/GacA signal transduction pathway (8, 17, 26, 29). The present study was undertaken to find out whether this enzyme contributes to the biocontrol properties of strain CHA0 in plant-nematode interactions.
Characterization of the aprA-aprI-aprD gene region involved in production of the major exoprotease of strain CHA0.
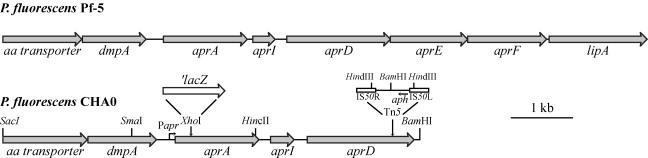
Strain CHA803, a Tn5 insertion mutant derivative of wild-type CHA0 (20), lacked proteolytic and lipolytic activities on indicator agar plates (17, 18) but showed wild-type production of antifungal metabolites, indicating that the Tn5 insertion was not in gacS or gacA (9). The Tn5 insertion was mapped to the 3′ end of the aprD gene (Fig. 1), whose deduced amino acid sequence has 56% identity with the ATP-driven translocator AprD, a component of the type I secretion machinery required for the secretion of alkaline protease AprA in P. aeruginosa (1, 3). By a chromosome walking approach (7), the genes located upstream of aprD, that is, an open reading frame coding for an amino acid transporter, dmpA (for a putative aminopeptidase), aprA (for extracellular protease), and aprI (for the cognate protease inhibitor), were cloned and sequenced in strain CHA0 (Fig. 1). The genomic sequence of P. fluorescens Pf-5, which is phenotypically and genotypically very similar to P. fluorescens CHA0 (4, 15), predicts that the aprAID genes are the proximal part of an aprAIDEF operon, which includes the lipA gene (for extracellular lipase) at the 3′ end (Fig. 1).
FIG. 1.
Genetic organization of the region surrounding aprA in P. fluorescens CHA0 and Pf-5. The 6.7-kb SacI-BamHI fragment of strain CHA0, which was sequenced in this study (GenBank accession no. AY644718), is aligned with the homologous region of strain Pf-5 (http://www.tigr.org) shown above. The sites where a translational ′lacZ fusion and a Tn5 element are inserted in the chromosome of strains CHA805 and CHA803, respectively, are shown above the aprA and aprD genes. Papr, promoter of aprA; aph, kanamycin resistance gene.
The deduced aprA gene product shows 62% identity with the AprA alkaline protease of P. aeruginosa (3) and contains Zn2+- and Ca2+-binding motifs. The calculated molecular mass of 49.9 kDa for the secreted form of AprA is in reasonable agreement with the value (47.1 kDa) previously determined for the EDTA-sensitive, major exoprotease of strain CHA0 (17). Between the aprA and aprD genes lies the aprI gene (Fig. 1) coding for a predicted 13.8-kDa protein which shows 40% amino acid sequence identity with the P. aeruginosa AprI protein, an AprA-specific inhibitor (3).
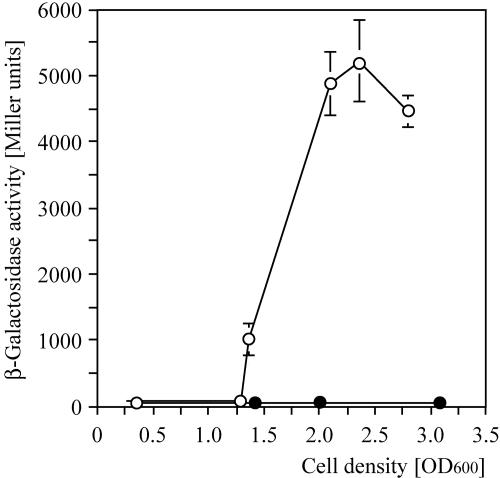
A nonpolar aprA mutation was constructed by the insertion of a ′lacZ cassette into the unique XhoI site of the chromosomal aprA gene (Fig. 1) in the wild type and in a gacS background, using the suicide plasmid pME6063 (Table 1). This resulted in strains CHA805 and CHA806 (Table 1), respectively. Strain CHA805 was exoprotease negative, as expected, but lipase positive, in keeping with the nonpolar nature of the ′lacZ insertion. β-Galactosidase activities of the aprA′-′lacZ translational fusion in strain CHA805 showed a marked cell density-dependent expression profile. In contrast, in the gacS mutant CHA806, almost no β-galactosidase activity was measured (Fig. 2).
