Abstract
The location and abundance of Campylobacter jejuni and Campylobacter lanienae in the intestines of beef cattle were investigated using real-time quantitative PCR in two studies. In an initial study, digesta and tissue samples were obtained along the digestive tract of two beef steers known to shed C. jejuni and C. lanienae (steers A and B). At the time of slaughter, steer B weighed 540 kg, compared to 600 kg for steer A, yet the intestine of steer B (40.5 m) was 36% longer than the intestine of steer A (26.1 m). In total, 323 digesta samples (20-cm intervals) and 998 tissue samples (3.3- to 6.7-cm intervals) were processed. Campylobacter DNA was detected in the digesta and in association with tissues throughout the small and large intestines of both animals. Although C. jejuni and C. lanienae DNA were detected in both animals, only steer A contained substantial quantities of C. jejuni DNA. In both digesta and tissues of steer A, C. jejuni was present in the duodenum and jejunum. Considerable quantities of C. jejuni DNA also were observed in the digesta obtained from the cecum and ascending colon, but minimal DNA was associated with tissues of these regions. In contrast, steer B contained substantial quantities of C. lanienae DNA, and DNA of this bacterium was limited to the large intestine (i.e., the cecum, proximal ascending colon, descending colon, and rectum); the majority of tissue-associated C. lanienae DNA was present in the cecum, descending colon, and rectum. In a second study, the location and abundance of C. jejuni and C. lanienae DNA were confirmed in the intestines of 20 arbitrarily selected beef cattle. DNA of C. jejuni and C. lanienae were detected in the digesta of 57% and 95% of the animals, respectively. C. jejuni associated with intestinal tissues was most abundant in the duodenum, ileum, and rectum. However, one animal contributed disproportionately to the abundance of C. jejuni DNA in the ileum and rectum. C. lanienae was most abundant in the large intestine, and the highest density of DNA of this bacterium was found in the cecum. Therefore, C. jejuni colonized the proximal small intestine of asymptomatic beef cattle, whereas C. lanienae primarily resided in the cecum, descending colon, and rectum. This information could be instrumental in developing efficacious strategies to manage the release of these bacteria from the gastrointestinal tracts of cattle.
Beef cattle production is predominant in the Chinook Health region in southern Alberta, Canada. The prevalence of Campylobacter infections in humans in this region is higher than the national average and has increased three times faster than the population growth (Paul Hasselback, Canadian Laboratory Medicine Congress, Calgary, Alberta, Canada, May 2002). Perhaps beef cattle are an important reservoir of Campylobacter species infecting humans in this region. Many Campylobacter species are present in the feces of beef cattle (17, 18, 19, 20, 26); in particular, Campylobacter lanienae and Campylobacter jejuni are frequently shed in large numbers (20). The frequency of campylobacteriosis in human populations is often not correlated with Campylobacter in poultry (25), and genotyping has suggested that cattle may be an important source of human-pathogenic campylobacters (9, 30, 31, 34, 36). Furthermore, waterborne Campylobacter species from a bovine source were implicated in the infection of a large number of people at Walkerton, Ontario, Canada, in 2000 (7), and passive surveillance information in the Chinook Health region of Alberta suggests that cattle production is linked to the transmission of Campylobacter to humans (Hasselback, Canadian Laboratory Medicine Congress, 2002).
Very limited information is available on the process of colonization of the gastrointestinal (GI) tracts of cattle by Campylobacter species. Although campylobacters have been isolated from the intestines of healthy calves and adult cattle (13, 28, 32, 37), as well as from calves exhibiting signs of enteritis (1, 2, 3, 4, 5, 39, 40), detailed examination of the site of colonization of the intestines of healthy cattle has not been undertaken. In this regard, real-time quantitative PCR (RTQ-PCR) allows quantification of DNA of specific taxa within the digestive tract (18). The objective of the current study was to use RTQ-PCR to measure the distribution and abundance of C. jejuni and C. lanienae in the intestines of beef cattle naturally colonized by Campylobacter species.
MATERIALS AND METHODS
Chronically shedding cattle.
Two beef animals (steers A and B) were selected from a previous trial in which the chronic shedding of Campylobacter species in feces was examined (20). These two steers shed substantial numbers of C. jejuni and C. lanienae for a prolonged time in the feedlot. They were fed a barley-based diet until slaughter. Each animal was euthanized humanely under the supervision of a licensed veterinarian on separate mornings (2 and 4 April 2003). The GI tract of each animal was removed approximately 10 min after death and placed on a clean sheet of plastic on a cool cement floor; tissue processing was started immediately, and tissues (and digesta) from the proximal duodenum to the rectum were obtained as shown in Fig. 1. Initially, the small and large intestines were tied at approximately 20- to 40- cm intervals (to prevent movement of digesta), anatomical landmarks were identified, and colored strings were used to distinguish the anterior and posterior ends. Preliminary removal of mesentery was conducted, the intestine was divided into “portions” that were 61 to 580 cm long (Fig. 1A), the lengths were measured, and each portion was placed in a plastic bag and transported on ice to the necropsy facility located at the Lethbridge Research Centre. In addition, the pancreas was removed from steer B and placed on ice until it was processed. Tissues were maintained on ice for ca. 2 to 11 h. In the necropsy room, the intestinal portions were cut into 20-cm “sections” (Fig. 1B). The “sections” were excised longitudinally using scissors that were free of Campylobacter DNA, and the digesta was aseptically removed with a pipette tip (Fig. 1C). Approximately 200 mg of digesta was placed in a DNA-free 5-ml tube, and samples were immediately placed at −20°C. The “sections” were then gently washed with sterile phosphate-buffered with saline (0.2 mol liter−1) (PBS) (10 mM sodium phosphate buffer with 130 mM sodium chloride [pH 7.2]); care was taken to remove digesta while minimizing disruption of mucus on the mucosal surface. Following washing, tissue plugs were collected at 3.3-cm intervals with a sterile 4-mm-diameter Biopsy Acu-Punch (CDMV, St. Hyacinthe, Quebec, Canada) free of Campylobacter DNA; five tissue samples were taken from each 20-cm “section” (Fig. 1C). Intestinal and pancreatic tissue samples were then removed from the punch with a clean pair of forceps, and the plugs were individually placed in DNA-free 2-ml microcentrifuge tubes and immediately placed at −20°C.
FIG. 1.
