Abstract
Microarrays with oligonucleotides of different lengths were used to monitor gene expression at a whole-genome level. To determine what length of oligonucleotide is a better alternative to PCR-generated probes, the performance of oligonucleotide probes was systematically compared to that of their PCR-generated counterparts for 96 genes from Shewanella oneidensis MR-1 in terms of overall signal intensity, numbers of genes detected, specificity, sensitivity, and differential gene expression under experimental conditions. Hybridizations conducted at 42°C, 45°C, 50°C, and 60°C indicated that good sensitivities were obtained at 45°C for oligonucleotide probes in the presence of 50% formamide, under which conditions specific signals were detected by both PCR and oligonucleotide probes. Signal intensity increased as the length of the oligonucleotide probe increased, and the 70-mer oligonucleotide probes produced signal intensities similar to the intensities obtained with the PCR probes and detected numbers of open reading frames similar to the numbers detected with the PCR probes. PCR amplicon, 70-mer, 60-mer, and 50-mer arrays had detection sensitivities of 5.0, 25, 100, and 100 ng of genomic DNA, which were equivalent to approximately 1.9 × 106, 9.2 × 106, 3.7 × 107, and 3.7 × 107 copies, respectively, when the array was hybridized with genomic DNA. To evaluate differential gene expression under experimental conditions, S. oneidensis MR-1 cells were exposed to low- or high-pH conditions for 30 and 60 min, and the transcriptional profiles detected by oligonucleotide probes (50-mer, 60-mer, and 70-mer) were closely correlated with those detected by the PCR probes. The results demonstrated that 70-mer oligonucleotides can provide the performance most comparable to the performance obtained with PCR-generated probes.
Microarrays are a powerful tool for monitoring gene expression changes under various conditions and have been widely used for genome-wide transcriptional analyses (4, 5, 15, 16, 22, 32, 33), discovery of gene functions (8), cancer studies (6, 14, 18, 19), neuroscience (17), discovery of drug targets (3, 10), and environmental studies (21, 28, 29, 34, 36, 38). Generally, microarrays have been constructed with two types of probes, PCR-generated probes that typically range in size from 200 to 2,000 bp and oligonucleotide probes that are typically 20 to 70 nucleotides (nt) long. Producing PCR product-based DNA arrays can be a time-consuming procedure that includes PCR primer design, amplification, size verification, product purification, and quantification. Also, some open reading frames (ORFs) are difficult to amplify, and thus the construction of comprehensive arrays can be a challenge. Recently, to alleviate some of the problems associated with PCR amplicon microarrays, oligonucleotide microarrays that contain probes longer than 40 nt have been evaluated and used for whole-genome expression studies (9, 13). These microarrays should have higher specificity and are easy to construct, and they can thus provide an important alternative approach for monitoring gene expression. However, due to the smaller probe size, it is expected that the detection sensitivity of oligonucleotide arrays will be lower than that of PCR probes.
Several previous studies indicated that a single 50-mer (11, 21, 29), 60-mer (9, 24), or 70-mer (2, 7) oligonucleotide probe per gene could produce hybridization signals comparable to those obtained with PCR amplicon arrays. Kane et al. (11) demonstrated that the sensitivity of microarrays was not substantially different for PCR probes (322 to 393 bases) and 50-mer oligonucleotide probes. Both probe types could reproducibly detect approximately 10 gene copies per cell and discern threefold changes in cDNA levels, but only three genes were targeted by three PCR probes and six oligonucleotide probes (two for each gene). Also, the PCR probes were 320 to 390 bp long, and it is not known how oligonucleotide probes compare to longer PCR probes (e.g., the 889- to 995-bp probes used in this study). In addition, various studies reported previously in the literature used oligonucleotide probes that ranged from 20 to 70 nt long, but a direct comparison of the performance of oligonucleotide probes of different lengths has not been reported previously.
In this study, microarrays that contained oligonucleotide probes of different lengths were compared in order to determine a better alternative to PCR amplicon microarrays. We systematically evaluated the hybridization conditions for oligonucleotide microarrays in terms of overall hybridization signal intensity, numbers of genes detected, specificity, sensitivity, and gene expression differentiation. The probes of different lengths were derived from the same 96 ORFs from Shewanella oneidensis MR-1. Our results indicated that the performance of the 70-mer probes was most comparable to the performance of PCR probes under the hybridization conditions used (45°C and 50% formamide), and thus a 70-mer oligonucleotide array is an alternative to a PCR amplicon microarray, although the sensitivity is lower than that of an array with PCR probes.