TABLE 1.
P. fluorescens strains and plasmids
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains of P. fluorescens | ||
| CHA0 | Wild type | 27 |
| CHA19 | ΔgacS | 29 |
| CHA89 | gacA::Kmr | 11 |
| CHA803 | aprD::Tn5 | 20 |
| CHA805 | aprA::′lacZ | This study |
| CHA806 | ΔgacS aprA::′lacZ | This study |
| Plasmids | ||
| pME3087 | Suicide vector; ColE1 replicon, IncP-1-Mob, Tcr | 27 |
| pME6063 | 5.0-kb aprA′-′lacZ-′aprA fragment inserted into pME3087, suicide plasmid for aprA::′lacZ chromosomal fusions | This study |
FIG. 2.
GacS control of an aprA::′lacZ fusion in P. fluorescens grown in liquid King's B medium. β-Galactosidase activities were determined by the Miller method (13) for aprA::′lacZ in the wild-type derivative CHA805 (○) and in the gacS mutant CHA806 (•). The growth rates of both strains were similar (data not shown). Each value is the average ± standard deviation from three different cultures.
Impact of the aprA gene product on nematode populations.
Meloidogyne spp., the root-knot nematodes, are sedentary endoparasites of a wide range of plants, including many of agronomical importance. Meloidogyne incognita belongs to a group of nematodes that cause important crop losses in developing countries (12, 19). Culture supernatants of wild-type strain CHA0 grown in 1/20-strength King's B medium (0.1% [wt/vol] Oxoid proteose peptone, 0.05% [wt/vol] glycerol, 0.3 mM MgSO4, 0.3 mM K2HPO4) inhibited egg hatching and caused mortality of the juveniles of M. incognita in vitro, in comparison with the uninoculated controls (P ≤ 0.05) (Table 2). The protease-negative mutants CHA805 (aprA) and CHA89 (gacA) failed to inhibit egg hatching and to kill M. incognita juveniles (Table 2). The addition of the protease inhibitor EDTA (4 mM) to a culture supernatant of strain CHA0 grown in King's B medium markedly reduced (P ≤ 0.05) the juvenile killing activity of strain CHA0 but had little effect on the supernatants of the mutants CHA805 and CHA89 (Table 3). These data support the involvement of AprA protease in the inhibition of egg hatching and in killing of juveniles. However, AprA protease may not be the only antinematode factor of strain CHA0, in that antibiotic compounds produced under GacA control may also have a role in nematode control (23; I. A. Siddiqui and S. S. Shaukat, unpublished data).
TABLE 2.
In vitro effects of culture filtrates of P. fluorescens strains on M. incognita egg hatching and juvenile mortality
| Strain or LSDa | Hatchingb | Mortalityb |
|---|---|---|
| Control | 83a | 18a |
| CHA0 | 51b | 56b |
| CHA89 | 77a | 25a |
| CHA805 | 70a | 31a |
| LSDc | 17 | 14 |
Strains CHA0 (wild type), CHA89 (gacA), and CHA805 (aprA′-′lacZ) were grown in 1/20 King's B medium for 48 h. The control consisted of sterile medium.
Hatching and mortality were tested as previously described (22) and are expressed as the percentage of eggs hatched after 72 h of exposure and as the percentage of juvenile mortality after 24 h of exposure, respectively. Data for each strain and medium were obtained from three different experiments with six repetitions. Means of the experiments in each column followed by different letters are significantly different (P ≤ 0.05) according to Fisher's test.
LSD, least significant difference.
TABLE 3.
In vitro effects of the addition of 4 mM EDTA to P. fluorescens culture filtrates on the mortality of M. incognita juveniles
Culture filtrates of strains CHA0 (wild type), CHA89 (gacA), and CHA805 (aprA′-′lacZ) were grown in 1/20 King's B medium for 48 h. The control was made with sterile medium.
Mortality is expressed as the percentage of dead juveniles after 24 h of exposure. Data for each strain and treatment were obtained from three different experiments with five repetitions. The mean of the experiment marked with an asterisk is significantly different (P ≤ 0.05) from all the other values according to Fisher's test (least significant difference = 20%).