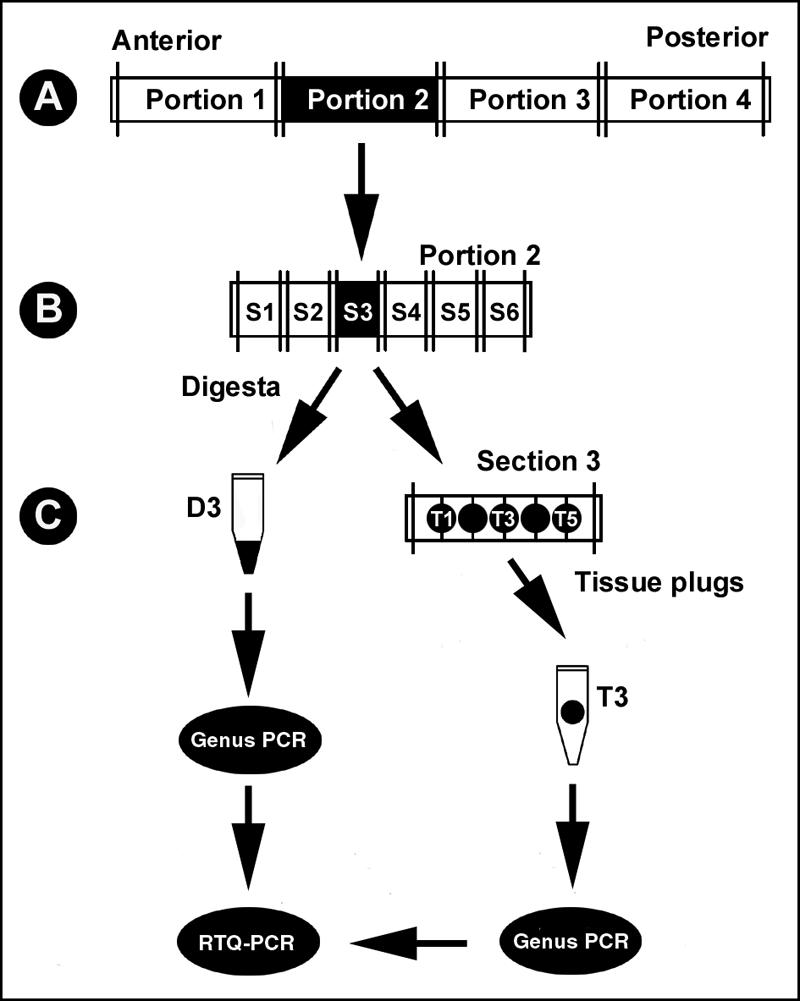
Schematic illustration of the method used for collection and processing of tissue and digesta samples from two chronically shedding beef cattle. (A) In the field, the digestive tract was divided into “portions” that were arbitrary lengths. (B) Subsequently, each portion was excised into 20-cm “sections” (designated S1, S2, S3, etc.). (C) Each section was longitudinally incised, and digesta was collected and frozen at −20°C. Following removal of digesta, tissue plugs (designated T1, T2, T3, etc.) were removed at 3.3-cm intervals, placed in microcentrifuge tubes, and frozen at −20°C. At designated locations, tissue plugs also were removed for microscopy. DNA was extracted from digesta and tissue plugs and subjected to conventional PCR for the genus Campylobacter and an internal amplification control and to real-time quantitative PCR to determine the numbers of C. jejuni and C. lanienae associated with digesta and each tissue plug.
Cattle survey.
In a subsequent study, the intestines of 20 arbitrarily selected beef cattle were obtained from an abattoir on six separate occasions (28 and 29 January 2004, 2, 9, and 12 February 2004, and 6 April 2004). These cattle ranged in age from 18 to 26 months. The cattle were humanely euthanized, and intestinal samples were obtained within ca. 15 to 30 min after death. Eleven gut sections (length, ∼20 cm) were obtained from each animal at the following locations: (i) proximal duodenum (i.e., following the cranial flexure), (ii) distal duodenum (following the caudal flexure), (iii) proximal jejunum, (iv) central jejunum, (v) distal jejunum, (vi) ileum (∼10 cm before the ileal-cecal junction), (vii) free end of the cecum, (viii) proximal loop of the ascending colon, (ix) central flexure of the ascending colon, (x) descending colon (∼20 cm before the sigmoid colon), and (xi) rectum. Before excision of the gut sections, bilateral ligatures were applied adjacent to the excision site to minimize external contamination of the tissues with digesta. Tissue samples were then placed in individual bags on ice and transported to the necropsy room (ca. 2 to 4 h). In the necropsy room, samples were excised longitudinally, digesta was aseptically removed and collected, and the mucosa was washed as described above. Only digesta from the rectum and descending colon (if adequate quantities of digesta could not be obtained from the rectum) were processed. Biopsy samples were obtained as described above, except that three samples (obtained in close proximity to each other) were obtained from each tissue. One biopsy sample was used for DNA extraction, and the other two were processed for microscopy.
DNA extraction.
For digesta, a QIAamp DNA stool mini kit (QIAGEN Inc., Mississauga, Ontario, Canada) was used to extract DNA from 200 ± 5 mg of feces from each sample by using the manufacturer's protocol for isolation of DNA from stools for pathogen detection, except that adjustments were made for the differential weights of digesta by adjusting the amount of ASL buffer used (17). For tissues, a QIAGEN DNeasy tissue kit (QIAGEN Inc.) was used according to the manufacturer's protocol. To determine whether PCR inhibitors had been sufficiently removed to allow amplification, an internal amplification control (IAC) was used (17); inclusion of an IAC was necessary to eliminate false-negative results (15). The IAC was constructed by deleting a fragment of the C. jejuni ATCC 49943 16S rRNA gene, and it was designed to amplify under the same PCR conditions as the genus Campylobacter primer set but to yield a 475-bp product instead of the 816-bp product. Prior to extraction, 15 μl (700 copies/μl) of the IAC was added to thawed digesta and tissues. All DNA samples were stored at −20°C until they were used.
PCR.
