Abstract
Six fluorescein-labeled peptide nucleic acid oligomers targeting Listeria-specific sequences on the 16S ribosomal subunit were evaluated for their abilities to hybridize to whole cells by fluorescence in situ hybridization (FISH). Four of these probes yielded weak or no fluorescent signals after hybridization and were not investigated further. The remaining two FISH-compatible probes, LisUn-3 and LisUn-11, were evaluated for their reactivities against 22 Listeria strains and 17 other bacterial strains belonging to 10 closely related genera. Hybridization with BacUni-1, a domain-specific eubacterial probe, was used as a positive control for target accessibility in both Listeria spp. and nontarget cells. RNase T1 treatment of select cell types was used to confirm that positive fluorescence responses were rRNA dependent and to examine the extent of nonspecific staining of nontarget cells. Both LisUn-3 and LisUn-11 yielded rapid, bright, and genus-specific hybridizations at probe concentrations of approximately 100 pmol ml−1. LisUn-11 was the brightest probe and stained all six Listeria species. LisUn-3 hybridized with all Listeria spp. except for L. grayi, for which it had two mismatched bases. A simple ethanolic fixation yielded superior results with Listeria spp. compared to fixation in 10% buffered formalin and was applicable to all cell types studied. This study highlights the advantages of peptide nucleic acid probes for FISH-based detection of gram-positive bacteria and provides new tools for the rapid detection of Listeria spp. These probes may be useful for the routine monitoring of food production environments in support of efforts to control L. monocytogenes.
The genus Listeria is comprised of six species, L. monocytogenes, L. grayi, L. innocua, L. ivanovii, L. seeligeri, and L. welshimeri (17). Of these species, only one, L. monocytogenes, is pathogenic to humans. The high mortality (∼25 to 30%) associated with food-borne cases of listeriosis underscores the need for control of this pathogen. It has been suggested that the presence of generic Listeria in a food production facility can serve as an indicator for conditions that may support the growth of L. monocytogenes (5). New rapid genotypic methods for the detection of generic Listeria may therefore allow more efficient monitoring of the plant sanitation practices aimed at reducing the incidence of listeriosis.
Fluorescence in situ hybridization (FISH) is a rapid and highly specific nucleic acid-based method for the whole-cell identification of bacteria (3, 13). In the FISH technique, fluorescently labeled nucleic acid probes complementary to genus- or species-specific rRNA sequences are hybridized to whole bacterial cells, resulting in the selective staining of target cells (13). As a whole-cell method, FISH allows the simultaneous collection of information on both cell morphology and molecular identity. Recently, two DNA-based FISH probes have been developed for the detection of Listeria spp. (20, 21). The first, Lis-1255 (Escherichia coli nucleotide positions 1255 through 1272), was originally reported for use as a PCR primer (26) but has been adapted for use as a FISH probe (20, 21). This probe is complementary to the 16S rRNA of all six species of Listeria but also reacts with Brochothrix spp. (20, 21, 25). The other probe, Lis-637 (E. coli nucleotide positions 637 through 658) (20, 21), reacts with all members of the genus Listeria except L. grayi. An ideal probe for the detection of generic Listeria would both be restricted to the genus and react with all six species.
Gram-positive bacteria present unique challenges to the use of DNA-based FISH probes due to the permeability barrier posed by their thick and highly anionic cell walls (8, 15). As a result, DNA-based FISH analysis of gram-positive cells often requires extensive preparatory steps, including lysozyme and proteinase K digestions (15, 25). Because an unknown sample may contain cells that differ markedly in their requirements for permeabilization, these steps may result in overdigestion and cell loss (25). Extensive processing may also result in the alteration of cellular light-scatter properties, which could interfere with analyses by fluorescence microscopy or flow cytometry that are often used in conjunction with FISH.
Peptide nucleic acid (PNA) is a pseudopeptide DNA mimic with an uncharged, achiral backbone (24). The unique chemical makeup of PNA probes confers a number of beneficial properties, including rapid hybridization kinetics, resistance to nucleases, and the ability to hybridize to positions on the ribosome that are inaccessible to DNA probes (24). PNA probes are also able to penetrate recalcitrant biological structures such as mycobacterial and gram-positive cell walls (22). In the present study, six Listeria-targeted PNA probes were evaluated for their suitability as FISH probes. Two probes were found to be FISH compatible and were evaluated for their specificities against a number of target and nontarget cells. This work adds two new probes to the set of tools available for the rapid molecular detection of Listeria spp. and clearly demonstrates the advantages of PNA probes for FISH-based detection of this genus.
MATERIALS AND METHODS
Chemicals.
RNase T1 (EC 3.1.27.3, 90 Kunitz units mg−1) was from Sigma-Aldrich (St. Louis, MO); Unless otherwise stated, all chemicals were from Sigma-Aldrich or from Fisher Scientific (Itasca, IL). Microbiological media were from Difco Laboratories (Detroit, MI).
Probe design and synthesis.