In comparison to nonbacterized controls, P. fluorescens CHA0 applied to unsterilized sandy loam soil suppressed (P ≤ 0.05) root-knot development and nematode final population densities on both tomato and soybean under greenhouse conditions (Table 4). Carbofuran (Furadan) treatment, however, was more effective in reducing nematode population densities in soil and roots and subsequent root-knot development in both crops (Table 4). Strains CHA805 and CHA89 had no significant impact on nematode population densities in soil and root-knot disease in either crop (Table 4). Application of strain CHA805 resulted in a reduction (P ≤ 0.05), but not a complete loss, of nematode final population densities in soybean roots (Table 4). In these experiments, bacterial colonization of the tomato and soybean rhizospheres was not significantly different between the three strains tested (data not shown).
TABLE 4.
Effects of carbofuran and P. fluorescens strains on gall formation caused by M. incognita and on soil and root populations in tomato and soybean grown under glasshouse conditionsa
| Strain or statistic | Galls/g rootb
|
M. incognita organisms/g rootc
|
M. incognita organisms/ 100 cm3 soil
|
|||
|---|---|---|---|---|---|---|
| Tomato | Soybean | Tomato | Soybean | Tomato | Soybean | |
| Control | 101a | 81a | 136a | 121a | 1,524a | 1,330a |
| Carbofuran | 34c | 29c | 62c | 52c | 625c | 380c |
| CHA0 | 62b | 46bc | 82bc | 80b | 1,238b | 1,070b |
| CHA89 | 86ab | 69ab | 116a | 107a | 1,461a | 1,196ab |
| CHA805 | 76ab | 55b | 107ab | 97ab | 1,456a | 1,218ab |
| LSDd | 26 | 23 | 30 | 25 | 164 | 153 |
Sandy loam soil (sand:silt:clay, 70:19:11; pH 8.1; moisture holding capacity, 39%) from Karachi was placed in plastic pots (8-cm diameter). After removing the upper 2-cm layer of soil, 2.2 × 108 to 3.1 × 108 CFU ml−1 of bacteria suspended in 25 ml sterile 100 mM MgSO4 or 120 mg kg−1 carbofuran (a granular nematocide purchased from Pak Agrochemicals, Karachi, Pakistan) was drenched. Soil drenched with 25 ml of sterile 100 mM MgSO4 was used as a control. After treatment, three tomato seedlings (cv. SUN 6002 PVP) were planted in each pot. After 1 week, 2 × 103 M. incognita juveniles were added to each pot. The suspension was adjusted to 500 juveniles ml−1 by adding tap water, and a total of 4 ml was pipetted into four holes made around the seedlings in the soil. In another series of pots, after bacterial and chemical treatments, eight soybean (Glycine max L. cv. NARC I) seeds were sown in each pot, and after germination four seedlings per pot were retained. One week after emergence, the seedlings were infested with M. incognita juveniles as described above.
Gall formation is expressed as the number of galls per gram of root after 45 days.
M. incognita populations are expressed as the number of individuals per gram of root or per 100 cm3 of soil (16, 22) after 45 days. Data for each strain and plant system were obtained from four different experiments with four repetitions. Means of the experiments in each column followed by different letters are significantly different (P ≤ 0.05) according to Fisher's test.
LSD, least significant difference.
In conclusion, these findings are consistent with the notion that AprA protease of strain CHA0 contributes, directly or indirectly, to biocontrol of M. incognita. This study also extends previous observations that P. fluorescens CHA0 has biological control activity against root-knot nematodes (23-25).
Nucleotide sequence accession number.
The 6.7-kb SacI-BamHI fragment of strain CHA0 was sequenced in this study and was deposited in GenBank under accession no. AY644718.
Acknowledgments
We thank Karin Heurlier for determining the lipase phenotype of P. fluorescens strains.
Support from the Swiss National Foundation for Scientific Research (project 3100A0-100180) is gratefully acknowledged.