All DNA was subjected to PCR for the genus Campylobacter as described previously (17). The presence of either a genus-specific or IAC amplicon indicated that there had been adequate removal of PCR inhibitors. Samples that were positive for Campylobacter DNA were then subjected to nested or nonnested RTQ-PCR, as described by Inglis and Kalischuk (18). Nested RTQ-PCR was used for C. lanienae (primer set 2), whereas nonnested RTQ-PCR was used for C. jejuni. To prepare the quantification standard, C. jejuni and C. lanienae cells were plated on brucella agar and Karmali agar, respectively, and scraped from the agar surface 48 h after plating. DNA was extracted from the harvested cell mass using a DNeasy kit (QIAGEN Inc.) according to the manufacturer's protocol. DNA was measured fluorimetrically using a Hoefer DyNA Quant 200 apparatus (Amersham Biosciences Corp., Piscataway, NJ); calf thymus DNA (Calbiochem, San Diego, CA) was used as a standard. The numbers of C. jejuni genome copies (based on a genome size of 1.6 Mbp) and C. lanienae genome copies (based on a genome size of 0.8 Mbp) in 1 ng of DNA were 5.6 × 105 and 1.1 × 106 copies, respectively. Genomic DNA standards for both bacteria were diluted in a 10-fold dilution series in 10 mM Tris (pH 8.5); standard DNA was thawed and frozen a maximum of two times. The log10 numbers of copies of C. jejuni and C. lanienae DNA in 2 μl of template were determined relative to a standard curve, the data were converted to the numbers of genome copies in 2 μl of template, and the mean of two observations per sample was calculated. If one of the duplicate samples was negative, it was entered as a missing value (i.e., the single positive value was used).
Microscopy.
Tissue samples were placed in a histocassette, transferred to 2% freshly prepared paraformaldehyde (2 g of paraformaldehyde in 90 ml of H2O was heated to 60°C in a fume hood, 1 drop of 1 M NaOH was added, and the preparation was cooled to 4°C). Tissues were fixed in paraformaldehyde for 6 to 24 h at room temperature in a fume hood, rinsed with PBS, dehydrated in ethanol, and cleared in Histoclear (Fisher Scientific, Edmonton, Alberta, Canada) for 2 h at 60°C in a vacuum oven; the Histoclear was replaced, and the tissues were incubated at 60°C in the vacuum oven for an additional 2 h. Tissues were then embedded in Paraplast Plus (Fisher Scientific) using a Shandon Histocentre III (Fisher Scientific, Edmonton, Alberta, Canada) and were sectioned using a Finesse 325 microtome (Fisher Scientific). Sections were stained with Hp Yellow and Hp Blue used according to the manufacturer's protocol (Anatech Ltd., Battle Creek, MI); mucus appeared yellow, and bacteria embedded with mucus and intestinal tissues appeared blue. Sections were examined with a Zeiss Axioskop III (Carl Zeiss Canada Ltd., Toronto, Ontario, Canada), and images were recorded digitally using an Axiocam camera (Carl Zeiss Canada Ltd.).
RESULTS
Chronically shedding cattle. (i) Genus Campylobacter.
The total lengths of the small and large intestines obtained from steers A and B were 26.1 and 40.5 m, respectively. Despite possessing small and large intestines that were 14.4 m shorter, steer A (600 kg) was 60 kg heavier than steer B (540 kg). DNA was extracted, and conventional PCR for the genus Campylobacter was conducted with DNA extracted from 323 digesta and 998 tissue samples (total, 1,321 samples). Fifteen (7.7%; n = 196) of the digesta samples obtained from steer B were negative for both the IAC and Campylobacter DNA. In contrast, all DNA samples extracted from the digesta obtained from steer A (n = 127) were positive for the IAC and/or Campylobacter DNA. For the digesta of steers A and B, 98.4% (n = 125) and 92.3% (n = 167) of the samples were positive for Campylobacter DNA, respectively. For tissues obtained from steers A and B, 21 (5.4%; n = 390) and 5 (0.8%; n = 608) samples were negative for both the internal control and Campylobacter DNA, respectively. In all instances, samples that were negative for the IAC without amplification of Campylobacter DNA were excluded from the experiment. Of the 369 tissue samples obtained from steer A, 80.5% (n = 297) were positive for Campylobacter DNA. In contrast, only 29.2% (n = 176) of the tissue samples obtained from steer B were positive. Campylobacter DNA was detected in digesta and tissues obtained throughout the GI tract of both animals (Fig. 2A and B and 3A and B). Three of four tissue samples taken from the pancreas of steer B were positive for Campylobacter DNA but did not contain either C. jejuni or C. lanienae.
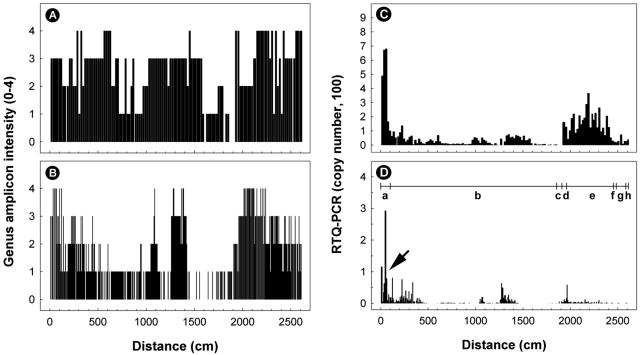
FIG. 2.
Distribution of samples positive for Campylobacter DNA obtained from digesta (A and C) and tissues (B and D) of the intestinal tract of steer A. (A and B) Genus Campylobacter DNA examined with conventional PCR. The intensity of the genus amplicon was assessed based on a scale from 0 to 4 relative to a standard sample of known DNA. (C and D) Abundance of C. jejuni (genome copies in 2 μl of template) determined by nonnested real-time quantitative PCR targeting the mapA gene. The horizontal line with vertical lines in panel D indicates the various regions of the small and large intestines, where “a” is the duodenum, “b” is the jejunum, “c” is the ileum, “d” is the cecum, “e” is the ascending colon, “f” is the transverse colon, “g” is the descending colon, and “h” is the rectum. The arrow indicates a region where there was abundant C. jejuni DNA. The total length of the small and large intestines was 26.1 m.
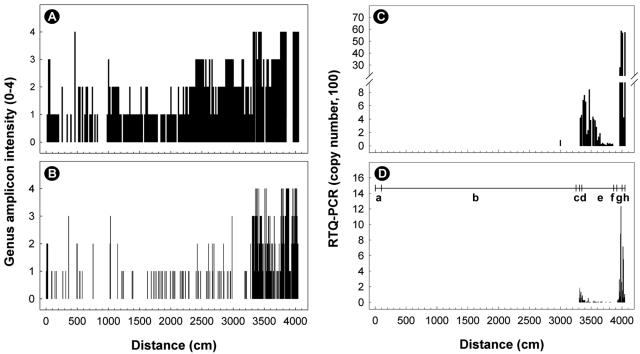
FIG. 3.
Distribution of samples positive for Campylobacter DNA obtained from digesta (A and C) and tissues (B and D) of the intestinal tract of steer B. (A and B) Genus Campylobacter DNA examined with conventional PCR. The intensity of the genus amplicon was assessed based on a scale from 0 to 4 relative to a standard sample of known DNA. (C and D) Abundance of C. lanienae (genome copies in 2 μl of template) determined by using nested real-time quantitative PCR targeting the 16S rRNA gene. The horizontal line with vertical lines in panel D indicates the various regions of the small and large intestines, where “a” is the duodenum, “b” is the jejunum, “c” is the ileum, “d” is the cecum, “e” is the ascending colon, “f” is the transverse colon, “g” is the descending colon, and “h” is the rectum. The total length of the small and large intestines was 40.5 m.