The common and systematic names, ribosomal locations (Escherichia coli base and helix numbering), and sequences of the PNA probes used in this study are given in Table 1. Probes were designed and synthesized at Boston Probes, Inc. (Bedford, MA), and supplied by Applied Biosystems, Inc. (Foster City, CA). Probe design was carried out as described previously (22). Briefly, 16S rRNA sequences representing all six species of Listeria and several closely related genera were aligned using MegAlign software (version 4.0; DNASTAR, Madison, WI). Choices regarding which genera should be represented in these alignments were informed by previous studies on the 16S rRNA-based phylogeny of the listeriae (4, 11, 19). From these alignments, potentially diagnostic target sequences were identified and checked against the GenBank database for significant similarities to nontarget sequences using BLAST (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov]) and GeneMan softwares (version 3.3; DNASTAR, Madison, WI). Candidate sequences were also screened for secondary structure using PrimerSelect software (version 4.03; DNASTAR, Madison, WI). Probes were synthesized according to the method previously described by Stender et al. (23), with the notable exception that solubility-enhancing groups (7) were not used, as these have been implicated in reduced hybridization efficiency against gram-positive bacteria (24). PNA probes (50 μl) were received suspended in 50% N,N-dimethylformamide (DMF) at an approximate concentration of 250 to 350 μM. Probes were diluted to a working concentration of approximately 100 μM in 50% DMF-water and stored in polypropylene microcentrifuge tubes at −20°C until needed.
TABLE 1.
Common names, systematic names, locations, and sequences of the peptide nucleic acid oligomers used in this study
| Probe name | Systematic namea | Probe location (base no., helix no.)b | Sequence (N terminus to C terminus)c |
|---|---|---|---|
| LisUn-2-1 | S-G-Lis-0085-a-A-15 | 85-99, H61 | AAC TTT GGA AGA GCA |
| LisUn-2-2 | S-S-Lg-0086-b-A-14 | 86-99, H61 | ACG ACC AAA GGA GC |
| LisUn-3 | S-G-Lis-0134-a-A-15 | 134-148, H122 and H144 | CCC CAA CTT ACA GGC |
| LisUn-11 | S-G-Lis-0466-a-A-14 | 466-480, H441 | AAG GGA CAA GCA GT |
| LisUn-19 | S-G-Lis-1433-a-A-17 | 1433-1449, H1399 | GGT TAC CCT ACC GAC TT |
| LisUn-20 | S-G-Lis-1437-a-A-17 | 1437-1453, H1399 | TAA AGG TTA CCC TAC CG |
| BacUni-1d | S-D-Bact-0340-a-A-15 | 340-354, H339 and H47 | CTG CCT CCC GTA GGA |
Bacterial strains.
A total of 39 bacterial strains from both the genus Listeria and 10 closely related genera were examined. Nontarget strains were chosen in light of previous studies on the 16S rRNA phylogeny of Listeria (4, 11, 19). The strains and their sources are listed in Tables 2 and 3. Each of the six Listeria species was represented by at least two strains, including the type strain for each species. Due to the paramount importance of detecting L. monocytogenes, this species was represented by 12 strains, including strains belonging to the three serotypes most often implicated in human listeriosis (1/2a, 1/2b, and 4b).
TABLE 2.
Inclusivity data for LisUn-3 and LisUn-11 PNA probesf
| Organism | Strain | Comment | Result of hybridization
|
||
|---|---|---|---|---|---|
| BacUni-1 | LisUn-3 | LisUn-11 | |||
| Listeria monocytogenes | ATCC 15313a | Type strain | + | + | + |
| Listeria monocytogenes | FSL-J2-020b | Serotype 1/2a | + | + | + |
| Listeria monocytogenes | FSL-J2-066b | Serotype 1/2a | + | + | + |
| Listeria monocytogenes | FSL-J1-177b | Serotype 1/2b | + | + | + |
| Listeria monocytogenes | FSL-J2-064b | Serotype 1/2b | + | + | + |
| Listeria monocytogenes | FSL-J1-031b | Serotype 4a | + | + | + |
| Listeria monocytogenes | DD6824b | Serotype 4a | + | + | + |
| Listeria monocytogenes | FSL-C1-122b | Serotype 4b | + | + | + |
| Listeria monocytogenes | FSL-J1-110b | Serotype 4b | + | + | + |
| Listeria monocytogenes | DD6821b | Serotype 4c | + | + | + |
| Listeria monocytogenes | ATCC 19118c | Serotype 4e | + | + | + |
| Listeria grayi | KC1773b,d | Type strain | + | − | + |
| Listeria grayi | ATCC 25401a,e | + | − | + | |
| Listeria grayi | ATCC 700545a | + | − | + | |
| Listeria innocua | ATCC 33090a | Type strain | + | + | + |
| Listeria innocua | ATCC 51742c | + | + | + | |
| Listeria ivanovii | ATCC 19119a,b | Type strain | + | + | + |
| Listeria ivanovii | —b,g | + | + | + | |
| Listeria seeligeri | ATCC 35967a,b | Type strain | + | + | + |
| Listeria seeligerii | —b,g | + | + | + | |
| Listeria welshimeri | ATCC 35897a | Type strain | + | + | + |
| Listeria welshimeri | JLJ-20f | + | + | + | |
From Microbiologics, Inc., St. Cloud, MN.