REFERENCES
- 1.Binet, R., S. Létoffé, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene 192:7-11. [DOI] [PubMed] [Google Scholar]
- 2.Bonants, P. J., P. F. Fitters, H. Thijs, E. den Belder, C. Waalwijk, and J. W. Henfling. 1995. A basic serine protease from Paecilomyces lilacinus with biological activity against Meloidogyne hapla eggs. Microbiology 141:775-784. [DOI] [PubMed] [Google Scholar]
- 3.Duong, F., A. Lazdunski, B. Cami, and M. Murgier. 1992. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene 121:47-54. [DOI] [PubMed] [Google Scholar]
- 4.Ellis, R. J., T. M. Timms-Wilson, and M. J. Bailey. 2000. Identification of conserved traits in fluorescent pseudomonads with antifungal activity. Environ. Microbiol. 2:274-284. [DOI] [PubMed] [Google Scholar]
- 5.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 6.Haas, D., and G. Défago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 7.Heeb, S. 2001. Regulation of exoproduct formation by the GacS/GacA two-component system in Pseudomonas fluorescens CHA0. Ph.D. thesis. University of Lausanne, Lausanne, Switzerland.
- 8.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 10.Kerry, B. R. 2000. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 38:423-441. [DOI] [PubMed] [Google Scholar]
- 11.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Défago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luc, M., R. A. Sikora, and J. Bridge. 1990. Plant parasitic nematodes in subtropical and tropical agriculture. CAB International, Oxford, United Kingdom.
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Raaijmakers, J. M., M. Vlami, and J. T. de Souza. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Leeuwenhoek 81:537-547. [DOI] [PubMed] [Google Scholar]
- 15.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2001. Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2,4-diacetylphloroglucinol and comparison of PhlD with plant polyketide synthases. Mol. Plant-Microbe Interact. 14:639-652. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-Kábana, R., and M. H. Pope. 1981. A simple incubation method for the extraction of nematodes from soil. Nematropica 11:175-185. [Google Scholar]
- 17.Sacherer, P., G. Défago, and D. Haas. 1994. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 116:155-160. [DOI] [PubMed] [Google Scholar]
- 18.Safarik, I., and M. Safarikova. 1994. A modified procedure for the detection of microbial producers of extracellular proteolytic enzymes. Biotechnol. Tech. 8:627-628. [Google Scholar]
- 19.Sasser, J. N., and D. W. Freckmann. 1987. A world perspective on nematology: the role of society, p. 7-14. In J. A. Veech and D. W. Dickson (ed.), Vistas on nematology. Society of Nematologists, Hyattsville, Md.
- 20.Schmidli-Sacherer, P. 1996. Mechanisms of suppression of plant diseases by Pseudomonas fluorescens CHA0: performance of gacA mutants, influence of salicylic acid overproduction, analysis of extracellular enzymes. Ph.D. thesis 11761. ETH, Zürich, Switzerland.
- 21.Segers, R., T. M. Butt, B. R. Kerry, and J. F. Peberdy. 1994. The nematophagous fungus Verticillium chlamydosporium produces a chymoelastase-like protease which hydrolyses host nematode proteins in situ. Microbiology 140:2715-2723. [DOI] [PubMed] [Google Scholar]
- 22.Siddiqui, I. A., and S. Ehteshamul-Haque. 2001. Suppression of the root rot-root knot disease complex by Pseudomonas aeruginosa in tomato: the influence of inoculum density, nematode populations, moisture and other plant-associated bacteria. Plant Soil 237:81-89. [Google Scholar]
- 23.Siddiqui, I. A., and S. S. Shaukat. 2003. Plant species, host age and host genotype effects on Meloidogyne incognita biocontrol by Pseudomonas fluorescens strain CHA0 and its genetically-modified derivatives. J. Phytopathol. 151:231-238. [Google Scholar]
- 24.Siddiqui, I. A., and S. S. Shaukat. 2002. Rhizobacteria-mediated induction of systemic resistance (ISR) in tomato against Meloidogyne javanica. J. Phytopathol. 150:469-473. [Google Scholar]
- 25.Siddiqui, I. A., and S. S. Shaukat. 2003. Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite, 2,4-diacetylphloroglucinol. Soil Biol. Biochem. 35:1615-1623. [Google Scholar]
- 26.Valverde, C., S. Heeb, C. Keel, and D. Haas. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361-1379. [DOI] [PubMed] [Google Scholar]
- 27.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, M. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, F. D. N. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Verlagsgesellschaft mbH, Weinheim, Germany.
- 28.Weller, D. M., J. M. Raaijmakers, B. B. M. Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 29.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]