(ii) C. jejuni.
Digesta and tissue samples that were positive for Campylobacter DNA were subsequently subjected to nonnested RTQ-PCR for C. jejuni. For digesta, 97.6% (n = 122) and 6.0% (n = 10) of the samples obtained from steers A and B were positive for the bacterium, respectively. Similarly, a much higher percentage of the tissue samples obtained from steer A (69.6%; n = 206) than of the tissue samples obtained from steer B (5.2%; n = 9) were positive for C. jejuni DNA. In the digesta from steer A, the majority of C. jejuni DNA (typically >50 genome copies) was observed in the proximal small intestine (i.e., duodenum and jejunum) and in the large intestine (i.e., cecum and ascending colon) (Fig. 2C). Although the density of C. jejuni DNA associated with tissues was typically less than the density in the digesta, a similar pattern of C. jejuni abundance associated with intestinal tissues was observed in the small intestine but not in the large intestine of steer A (Fig. 2D). In particular, an abundance of C. jejuni DNA (3 to 292 genome copies from ∼0 to 1.4 m and 4 to 76 genome copies from ∼2.1 to 3.4 m) was observed in the duodenum and proximal jejunum (Fig. 2D). Appreciable quantities of C. jejuni DNA also were observed in the mid-jejunum (11 to 64 genome copies from ∼12.6 to 13.5 m) and in the proximal ascending colon (6 to 58 genome copies from ∼19.4 to 19.7 m).
(iii) C. lanienae.
Nested RTQ-PCR for C. lanienae was applied to DNA extracted from digesta and intestinal tissues. For digesta, 85.6% (n = 107) and 88.6% (n = 148) of the samples obtained from steers A and B were positive for C. lanienae DNA, respectively. High percentages of tissue samples obtained from both steers also were positive for this bacterium; DNA was detected in 55.6% (n = 165) and 65.9% (n = 116) of the samples obtained from steers A and B, respectively. Although similar percentages of the samples were positive for C. lanienae DNA for the two steers, relatively small quantities of DNA were associated with digesta and tissues obtained from steer A (typically ≤5 genome copies); in contrast, substantial amounts of C. lanienae DNA (typically >50 genome copies) were detected in samples from steer B. Similar distribution patterns of C. lanienae were observed for both the digesta and the tissues obtained from steer B. In digesta, C. lanienae DNA was primarily concentrated in the cecum (420 to 460 genome copies from ∼33.1 to 33.5 m), in the proximal ascending colon (83 to 768 genome copies from ∼33.5 to 36.4 m), and in the descending colon and rectum (Fig. 3C and D), and the greatest quantities of DNA were observed in the descending colon and rectum (423 to 5,876 genome copies from ∼39.7 to 40.5 m). In association with tissues, 6 to 188 genome copies were observed in the cecum (∼33.1 to 33.4 m), 19 to 86 genome copies were observed in the ascending colon (∼33.5 to 34.0 m), 5 to 1,234 genome copies were observed in the descending colon (∼39.2 to 40.1 m), and 32 to 716 genome copies were observed in the rectum (∼40.1 to 40.4 m).
DNA of both C. lanienae and C. jejuni were obtained from 84.0% and 6.0% of the digesta samples obtained from steers A and B, respectively. For the tissues, 42.6% and 2.3% of the samples obtained from steers A and B were positive for DNA of both bacteria, respectively.
Cattle survey. (i) Digesta.
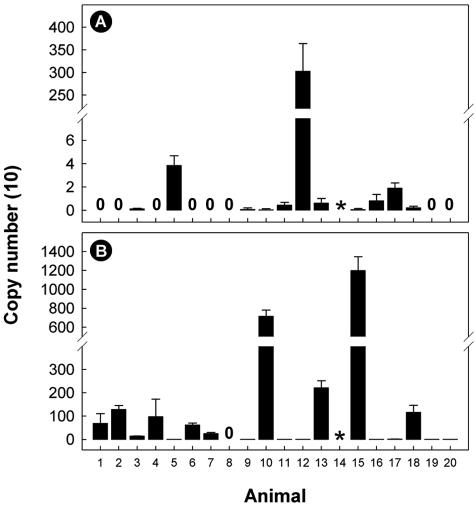
To confirm that the colonization sites described above occur in other beef cattle, the intestines of 20 arbitrarily selected individuals were subsequently obtained and examined by PCR. Campylobacter DNA was detected in the digesta obtained from 18 of 19 animals. An IAC or Campylobacter amplicon was not obtained from digesta from one animal. Using RTQ-PCR, DNA of C. jejuni was detected in 57.9% (n = 11) of the digesta samples (Fig. 4A). In particular, substantial quantities (3,033 genome copies) of C. jejuni DNA were observed in the digesta of animal 12. DNA of C. lanienae was detected in 94.7% (n = 18) of the digesta samples (Fig. 4B). Relatively large quantities of C. lanienae DNA were observed in the digesta of animals 10 (7,169 genome copies) and 15 (12,001 genome copies).
FIG. 4.
Quantities of C. jejuni (A) and C. lanienae (B) DNA in digesta of 20 arbitrarily selected beef cattle. Quantities of DNA were determined using real-time quantitative PCR targeting the mapA (nonnested) and 16S rRNA (nested) genes for C. jejuni and C. lanienae, respectively. Most of the digesta samples were obtained from the rectum; the exceptions were animals 11 and 18, where samples were obtained from the descending colon. Neither an internal control amplicon nor an amplicon for the genus Campylobacter was obtained from the digesta sample indicated by the asterisk. The error bars indicate standard deviations (n = 2).
(ii) Intestinal tissues.