From Martin Wiedmann, Cornell University, Ithaca, NY.
From the American Type Culture Collection, Manassas, VA.
Strain is identical to ATCC 19120.
Type strain for L. murrayi.
From Kathy Glass, University of Wisconsin Food Research Institute, Madison, WI.
—, no strain designation.
TABLE 3.
Exclusivity data for LisUn-3 and LisUn-11 PNA probes
| Organism | Strain | Result of hybridization
|
||
|---|---|---|---|---|
| BacUni-1 | LisUn-3 | LisUn-11 | ||
| Bacillus cereus | ATCC 11778a | + | − | − |
| Bacillus licheniformis | ATCC 12759a | + | − | − |
| Bacillus subtilis | ATCC 33608a | + | − | − |
| Brochothrix campestris | ATCC 43754a | + | − | − |
| Brochothrix thermosphacta | ATCC 11509a | + | − | − |
| Carnobacterium divergens | NRRL B-14830b | + | − | − |
| Carnobacterium piscicola | NRRL B-14829b | + | − | − |
| Enterococcus faecalis | DSCC 4025c | + | − | − |
| Erysipelothrix rhusiopathiae | ATCC 19414a | + | − | − |
| Gemella haemolysans | ATCC 10379a | + | − | − |
| Kurthia sp. | DSCC 7003c | + | − | − |
| Lactobacillus fermentum | ATCC 14931a | + | − | − |
| Staphylococcus aureus | ATCC 29123a | + | − | − |
| Staphylococcus carnosus | NRRL B-14760b | + | − | − |
| Staphylococcus schleiferi subsp. schleiferi | NRRL B-14775b | + | − | − |
| Staphylococcus xylosus | ATCC 29971a | + | − | − |
| Streptococcus vestibularis | ATCC 49124a | + | − | − |
From the American Type Culture Collection, Manassas, VA.
From the Agricultural Research Service (ARS) Culture Collection, Peoria, IL.
From the University of Wisconsin—Madison Bacteriology Departmental Stock Culture Collection.
Cell growth and fixation.
Cells grown for 18 to 24 h at 30°C (or at 25°C for Brochothrix spp.) in appropriate media were harvested by centrifugation (2,000 × g, 5 min). All strains except for Erysipelothrix rhusiopathiae, Gemella haemolysans, Carnobacterium divergens, Carnobacterium piscicola, and Lactobacillus fermentum were grown in Columbia broth. E. rhusiopathiae and G. haemolysans were grown in filter-sterilized Columbia broth plus 5% bovine calf serum (Serum Supreme; Cambrex Bio Science Walkersville, Inc., Walkersville, MD). The remaining strains were grown in MRS broth. Cells were washed once in phosphate-buffered saline (PBS) and fixed in either 10% buffered formalin or a 50% solution of absolute ethanol in PBS. For formalin fixation, washed cells were resuspended in one milliliter of 10% buffered formalin and fixed for 1 h at room temperature. Cells were centrifuged (2,000 × g, 5 min), and the fixative was removed. Fixed cells were washed again in PBS, resuspended in a 50:50 mixture of absolute ethanol/RNase-free distilled water, and stored at −20°C until use. For ethanol-based fixation, cells were harvested as described above, washed once, resuspended in a 50:50 mixture of absolute ethanol and PBS, and stored at −20°C. As a matter of convenience, most cell preparations were made in advance and stored under these conditions for up to a week prior to hybridization experiments.
Sequence-based strain identification.
In order to unambiguously demonstrate the specificities of the probes tested and to verify the taxonomic identities of the cultures used, all bacterial strains were identified through the sequencing of PCR products generated from 16S and/or 23S rDNA. Sequences were compared against the GenBank database using the BLAST program, and results were compared with nominal strain identities (data not shown).
Hybridization and microscopy.
Approximately 108 cells (100-μl aliquots of previously prepared cells) were used per hybridization reaction. Cell preparations were centrifuged (2,000 × g, 5 min) and the supernatant removed. Cell pellets were resuspended in 50 μl room temperature PNA hybridization buffer (20 mM Tris [pH 9.0], 100 mM NaCl, 0.5% sodium dodecyl sulfate) containing approximately 100 pmol ml−1 of each probe being tested or 300 pmol ml−1 of a universal bacterial probe, BacUni-1 (16). Hybridization reactions were performed on a PCR block set to a constant temperature of 55°C (DNA ThermalCycler 480; Applied Biosystems, Foster City, CA) in 0.5-ml thin-walled PCR tubes (Corning Life Sciences, Acton, MA). Cells were hybridized for up to 1 h, and then 500 μl PNA wash solution (10 mM Tris [pH 9.0], 1 mM EDTA) preheated to the hybridization temperature was added to each reaction. Cells were incubated in wash solution for another 10 min, pelleted (2,000 × g, 7 min), resuspended in 500 μl fresh, preheated wash solution, and incubated for another 20 min at the same temperature. Tubes were thoroughly vortexed whenever PNA wash buffer was added. At the end of this second wash period, hybridized cells were pelleted (2,000 × g, 7 min) and resuspended in a small amount (ca. 25 to 30 μl) of the remaining supernatant. Cell suspensions (2 μl) were smeared onto clean glass microscope slides and either air dried or dried on a PCR block set to 70°C. Bacterial smears were then mounted in VECTASHIELD mounting medium (Vector Labs, Burlingame, CA) and viewed with a fluorescence microscope. Hybridization results were scored as positive or negative.