MATERIALS AND METHODS
Preparation of PCR probes.
A plate containing 96 genes was randomly selected for this study from the 50 96-well plates used for constructing whole-genome arrays of S. oneidensis MR-1 (5, 32). PCR primer pairs designed with the PRIMEGENS software (35) were used to amplify each of the 96 ORFs. A complete or nearly complete sequence of a gene was selected as a probe if it had a maximum level of identity to all other genes of ≤75%. For genes that had a level of identity to other genes of >75%, a maximum internal region that had <75% identity was selected as a probe, or the cutoff value was increased to 85%. All 96 primer pairs were synthesized and then arrayed in 96-well plates by MWG Biotech Inc. (High Point, NC). The information about 96 S. oneidensis MR-1 ORFs, the PCR primer pairs, and the sizes of PCR products is summarized in Table S1 in the supplemental material. Each gene was amplified eight times in a 96-well plate. The amplified PCR products were pooled and purified using a QIAquick PCR purification kit (QIAGEN Inc., California) according to the protocol of the manufacturer. The concentrations of pooled PCR products were 200 to 500 ng/μl. The purified PCR fragments were visualized and checked for size by agarose gel electrophoresis in the presence of ethidium bromide.
Oligonucleotide probe design.
Oligonucleotide probes (30-, 40-, 50-, 60-, and 70-mers) for the 96 ORFs were designed using a modified version of the PRIMEGENS software (35). Each of the PCR-amplified DNA probe sequences selected was compared with the entire sequence database using BLAST (1) and was aligned with other sequences that exhibited more than 85% identity using dynamic programming. Based on the global optimal alignments, segments exhibiting <85% nucleotide identity to the corresponding aligned regions of any BLAST hit sequences were selected as potential probes. Among the segments identified as potential probes, one probe for each gene was selected based on the G+C content, melting temperature (Tm), and lack of self-complementary. The information about the 70-mer oligonucleotide sequences, melting temperatures, and G+C contents are summarized in Table S2 in the supplemental material. For comparison, the longer probe sequences contained the sequences of shorter probes. All of the oligonucleotides designed were commercially synthesized without modification by MWG Biotech Inc. (High Point, NC). The concentration of oligonucleotides was adjusted to 100 pmol/μl.
Microarray construction.
PCR probes and oligonucleotide probes prepared in 50% dimethyl sulfoxide (Sigma Chemical Co., Missouri) were spotted onto SuperAmine glass slides (Telechem International, California) using a PixSys 5500 robotic printer (Cartesian Technologies Inc., California). For each gene there were two replicates on a single slide. The PCR-oligonucleotide microarrays contained PCR products and 70-, 60-, 50-, 40-, and 30-mer oligonucleotides, and in total, there were 1,152 spots on a single slide. UV cross-linking (300 mJ) and washing were carried out according to the protocol of the manufacturer (Telechem International, California).
Preparation of PCR and artificial targets.
PCR targets were prepared as described above and labeled with random primers and the Klenow fragment (genomic DNA [gDNA] labeling methods). Different amounts of a PCR target mixture were used according to the experimental needs. Four artificial oligonucleotide targets (T1-SO1679, T2-SO1744, T3-SO2680, and T4-SO0848) that were complementary to the 70-mer perfect match oligonucleotide probes were synthesized at the Molecular Structure Facility at Michigan State University (East Lansing, MI). The oligonucleotides were labeled at the 5′ end with Cy5 (T1-SO1679, T2-SO1744, and T3-SO2680) or Cy3 (T4-SO0848) fluorescent dye during synthesis. The different labeled oligonucleotide targets were normalized based on fluorescent dye concentration and mixed at equal ratios.
S. oneidensis MR-1 growth.
For general extraction of genomic DNA and RNA, S. oneidensis MR-1 cells were grown in LB medium at 30°C overnight in a shaker (180 rpm) and collected by centrifugation at 3,500 × g for 5 min. For the pH treatment experiment, S. oneidensis MR-1 cells were grown in LB medium at 30°C overnight in a shaker (180 rpm), and 1.0 ml of the cells was inoculated into 250 ml of new LB medium (pH 7) and grown for 5 h (at the mid-logarithmic phase with an optical density of ∼0.6). Cells were centrifuged and resuspended in 25 ml of LB medium at pH 4.0, 7.0 (control), or 10.0. There were three replicates for each pH treatment. Cultures (10 ml) were sampled at 30 min and 60 min, and cells were harvested by centrifugation at 3,500 × g for 5 min. Cell pellets were immediately frozen in liquid nitrogen and stored at −80°C until RNA was extracted.