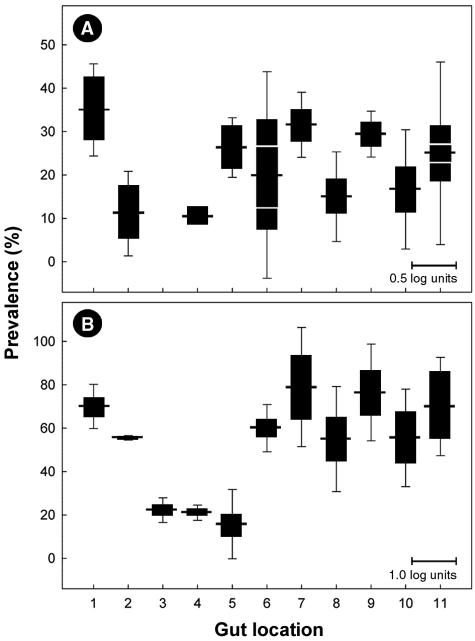
DNA extracted from intestinal tissues washed with PBS was also subjected to conventional PCR and RTQ-PCR. An IAC or Campylobacter genus-specific amplicon was not detected in 10 of 218 (4.6%) tissue samples. Of the remaining samples, 20.2% (n = 42) were positive for C. jejuni. Two animals that possessed C. jejuni DNA in digesta were deemed negative for the bacterium in tissues. Conversely, for three animals that were negative for C. jejuni in digesta, tissue samples were positive for the bacterium. Altogether (i.e., digesta and tissue samples combined), 70.0% of the 20 beef animals sampled were positive for C. jejuni. On average, C. jejuni DNA was most frequently detected in the proximal duodenum, but DNA of this bacterium also was observed at various frequencies in all regions except the proximal jejunum (Fig. 5A). C. jejuni was most abundant in the duodenum, ileum, and rectum (Fig. 5A). However, one animal contributed disproportionately to the mean value for samples obtained from the ileum and rectum; 878 and 318 copies of the C. jejuni mapA gene were detected in 2 μl of template from the ileum and rectum, respectively. This animal was a 24-month-old heifer with pneumonia at the time of euthanasia. In support of the RTQ-PCR data, Campylobacter cells were associated with mucus on the surface of the intestinal epithelium; most cells occurred singly or in accumulations of a limited number of cells that were either embedded within mucus or associated with mucus strands (Fig. 6A and B).
FIG. 5.
Prevalence of C. jejuni (A) and C. lanienae (B) associated with intestinal tissues of 20 beef cattle. The horizontal lines extending from the black bars indicate the percentages of positive animals (n = 20) for 11 locations in the small and large intestines. The locations are as follows: 1, proximal duodenum; 2, distal duodenum; 3, proximal jejunum; 4, central jejunum; 5, distal jejunum; 6, ileum; 7, free end of the cecum; 8, proximal loop of the ascending colon; 9, central flexure of the ascending colon; 10, descending colon; and 11, rectum. The solid bars indicate the relative abundance of each bacterium (mean log10 copy number in 2 μl of template). The vertical lines extending from the solid bars indicate standard deviations. The scale bars at the bottom right in the panels indicate mean template abundance (panel A, 0.5 log unit; panel B, 1.0 log unit). In panel A for gut locations 6 and 11, the areas delineated by the white lines in the bars indicate the mean values for C. jejuni template abundance minus the value for an animal with pneumonia.
FIG. 6.
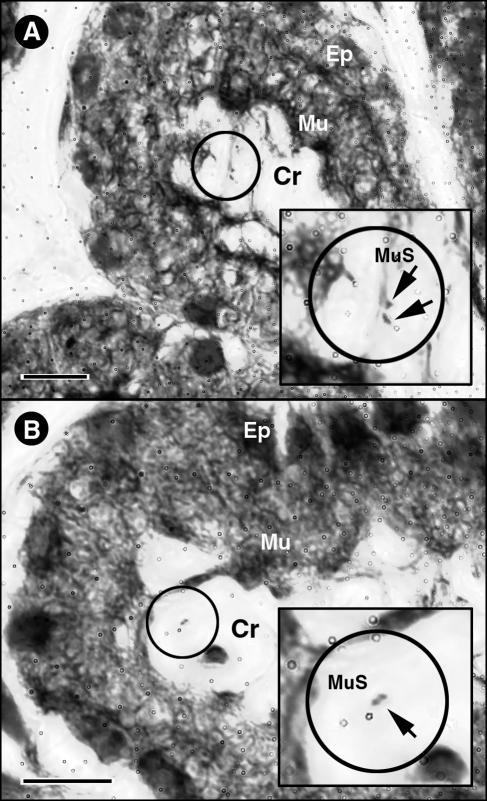
Light micrographs showing the Campylobacter cells in association with mucosa of the distal duodenum of animal 13 obtained using the Hp Yellow and Hp Blue staining method (Anatech Ltd.). With this staining method, mucus appeared yellow, and bacteria within mucus and intestinal tissues appeared blue. (A) Crypt (Cr) with a layer of mucus stained yellow (Mu) coating the epithelium (Ep) stained blue with two Campylobacter cells also stained blue (arrows) associated with a strand of mucus (MuS). (B) Single Campylobacter cell (arrow; note the spiral morphology) associated with a thin strand of mucus within a duodenal crypt. Bars = 10 μm.
Tissue samples taken from all animals were positive for C. lanienae DNA. Overall, 52.9% (n = 110) of the tissue samples were positive for the bacterium. The highest frequency of samples positive for C. lanienae was obtained for the large intestine, but a relatively high percentage of samples taken from the duodenum also were positive for this bacterium (Fig. 5B). C. lanienae DNA was most abundant in tissue samples from the cecum, ascending colon, descending colon, and rectum on average (Fig. 5B).
DISCUSSION
Elucidating the site of colonization, the mechanisms utilized by campylobacters to colonize and persist within the intestine of cattle, and factors that interfere with this process is fundamental if efficacious management strategies are to be developed. In the current study, both C. jejuni (70% of the samples examined) and C. lanienae (56%) were commonly associated with intestinal tissues obtained from steer A. In contrast, DNA of C. lanienae (66%) but not DNA of C. jejuni (5%) was commonly detected in tissues obtained from steer B. Although DNA of both taxa were abundant, appreciable quantities of each bacterium occurred in relatively restricted sites along the intestinal tract. C. jejuni was primarily concentrated in the proximal region of the small intestine. In contrast, C. lanienae populations were concentrated in the large intestine, including the cecum, proximal ascending colon, distal descending colon, and rectum. In a second study, C. jejuni and C. lanienae abundance was examined at 11 intestinal sites in 20 arbitrarily selected beef cattle, and the data obtained supported the conclusion that C. jejuni primarily colonizes the small intestine, whereas C. lanienae dwells in the large intestine. Other workers have isolated Campylobacter species from various regions of the GI tract of cattle (1, 2, 3, 4, 5, 13, 28, 32, 37, 39, 40). However, all previous studies have relied on relatively restricted and/or undefined sampling sites in healthy or diseased tissues. For example, Stanley et al. (37) cultured thermophilic campylobacters from digesta collected at single locations in the true stomach (i.e., omasum), small intestine, cecum, and colon of healthy cattle, but they did not disclose the specific locations from which samples were obtained.