RNase treatment.
In order to verify that PNA probes were targeting rRNA and to investigate the potential for nonspecific binding due to probe interactions with cell surfaces, suspensions of select cells were treated with RNase T1 prior to hybridization. Ethanol-fixed cells (100 μl) of L. monocytogenes ATCC 15313, G. haemolysans ATCC 10379, and Staphylococcus aureus ATCC 29123 were pelleted (2,000 × g, 7 min) and resuspended in 1 volume (100 μl) RNase T1 solution (4 mg ml−1 in 0.1 M Tris [pH 8.0], 1 mM EDTA; final enzyme concentration, 36 Kunitz units). Cell suspensions were digested for 1 h at room temperature, pelleted (2,000 × g, 7 min), resuspended in probe-containing hybridization buffer, and hybridized as described above. To facilitate comparison between treatments, RNase-digested cells were hybridized in parallel with untreated cell suspensions.
Flow cytometry.
Flow cytometry was used to examine the impact of different fixation methods on hybridization quality and to selectively identify Listeria subpopulations in mixed cultures after FISH. Samples were hybridized and washed as described above and then resuspended and diluted further (1:10) in 0.5 ml 10× TE buffer (0.1 M Tris [pH 8.0], 10 mM EDTA) prior to analysis with a FACSCalibur flow cytometer (BD Biosystems). Data were analyzed using FlowJo software (version 3.4; Tree Star, Inc., Ashland, OR).
RESULTS
Identification of FISH-suitable probes and effect of fixation method on hybridization quality.
All bacterial strains hybridized with BacUni-1, indicating that they were fully permeable to peptide nucleic acid probes. In an initial screen against the formalin-fixed cells of all six Listeria type strains, hybridization with either LisUn-2-1 or LisUn-2-2 did not result in a detectable signal and signals from LisUn-19 and LisUn-20 were weak. These probes were not investigated further. Both LisUn-3 and LisUn-11 were found to be suitable for FISH analysis. As expected from sequence alignments, LisUn-3 hybridized with all Listeria spp. except L. grayi, for which it had two mismatched bases. LisUn-11 hybridized with all Listeria spp.
The type strains of L. grayi and L. welshimeri yielded consistently lower staining with FISH probes than did the other formalin-fixed Listeria spp. These results suggested that either these cells had lower rRNA content or they were poorly permeabilized by formalin fixation. In an effort to obtain higher quality hybridizations, an alternative, ethanol-based fixation step was investigated. This involved simply resuspending freshly harvested, washed cells in a 50:50 mixture of ethanol and PBS, followed by storage at −20°C prior to use. This method yielded stronger, more vivid hybridizations for Listeria spp. and was found to be suitable for all cell types investigated. This fixation protocol was used throughout the rest of this work.
Relative probe hybridization intensities.
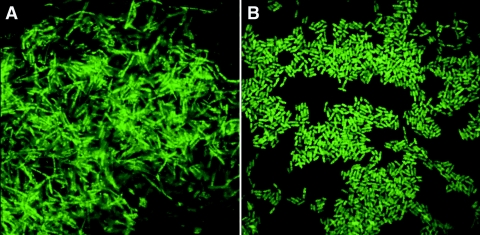
Although all cells yielded positive hybridizations with BacUni-1, the brightness of these hybridizations varied with each strain. Bacillus cereus, Kurthia spp., Enterococcus faecalis, C. divergens, and some Listeria spp. yielded exceptional results. The dimmest positive hybridization of BacUni-1 occurred with E. rhusiopathiae. Both LisUn-3 and LisUn-11 led to bright staining of stationary-phase target cells. However, LisUn-11 yielded uniformly brighter results than did LisUn-3, despite being present at a lower concentration (80 pmol ml−1 versus 95 pmol ml−1). For most nontarget cells, background staining was either undetectable or very low. Nonspecific staining was highest for G. haemolysans but was still too low to be mistaken for a positive reaction. A photograph depicting typical positive hybridization results for BacUni-1 is shown in Fig. 1A. A photograph depicting typical positive hybridization results for LisUn-11 is shown in Fig. 1B.
FIG. 1.
Typical PNA hybridization results. (A) Cells of Bacillus cereus ATCC 11778 hybridized with the universal bacterial probe BacUni-1. (B) Cells of Listeria monocytogenes FSL-C1-122 hybridized with the Listeria-specific probe LisUn-11. Cells were fixed in a 50:50 mixture of absolute ethanol and PBS. Hybridizations were carried out as described in Materials and Methods.
Probe inclusivity.