It is widely thought that campylobacters are nonpathogenic in adult ruminants (38). Other animals, such as avians, rodents, and dogs, also appear to contain C. jejuni as part of their normal gut flora (16). Although cattle are putatively asymptomatic carriers of campylobacters, campylobacters can incite enteritis in calves (1, 2, 3, 4, 5, 8, 11, 39, 40), in which pathological changes are typically observed in the ileum and large intestine (3, 39, 40). In other animals suffering from enteritis caused by Campylobacter species, the ileum and colon are typically infected, and the bacteria interfere with the absorptive capacity of the intestine (16). We observed conspicuously large quantities of C. jejuni DNA in the ileum of one animal, a 24-month-old heifer suffering from pneumonia at the time of slaughter. This animal also was shedding conspicuously large numbers of C. jejuni in its feces (∼106 CFU g−1), but we noted no conspicuous evidence of infection (e.g., inflammation) or diarrhea in this animal. When gut loops of the jejunum and anterior ileum of calves were used, none of 15 C. jejuni strains induced abnormal fluid accumulation or histopathological changes (23). Although in this study evidence of diarrhea was not observed in small intestinal gut loops, the absence of diarrhea in adult cattle colonized by Campylobacter species may not be a good indicator of nonpathogenesis. Diarrhea in humans and other mammals is often malabsoptive in nature, but adult cattle are able to absorb enormous quantities of water in their colons (14), which may explain why they remain asymptomatic. Whether the abundance of C. jejuni in the ileum of the animal suffering from pneumonia was related to pathogenesis is not known and warrants study. Furthermore, the influence of the health status of adult cattle on pathogenesis caused by C. jejuni may be important. Immunodeficient humans may be more prone to infections by Campylobacter species (35), but the role of the physiological status of humans in infection by campylobacters is uncertain. Some anecdotal evidence suggests that shedding of campylobacters is increased in stressed livestock (41); it is possible that physiologically stressed livestock are more susceptible to infection by campylobacters, and this warrants study.
The large and small intestines are diverse organs physiologically and microbiologically, and studying the colonization of the GI tract of cattle by Campylobacter species presented a number of logistical problems. Two of the most salient difficulties were the substantial length of the intestines of adult cattle and the fastidiousness of Campylobacter species. The intestinal tract of a full-grown ox is typically 33 to 63 m long (29). The two animals that we examined in detail in the current study were housed in adjacent stalls during the experimental period (ca. 170 days), and they were young adults of similar weight, age, genetics, and nutrition (within the feedlot). Yet surprisingly they possessed intestinal tracts that differed greatly in length. Despite being 60 kg heavier, steer A had a much shorter intestinal tract (∼26 m) than steer B (∼40 m); the major difference between the two animals was primarily in the length of the jejunum (19 m compared to 33 m). Very limited research has addressed intestinal lengths in cattle, and the reasons for the tremendous discrepancy in the lengths of the intestinal tracts of the two cattle are currently unknown. Regardless of the reasons for the differences in length, both intestinal tracts were extensive, which presented a major obstacle in elucidating the site of colonization. The second obstacle faced was the need to enumerate Campylobacter species for a large number of samples. Tissue samples were obtained at a maximum of 6.7-cm intervals along the entire length of the intestinal tract of both steers examined, which resulted in a large number of tissue samples (∼1,000) obtained in 2 days. Because of the inherent limitations of culture-based enumeration methods combined with the extensive lengths of the intestinal tracts of adult cattle, we employed PCR detection and quantification methods. Furthermore, a decision was made to target both C. jejuni and C. lanienae. C. jejuni was investigated because it is currently recognized as the primary species that incites gastroenteritis in humans (24). Although very little is known about the pathogenicity of C. lanienae, this bacterium was initially isolated from the feces of healthy abattoir workers exposed to pigs and cattle (22). C. lanienae is commonly shed in the feces of beef cattle (17, 18, 19, 20). Although many cattle-associated strains of C. lanienae cannot be cultured on charcoal cefoperazone desoxycholate agar (17), the high frequency of occurrence of this organism in cattle makes it a candidate for an indicator microorganism (e.g., for antimicrobial resistance development in campylobacters). In the current study, we expressed the densities of C. jejuni and C. lanienae as the numbers of genome copies present in 2 μl of template. In some instances, very high densities of cells were detected. For example, we observed densities of C. jejuni and C. lanienae as high as ∼3,000 and ∼27,000 genome copies in templates extracted from digesta, respectively; these densities convert to ∼7 × 105 CFU g−1 for C. jejuni and ∼7 × 106 CFU g−1 for C. lanienae (18). However, cell densities in the ranges from 103 to 104 CFU g−1 and from 104 to 105 CFU g−1 were more common for C. jejuni and C. lanienae, respectively.
PCR-based technologies are not without their logistical problems. The first obstacle is the adequate removal of PCR inhibitors; feces and their constituents contain a number of inhibitors (21, 27, 42). Inclusion of an IAC is considered a prerequisite for diagnostic PCR (15), and we used an IAC designed so that it was amplified with a Campylobacter genus-specific primer set (17). Using this method, 3% of the tissue samples obtained from the intestinal tracts of the two chronically shedding steers did not produce either a genus or IAC amplicon, which was indicative of the presence of PCR inhibitors or inadequate extraction. A large percentage of the remaining samples were positive for campylobacters when a genus-specific primer set was used; for steers A and B, 81% and 29% of the tissue samples were positive for Campylobacter DNA, respectively. Furthermore, Campylobacter DNA was detected throughout the intestinal tracts of both animals, thereby limiting any useful conclusions concerning the site of colonization based on the genus-specific primer set. The pancreas of steer B also was positive for Campylobacter DNA but did not contain either C. jejuni or C. lanienae. To our knowledge, infection of the bovine pancreas has not been documented, but pancreatitis in humans infected with Campylobacter species has been reported (6, 10, 12, 33).
Intestinal samples that were positive for Campylobacter DNA were subsequently subjected to nested or nonnested RTQ-PCR (18). Nonnested RTQ-PCR targeting the mapA gene of C. jejuni was shown to have adequate sensitivity and specificity, but nested RTQ-PCR targeting the 16S rRNA gene was required to quantify C. lanienae. To our knowledge, this is the first time that this technology has been used to quantify Campylobacter populations in GI tracts. The two steers selected for study were identified as chronic shedders of both C. jejuni and C. lanienae (20), and the reasons that only one bacterium was abundant in each animal are not known. However, steer A was shedding particularly large quantities (≥104 CFU g−1) of C. jejuni just prior to slaughter, whereas steer B was not (20). Conversely, steer B but not steer A was shedding a large number of C. lanienae cells just before slaughter (20). The shedding data for the steers before slaughter also correspond to the low densities of C. jejuni and C. lanienae cells that we found in the digesta of steers B and A, respectively, in the current study. This suggests that the quantities of Campylobacter cells shed in feces are correlated with the population densities of the bacteria associated with intestinal tissues and raises questions about what factors influence population increases in the GI tract and thus shedding of campylobacters in feces.
In the current study, we carefully washed the intestinal surface with phosphate-buffered saline in order to remove digesta but maintain as much mucus integrity as possible. Not surprisingly, the viscosity of the digesta and the quantity of mucus varied tremendously in the different regions of the intestinal tract. For example, digesta in the rectum was very viscous compared to the liquid digesta found in the duodenum and proximal jejunum. There were two potential artifacts resulting from the tissue washing step: (i) inadequate removal of digesta, particularly viscous digesta in the colon, resulting in contamination of the mucosa with Campylobacter cells present in the digesta; and (ii) overexuberant washing, resulting in the loss of campylobacters associated with the mucosal surface. Although it was not possible to ensure complete removal of the digesta from the surface of the mucosa and thus eliminate all Campylobacter cells present in the digesta, the use of RTQ-PCR to quantify campylobacters obtained from washed tissue samples collected in close proximity to each other indicated that we were indeed measuring tissue-associated bacteria (i.e., inadequate washing of a tissue sample would be detected by comparison to adjacent samples that were properly washed). The second potential artifact was overwashing of the intestinal tissues. However, a comparison of C. jejuni and C. lanienae populations present in the large intestine did not support this possibility. Considerable quantities of C. jejuni DNA were observed in the digesta but not in washed tissues of the large intestine. In contrast, substantial quantities of C. lanienae DNA were observed both in the digesta and in association with tissues obtained from the large intestine; the same digesta and tissues were processed for both bacteria. Furthermore, microscopic examination of washed tissues revealed the presence of spiral-shaped bacteria in association with intestinal mucosa.
The vast majority of previous studies examining the colonization of the GI tracts of healthy cattle by Campylobacter species have relied on microbiological assessments of campylobacters present in digesta (13, 28, 32, 37). The findings of our comparison of Campylobacter abundance in digesta and Campylobacter abundance associated with intestinal tissues illustrate the potential pitfalls of relying on digesta to elucidate colonization sites within the intestinal tract. For example, by sampling digesta within the large intestine, a researcher may erroneous conclude that C. jejuni readily colonizes the ascending colon of healthy cattle. This would be consistent with data for other animals, in which the mucus layer and crypts of the intestinal mucosa of the colon and cecum are colonized (16). However, our results demonstrate that this is not the case in presumably healthy adult cattle, in which C. jejuni primarily colonizes the small intestine and bacterial cells released from this site subsequently accumulate in digesta within the colon. Interestingly, our results also clearly show that C. lanienae colonizes the large intestine and is commonly associated with the cecum, descending colon, and rectum. Conditions (e.g., oxygen tension, pH, host receptors, microflora) vary substantially in different parts of the intestinal tract, which may explain the different colonization sites for these two bacteria within the intestinal tract of cattle.
In conclusion, we utilized conventional and quantitative PCR to determine where C. jejuni and C. lanienae colonize the intestinal tract of cattle. Our results suggest that C. jejuni primarily colonizes the small intestine (i.e., duodenum and jejunum) of healthy cattle, whereas C. lanienae is primarily a large intestine dweller. Thus, these two bacteria occur in distinctly different locations in the intestines of cattle. One aspect of on-farm food safety is implementation of methods that prevent colonization of the GI tract and thereby reduce shedding of human-pathogenic bacteria in feces. Knowledge of the site and process of colonization of the GI tract of cattle by Campylobacter species should facilitate the development of efficacious on-farm management strategies. For example, if an efficacious “probiotic” is to be developed, it is important to understand the microbial ecology of the intestinal tract where the Campylobacter species reside.
Acknowledgments
We thank the following people: Darryl Gibb and Tim McAllister for providing the two chronically shedding beef steers; Fred Van Herk and Ben Van Herk for providing slaughter facilities and expertise; the owner and staff of Ben's Quality Meats, Picture Butte, Alberta, Canada, for providing intestines of beef cattle; and Susan Bach for her assistance with intestinal dissections. We extend special thanks to Grant Duke, Jenny Gusse, Kathaleen House, Lauri Lintott, and Wilco Tymensen for volunteering their time to assist with the processing of intestinal samples of the two chronically shedding steers.
This study was supported by a grant from the Canada-Alberta Beef Industry Development Fund (CABIDF).
Footnotes
Contribution 04041 from the Agriculture and Agri-Food Canada Research Centre, Lethbridge, Alberta, Canada.