Inclusivity is a measure of how comprehensively a probe reacts within its target group. A fully inclusive probe will react with all members of its target group (e.g., the genus Listeria). In order to determine the properties of inclusivity for each probe, LisUn-3 and LisUn-11 were screened against 22 Listeria strains, including the type strains of all six species and representatives of the L. monocytogenes serotypes most often implicated in human disease (1/2a, 1/2b, and 4b). LisUn-3 hybridized with all Listeria spp. except L. grayi. LisUn-11 hybridized with all Listeria spp., including L. grayi. Table 2 summarizes these data. Alignments for the region targeted by LisUn-3 (E. coli positions 134 through 148, Fig. 2a) show that nontarget strains have at least three mismatches to the probe sequence, L. grayi has two mismatches, and the other Listeria spp. are fully complementary to the probe. Alignments for the region targeted by LisUn-11 (E. coli positions 466 through 480) (Fig. 2b) show that most nontarget strains have at least four mismatches to the probe sequence and several strains have base insertions, deletions, or both. All Listeria spp. contain target sequences that are fully complementary to LisUn-11.
FIG. 2.
Sequence variation in the 16S rRNA genes of Listeria and related genera. Alignments of partial sequences corresponding to the regions targeted by LisUn-3 (a) and LisUn-11 (b) are shown. The sequence of each probe is provided above the corresponding alignment. Residues differing from those found in the sequence of L. monocytogenes are boxed. For LisUn-3, the following GenBank accession numbers were aligned: X56153 (L. monocytogenes), X56151 (L. ivanovii), X56152 (L. innocua), X56148 (L. seeligeri), X56149 (L. welshimeri), X56150 (L. grayi), M58798 (Brochothrix thermosphacta), X60646 (Bacillus subtilis), X68417 (S. aureus), L14326 (G. haemolysans), X70321 (Kurthia zopfii), AJ301831 (E. faecalis), Z73313 (Carnobacterium sp.), and AF302116 (L. fermentum). The same sequences were aligned for LisUn-11, with the following exceptions: U84150 (L. monocytogenes), X98531 (L. seeligeri), and X56155 (B. thermosphacta). These alternate sequences were used to avoid sequence errors in X56153, X56148, and M58798 occurring in the region targeted by this probe. Abbreviations: Cbx, carboxyl terminus of probe, analogous to the 3′ terminus of DNA; Am, amino terminus of probe, analogous to the 5′ terminus of DNA; O, 8-amino-3,6-dioxaoctanoic acid linker; Flu, fluorescein.
Probe exclusivity.
Exclusivity is a measure of a probe's restriction to its target group. An exclusive probe will not react with cells outside its target group. In order to determine the properties of exclusivity for each probe, LisUn-3 and LisUn-11 were screened against 17 nontarget organisms from 10 closely related genera. As expected from alignments of probe target regions (Fig. 2), neither probe hybridized to any cell type outside the genus Listeria. Table 3 summarizes these data.
RNase treatment.
Positive hybridizations with the universal bacterial probe were interpreted as proof that nontarget cells were sufficiently permeabilized to allow access of PNA probes to target rRNA. However, because BacUni-1 is expected to hybridize with nearly all bacteria, it was recognized that false hybridizations due to nonspecific binding would not be immediately obvious. To address this potential pitfall and to verify the dependence on rRNA of the LisUn-3- and LisUn-11-conferred fluorescence in target cells, RNase-digested controls were examined. For the RNase-treated cells of L. monocytogenes strain ATCC 15313, no signal was detected after hybridization with BacUni-1, LisUn-3, or LisUn-11, although these probes yielded bright signals in parallel hybridizations with untreated controls. RNase-treated cells of G. haemolysans ATCC 10379 that were hybridized with both LisUn-3 and LisUn-11 showed the same degree of background signal as did nontreated controls hybridized with the same probes. Therefore, the higher level of fluorescence seen with these cells did not arise from cross-hybridization but was instead nonspecific in nature. Some RNase-treated cells of S. aureus ATCC 29123 were fluorescent after hybridization with BacUni-1, but the signal was drastically diminished compared to that of untreated control cells. Taken together, these data show that positive hybridizations with BacUni-1 were an accurate indication of cell permeability to PNA probes and that rRNA within Listeria spp. was the target for both LisUn-3 and LisUn-11.
Flow cytometry.
Flow cytometry was used to study the effects of fixative choice on hybridization quality. Figure 3 shows that ethanol-fixed cells (solid histogram) were brighter and more uniformly hybridized than formalin-fixed cells (dashed histogram), confirming microscopic observations. The geometric mean fluorescence was used to provide a measure of hybridization intensity. Here, the log of the fluorescence was averaged and reported as a scaled value in fluorescence units. The geometric mean fluorescence was 35.9 for ethanol-fixed cell populations and 27.8 for formalin-fixed cell populations. The coefficient of variation was used to provide a measure of hybridization spread, or uniformity. Coefficients of variation for the ethanol-fixed and formalin-fixed populations were 59.5 and 68.3, respectively.
FIG. 3.
Flow cytometric comparison of cell fixation protocols. Cells from the same overnight culture of L. monocytogenes strain Scott A were fixed using either formalin or ethanol, as described in the text. In these histograms, cell number is plotted against probe-conferred fluorescence. The rightward shift and narrower distribution for the ethanol-fixed population (solid histogram) shows that this method resulted in brighter and more homogeneous hybridizations than did formalin-based fixation (dashed histogram).
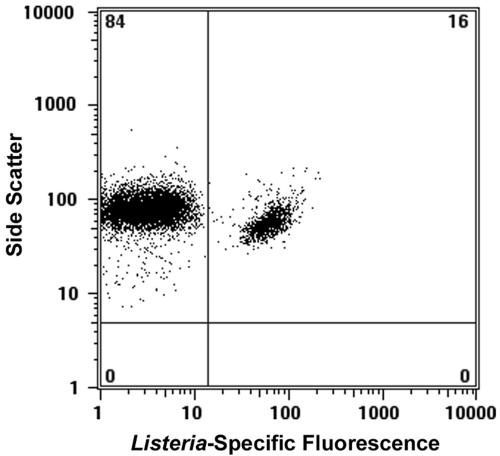
Flow cytometry was also used to demonstrate the ability of these probes to clearly differentiate Listeria spp. from nontarget flora in cell mixtures. The dot plot shown in Fig. 4 illustrates the probe-based detection of a subpopulation of L. monocytogenes (16% of the total population) against a high background of nontarget bacteria (L. fermentum, 84% of the total population).
FIG. 4.
Combination of PNA-FISH and flow cytometry for the detection of Listeria in mixed culture. This experiment demonstrates the ability of these probes to clearly differentiate Listeria spp. from nontarget flora in cell mixtures. A 1-ml sample was removed from a mixed culture of L. monocytogenes Scott A and Lactobacillus fermentum growing at 30°C in a nonselective medium (MRS broth). The sample was pelleted and then fixed for 15 min in a 50:50 mixture of ethanol and PBS. A 100-μl portion of this sample was hybridized for 30 min with 200 pmol ml−1 LisUn-11 and then analyzed by flow cytometry as described in Materials and Methods. Combined PNA-FISH and flow cytometry allowed a small number of L. monocytogenes cells (16% of total events collected) to be detected against a large background of L. fermentum (84% of total events collected).
DISCUSSION
Traditional media-based methods for the detection of Listeria spp. are time and labor intensive. Positive detection of Listeria in foods or environmental samples can take as long as 5 to 7 days using cultural approaches (14). New tools for the rapid and direct detection of Listeria are needed to help ensure the safety of foods or aid efforts to identify and control Listeria in food production environments. In this work, two rRNA-targeted peptide nucleic acid probes were used to achieve bright whole-cell hybridizations against stationary-phase Listeria spp. Neither of these probes showed any cross-hybridization with nontarget cells from 10 closely related genera. Probe specificities were not dependent on the use of toxic denaturants such as formamide, and combined hybridization and washing steps took only a fraction of the time required for DNA-based FISH of Listeria. Cell preparation was simple, and the use of highly toxic fixatives was avoided. No special preparatory treatments, such as acid hydrolysis or digestions with lysozyme, lysostaphin, or proteinase K, were required prior to hybridization. To our knowledge, this is the first report of peptide nucleic acid probes for use in the whole-cell detection of Listeria spp. and the first report of any single genus-specific FISH probe that reacts with all six species of Listeria.
Peptide nucleic acid probes have several advantages over DNA probes for the FISH-based detection of bacteria, especially gram-positive bacteria. Apart from fast reaction kinetics and resistance to nucleases, the two advantages most important to this application are the ability of PNA probes to penetrate recalcitrant biological structures such as the gram-positive cell wall and the capacity for these probes to bind regions on the ribosome that may be physically inaccessible to DNA-based FISH probes (24). The advantages of these two properties are discussed more thoroughly below.
The main obstacle to entry of rRNA-targeted nucleic acid probes into fixed cells is the cell wall. The gram-positive cell wall is considerably thicker than that of gram-negative bacteria and may contain large amounts of teichoic acids (up to 50% by cell wall mass) (8). These surface-displayed anionic polymers likely retard the passage of negatively charged polymers such as DNA probes through the cell wall. Although the gram-positive cell wall displays an intrinsically poor permeability to DNA probes, results can be improved through the use of alternative (e.g., nonaldehyde) fixation protocols, including heat or alcohol-based methods (13, 18, 25). Because these methods yield improved results with DNA probes, it was reasonable to believe that they might also enhance cell wall permeability to PNA probes. In the present study, 10% buffered formalin was used initially, but fixation in 50% ethanol was found to be simpler, lessen concerns regarding fixative toxicity, and produce brighter, more uniform hybridizations.
In their work with the DNA-FISH probe Lis-1255, Wagner et al. (25) also used ethanol as a fixative. However, these authors reported that cultures of L. monocytogenes grown beyond 9 h in brain heart infusion broth were undetectable using ethanol-based fixation alone. Lysozyme and proteinase K digestions were required to fully permeabilize these cells to the probe. In contrast, the PNA probes studied here yielded rapid (10 to 30 min) and bright hybridizations with stationary-phase cells (18 to 24 h growth) without any need for permeabilization beyond the initial ethanol fixation. It is also worth noting that all of the PNA work done here was accomplished in solution, whereas most literature reports of FISH-based detection of gram-positive bacteria are performed with cells adhered to microscope slides or membrane filters (10, 12, 13, 25). The effects of multiple enzyme digestions on the solution-phase behavior of Listeria cells are unknown, but cell clumping could be problematic. In the present study, PNA-hybridized Listeria spp. formed even suspensions of individual cells and results of PNA-FISH experiments were readily analyzed using flow cytometry.