REFERENCES
- 1.Al-Mashat, R. R., and D. J. Taylor. 1980. Campylobacter spp. in enteric lesions in cattle. Vet. Rec. 107:31-34. [DOI] [PubMed] [Google Scholar]
- 2.Al-Mashat, R. R., and D. J. Taylor. 1980. Production of diarrhoea and dysentery in experimental calves by feeding pure cultures of Campylobacter fetus subspecies jejuni. Vet. Rec. 107:459-464. [DOI] [PubMed] [Google Scholar]
- 3.Al-Mashat, R. R., and D. J. Taylor. 1981. Production of enteritis in calves by the oral inoculation of pure cultures of Campylobacter fecalis. Vet. Rec. 109:97-101. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mashat, R. R., and D. J. Taylor. 1983. Bacteria in enteric lesions of cattle. Vet. Rec. 112:5-10. [DOI] [PubMed] [Google Scholar]
- 5.Al-Mashat, R. R., and D. J. Taylor. 1983. Production of enteritis in calves by the oral inoculation of pure cultures of Campylobacter fetus subspecies intestinalis. Vet. Rec. 112:54-58. [DOI] [PubMed] [Google Scholar]
- 6.Castilla-Higuero, L., M. Castro-Fernandez, and P. Guerrero-Jimenez. 1989. Acute pancreatitis associated with Campylobacter enteritis. Dig. Dis. Sci. 34:961-962. [DOI] [PubMed] [Google Scholar]
- 7.Clark, C. G., L Price, R. Ahmed, D. L. Woodward, P. L. Melito, F. G. Rodger, F. Jamieson, B. Cieben, A. Li, and A. Ellis. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walketon, Ontario. Emerg. Infect. Dis. 9:1232-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diker, K. S., S. Diker, and M. B. Ozlem. 1990. Bovine diarrhea associated with Campylobacter hyointestinalis. Zentbl. Vetmed. Reihe B 37:158-160. [DOI] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezpeleta, C., P. R. de Ursua, F. Obregon, F. Goni, and R. Cisterna. 1992. Acute pancreatitis associated with Campylobacter jejuni bacteremia. Clin. Infect. Dis. 15:1050. [DOI] [PubMed] [Google Scholar]
- 11.Firehammer, B. D., and L. L. Myers. 1981. Campylobacter fetus subsp jejuni: its possible significance in enteric disease of calves and lambs. Am. J. Vet. Res. 42:918-922. [PubMed] [Google Scholar]
- 12.Gallagher, P., P. Chadwick, D. M. Jones, and L. Turner. 1981. Acute pancreatitis associated with Campylobacter infection. Br. J. Surg. 68:383. [DOI] [PubMed] [Google Scholar]
- 13.Garcia, M. M., H. Lior, R. B. Stewart, G. M. Ruckerbauer, J. R. R. Trudel, and A. Skljarevski. 1985. Isolation, characterization, and serotyping of Campylobacter jejuni and Campylobacter coli from slaughter cattle. Appl. Environ. Microbiol. 49:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecker, J. F., and W. L. Grovum. 1975. Rates of passage of digesta and water absorption along the large intestines of sheep, cows, and pigs. Aust. J. Biol. Sci. 28:161-167. [DOI] [PubMed] [Google Scholar]
- 15.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Making internal amplification control mandatory for diagnostic PCR. J. Clin. Microbiol. 41:5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, L., and D. J. Kopecko. 2000. Interactions of Campylobacter with eukaryotic cells: gut luminal colonization and mucosal invasion mechanisms, p. 191-215. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, D.C.
- 17.Inglis, G. D., and L. D. Kalischuk. 2003. Use of PCR for direct detection of Campylobacter species in bovine feces. Appl. Environ. Microbiol. 69:3435-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inglis, G. D., and L. D. Kalischuk. 2004. Quantification of Campylobacter jejuni and Campylobacter lanienae in bovine feces by real-time quantitative PCR. Appl. Environ. Microbiol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inglis, G. D., L. D. Kalischuk, and H. W. Busz. 2003. A survey of Campylobacter species shed in faeces of beef cattle using polymerase chain reaction. Can. J. Microbiol. 49:655-661. [DOI] [PubMed] [Google Scholar]
- 20.Inglis, G. D., L. D. Kalischuk, and H. W. Busz. 2004. Chronic shedding of Campylobacter species in beef cattle. J. Appl. Microbiol. 97:410-420. [DOI] [PubMed] [Google Scholar]
- 21.Koonjul, P. K., W. F. Brandt, J. M. Farrant, and G. G. Lindsey. 1999. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res. 27:915-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logan, J. M., A. Burnens, D. Linton, A. J. Lawson, and J. Stanley. 2000. Campylobacter lanienae sp. nov., a new species isolated from workers in an abattoir. Int. J. Syst. Evol. Microbiol. 50:865-872. [DOI] [PubMed] [Google Scholar]
- 23.Manninen, K. I., J. F. Prescott, and I. R. Dohoo. 1982. Pathogenicity of Campylobacter jejuni isolates from animals and humans. Infect. Immun. 38:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaud, S., S. Ménard, R. D. Arbeit. 2004. Campylobacteriosis, Eastern Townships, Québec. Emerg. Infect. Dis. 10:1844-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minihan, D., P. Whyte, M. O'Mahony, S. Fanning, K. McGill, and J. D. Collins. 2004. Campylobacter spp. in Irish feedlot cattle: a longitudinal study involving pre-harvest and harvest phases of the food chain. J. Vet. Med. Ser. B 51:28-33. [DOI] [PubMed] [Google Scholar]
- 27.Monteiro, L., D. Bonnemaison, A. Vekris, K. G. Petry, J. Bonnett, R. Vidal, J. Cabrita, and F. Mégraud. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng, L. K., D. E. Taylor, and M. E. Stiles. 1988. Characterization of freshly isolated Campylobacter coli strains and suitability of selective media for their growth. J. Clin. Microbiol. 26:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickel, R., A. Schummer, E. Seiferle, and W. O. Sack. 1973. The viscera of the domestic mammals. Springer-Verlag, New York, N.Y.
- 30.Nielsen, E. M., J. Engberg, V. Fussing, L. Petersen, C. H. Brogren, and S. L. On. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J. Clin. Microbiol. 38:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.On, S. L. W., E. M. Nielsen, J. Engberg, and M. Madsen. 1998. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, EpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol. Infect. 20:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, K., H. Masaki, and Y. Tokumaru. 1995. Isolation of Campylobacter spp. from slaughtered cattle and swine on blood-free selective medium. J. Vet. Med. Sci. 57:1085-1087. [DOI] [PubMed] [Google Scholar]
- 33.Ponka, A., and T. U. Kosunen. 1981. Pancreas affection in association with enteritis due to Campylobacter fetus ssp. jejuni. Acta Med Scand. 209:239-240. [DOI] [PubMed] [Google Scholar]
- 34.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. L. Willems, K. E. Dingle, F. M. Colles, and D. A. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology Press, Washington, DC.
- 36.Stanley, K., and K. Jones. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94(Suppl.):104S-113S. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472-480. [DOI] [PubMed] [Google Scholar]
- 38.Stern, N. J. 1992. Reservoirs for Campylobacter jejuni and approaches for intervention in poultry, p. 49-60. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, DC.
- 39.Terzolo, H. R., and G. H. K. Lawson. 1987. Enteric campylobacter infection in gnotobiotic calves and lambs. Res. Vet. Sci. 43:72-77. [PubMed] [Google Scholar]
- 40.Warner, D. P., and J. H. Bryner. 1984. Campylobacter jejuni and Campylobacter coli inoculation of neonatal calves. Am. J. Vet. Res. 45:1822-1824. [PubMed] [Google Scholar]
- 41.Whyte, P., J. D. Collins, K. McGill, C. Monahan, and H. O'Mahony. 2001. The effect of transportation stress on excretion rates of campylobacters in market-age broilers. Poult. Sci. 80:817-820. [DOI] [PubMed] [Google Scholar]
- 42.Widjojoatmodjo, M. N., A. C. Fluit, R. Torenma, G. P. H. T. Verdonk, and J. Verhoef. 1992. The magnetic immunopolymerase chain reaction assay for direct detection of salmonellae in fecal samples. J. Clin. Microbiol. 30:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]