The second major advantage to the use of PNA for FISH-based detection of bacteria is that these probes are able to bind areas of the ribosome that are inaccessible to DNA probes. PNA-FISH probes are hybridized under substantially different conditions than are their DNA counterparts. Typically, these hybridizations are carried out under low salt (100 mM NaCl), high-temperature (55°C), and high-pH (pH 9.0) conditions. These conditions contrast with those commonly used in DNA-based FISH protocols (900 mM NaCl, 46°C, and a pH of 7.0 to 8.0) (3, 6, 18). Nucleic acid secondary structures are stabilized at high salt conditions through charge-shielding effects. Low salt conditions are therefore destabilizing for these structures. Alkaline or high-temperature conditions are also common means for nucleic acid denaturation (2). The combined effects of low salt, high pH, and high temperature suggest that target nucleic acids (rRNA) are likely to be in nonnative form (e.g., denatured) under the conditions of PNA hybridization. These ribosome-denaturing conditions are thought to alleviate the influence of a higher-order ribosomal structure on probe accessibility, rendering highly structured regions of the ribosome accessible for detection by FISH (23). The bright hybridization for LisUn-11 (E. coli nucleotide positions 466 through 480) highlights this effect, as previous work using DNA-FISH probes has characterized the region spanning E. coli nucleotide positions 468 through 486 as the least accessible on the entire 16S subunit (6). Although LisUn-3 targets a supposedly more accessible region of the ribosome (6), hybridizations with this probe were not as bright as those with LisUn-11. This result likely reflects differences in melting temperature between the two probes. The estimated melting temperature of LisUn3 is 70°C, while that of LisUn-11 is 82°C. These data suggest that results with LisUn-3 could be improved by adjusting the hybridization temperature to a lower value (e.g., <55°C).
Sequence comparisons of both 16S and 23S rRNA have demonstrated that the genus Listeria can be divided into two relatedness groups: one containing both L. grayi subsp. grayi and L. grayi subsp. murrayi and the other containing the rest of the genus (4, 19). This split in sequence homology is reflected in the difficulty of identifying rRNA-targeted probes that both selectively hybridize to the genus Listeria and encompass all six Listeria species. However, for applications such as environmental monitoring of Listeria in food-processing plants, such probes are required. From all appearances, LisUn-11 is an ideal probe for the FISH-based detection of generic Listeria. This probe was very bright and hybridized to all Listeria spp. tested, but not to cells from closely related genera. Like LisUn-3, no special preparatory steps were required to facilitate its passage through the cell wall. These properties suggest that LisUn-11 could become a valuable tool in applications such as environmental monitoring of food-processing plants. Although LisUn-3 did not react with L. grayi, it remains a valuable diagnostic tool, yielding bright hybridizations with the five other Listeria spp., including L. monocytogenes. The ability of PNA probes to more freely penetrate the gram-positive cell wall, combined with their enhanced abilities to target rRNA in regions of high structural complexity, extends the possibilities of FISH for the detection of Listeria beyond that which has been previously reported using DNA probes.
The pace and volume of today's food production and distribution networks places new emphasis on the need for rapid detection methods for organisms such as Listeria. The PNA probes described here provide the food safety community with a highly specific and rapid genotypic means for the detection or identification of this genus. Their development is especially timely in light of recently proposed environmental testing requirements for generic Listeria (5). Apart from the use described here, these probes may also be adapted for additional assay formats (membrane-based detection of microcolonies, RNA dot blots, as reporter probes for real-time PCR, etc.), although their use will need to be optimized for each application. As part of a Listeria testing program, use of these probes may help prevent costly product recalls and reduce the incidence of disease.
Acknowledgments
We thank Martin Wiedmann at Cornell University for providing many of the Listeria strains used.
This work was supported in part by a grant from the North American Branch of the International Life Sciences Institute (ILSI N.A.). Additional support was provided by grants from sponsors of the Food Research Institute and by the College of Agricultural and Life Sciences, University of Wisconsin—Madison.
The opinions expressed herein are those of the authors and do not necessarily represent the views of ILSI.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. L. M. 1999. Solution hybridization: reaction conditions, p. 51. In Nucleic acid hybridization. BIOS Scientific Publishers, Oxford, United Kingdom.
- 3.Bohnert, J., B. Hübner, and K. Botzenhart. 2000. Rapid identification of Enterobacteriaceae using a novel 23S rRNA-targeted oligonucleotide probe. Int. J. Hyg. Environ. Health 203:77-82. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D., S. Wallbanks, D. J. Lane, J. Shah, R. Nietupski, J. Smida, M. Dorsch, and E. Stackebrandt. 1991. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:240-246. [DOI] [PubMed] [Google Scholar]
- 5.Federal Register. 2001. Performance standards for the production of processed meat and poultry products. Fed. Regist. 66:12590-12636. [Google Scholar]
- 6.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gildea, B. D., S. Casey, J. MacNeill, H. Perry-O'Keefe, D. Sørensen, and J. M. Coull. 1998. PNA solubility enhancers. Tetrahedron Lett. 39:7255-7258. [Google Scholar]
- 8.Gottschalk, G. 1979. Metabolic diversity of aerobic heterotrophs: biosynthesis of monomers and polymers, p. 110-111. In Bacterial metabolism, 1st ed. Springer-Verlag, New York, N.Y.
- 9.Gutell, R. R., J. J. Cannone, Z. Shang, Y. Du, and M. J. Serra. 2000. A story: unpaired adenosine bases in ribosomal RNA. J. Mol. Biol. 304:335-354. [DOI] [PubMed] [Google Scholar]
- 10.Krimmer, V., H. Merkert, C. von Eiff, M. Frosch, J. Eulert, J. F. Löhr, J. Hacker, and W. Ziebuhr. 1999. Detection of Staphylococcus aureus and Staphylococcus epidermidis in clinical samples by 16S rRNA-directed in situ hybridization. J. Clin. Microbiol. 37:2667-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludwig, W., K.-H. Schleifer, and E. Stackebrandt. 1984. 16S rRNA analysis of Listeria monocytogenes and Brochothrix thermosphacta. FEMS Microbiol. Lett. 25:199-204. [Google Scholar]
- 12.Matte-Tailliez, O., P. Quénée, R. Çibik, K. van Opstal, F. Dessevre, O. Firmesse, and P. Tailliez. 2001. Detection and identification of lactic acid bacteria in milk and industrial starter culture with fluorescently labeled rRNA-targeted peptide nucleic acid probes. Lait 81:237-248. [Google Scholar]
- 13.Moter, A., and U. B. Göbel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85-112. [DOI] [PubMed] [Google Scholar]
- 14.Norton, D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. AOAC Int. 85:505-515. [PubMed] [Google Scholar]
- 15.O'Donnell, A. G., and A. S. Whiteley. 1999. Fluorescent in situ hybridization and the analysis of the single cells, p. 221-235. In C. Edwards (ed.), Methods in biotechnology, vol. 12. Humana Press, Totowa, N.J. [Google Scholar]
- 16.Perry-O'Keefe, H., H. Stender, A. Broomer, K. Oliveira, J. Coull, and J. J. Hyldig-Nielsen. 2001. Filter-based PNA in situ hybridization for rapid detection, identification and enumeration of specific micro-organisms. J. Appl. Microbiol. 90:180-189. [DOI] [PubMed] [Google Scholar]
- 17.Rocourt, J. 1999. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification, p. 1-20. In E. T. Ryser and E. H. Marth (ed.) Listeria, listeriosis and food safety. Marcel Dekker, New York, N.Y.
- 18.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 19.Sallen, B., A. Rajoharison, S. Desvarenne, F. Quinn, and C. Mabilat. 1996. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int. J. Syst. Bact. 46:669-674. [DOI] [PubMed] [Google Scholar]
- 20.Schmid, M. W. 2000. Ph.D. thesis. Universität München, Munich, Germany.
- 21.Schmid, M. W., M. Walcher, A. Bubert, M. Wagner, M. Wagner, and K.-H. Schleifer. 2003. Nucleic acid-based, cultivation-independent detection of Listeria spp. and genotypes of L. monocytogenes. FEMS Immunol. Med. Microbiol. 35:215-225. [DOI] [PubMed] [Google Scholar]
- 22.Stender, H., A. J. Broomer, K. Oliveira, H. Perry-O'Keefe, J. J. Hyldig-Nielsen, A. Sage, and J. Coull. 2001. Rapid detection, identification, and enumeration of Escherichia coli cells in municipal water by chemiluminescent in situ hybridization. Appl. Environ. Microbiol. 67:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stender, H., C. Kurtzman, J. J. Hyldig-Nielsen, D. Sørensen, A. J. Broomer, K. Oliveira, H. Perry-O'Keefe, A. Sage, B. Young, and J. Coull. 2001. Identification of Dekkera bruxellensis (Brettanomyces) from wine by fluorescence in situ hybridization using peptide nucleic acid probes. Appl. Environ. Microbiol. 67:938-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stender, H., M. Fiandaca, J. J. Hyldig-Nielsen, and J. Coull. 2002. PNA for rapid microbiology. J. Microbiol. Methods 48:1-17. [DOI] [PubMed] [Google Scholar]
- 25.Wagner, M., M. Schmid, S. Juretschko, K.-H. Trebesius, A. Bubert, W. Goebel, and K.-H. Schleifer. 1998. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol. Lett. 160:159-168. [DOI] [PubMed] [Google Scholar]
- 26.Wang, R.-F., W.-W. Cao, and M. G. Johnson. 1992. 16S rRNA-based probes and polymerase chain reaction method to detect Listeria monocytogenes cells added to foods. Appl. Environ. Microbiol. 58:2827-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]