Genomic DNA extraction, purification, and labeling.
Genomic DNA was isolated and purified from S. oneidensis MR-1 as described previously (37). The purified genomic DNA was fluorescently labeled by random priming using the Klenow fragment of DNA polymerase. Thirty-five microliters of mixture I containing 500 ng of genomic DNA and 20 μl of random primers (Invitrogen Life Technologies, California) was heated at 98°C for 3 to 5 min, quickly cooled on ice, and then centrifuged. Fifteen microliters of mixture II containing 1 μl of a solution containing 5 mM dATP, 5 mM dGTP, 5 mM dTTP, and 2.5 mM dCTP, 2 μl (80 U) of the Klenow fragment (Invitrogen Life Technologies, California), and 0.5 μl of Cy3-dUTP fluorescent dye (Amersham BioSciences, United Kingdom) was added to mixture I. A 50-μl (total volume) labeling reaction solution was incubated for 3 h at 42°C. The labeling reaction was terminated by heating the solution at 98°C for 3 min. The tubes were removed and quickly placed on ice. After a quick centrifugation, the sample was hydrolyzed in 50 mM NaOH at 37°C for 10 min and then neutralized with the same amount of HCl. The labeled cDNA probe was purified immediately using a QIAquick PCR purification column and was concentrated in a Savant Speedvac centrifuge (Savant Instruments Inc., Holbrook, NY).
RNA extraction, purification, and labeling.
Total cellular RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, California). RNA samples were treated with RNase-free DNase I (Ambion, Austin, TX) and were purified using a Mini RNeasy kit (QIAGEN, Chatsworth, CA). The concentration and purity of RNA samples were estimated with a spectrophotometer using A260/A230 and A260/A280 ratios of >1.85, as well as by agarose gel electrophoresis. Total cellular RNA (10 μg) was incubated at 70°C for 10 min in the presence of 10 μg of random primers (Invitrogen Life Technologies, California). The labeling reaction was catalyzed by 200 U of Superscript II RNase H− reverse transcriptase (Invitrogen Life Technologies, California) in the presence of 500 μM dATP, 500 μM dGTP, 500 μM dCTP, 25 μM dTTP, and the fluorophor Cy5-dUTP or Cy3-dUTP (Amersham BioSciences, United Kingdom) at a concentration of 25 μM. The reverse transcription reaction was allowed to proceed for 2 h at 42°C, and this was followed by RNA hydrolysis in 1 N NaOH at 37°C for 10 min. The labeled cDNA probe was purified immediately using a QIAquick PCR purification column and was concentrated in a Savant Speedvac centrifuge (Savant Instruments Inc., Holbrook, NY). For each biological sample, three slides were used. In addition to the duplicates of arrays on the same slide, three biological cell samples produced a total of 18 possible spots for each gene.
Microarray hybridization, washing, and scanning.
For determination of the overall hybridization signals, sensitivity, and the number of detected genes, a gDNA or RNA sample labeled with a single dye was used. For the pH stress study, the dye-swap method was used. Hybridization was performed using three replicates, with each slide containing two replicates on microarrays so that for each gene there was a total of six data points. The microarrays were hybridized at 45°C overnight in the presence of 50% formamide. The labeled cDNAs were resuspended in 20 to 25 μl of hybridization solution that contained 50% formamide, 1 mM dithiothreitol, 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.3% sodium dodecyl sulfate (SDS), and 0.8 μg/μl of herring sperm DNA (Invitrogen Life Technologies, California). The sample was incubated at 98°C for 5 min, centrifuged to collect condensation, and kept at 50 to 60°C. The sample was immediately applied to a microarray slide, and hybridization was carried out in a waterproof Corning hybridization chamber (Corning Life Science, New York) submerged in a 45°C water bath in the dark for 16 h. After hybridization, the coverslips were immediately removed, and the slides were washed in a buffer containing 1× SSC and 0.1% SDS for 5 min at 37°C. The microarrays were washed in a new buffer with 0.1× SSC and 0.1% SDS for another 5 min at room temperature. Finally, the microarrays were washed with distilled water for 30 s at room temperature and dried with compressed air or by centrifugation. Microarrays were scanned using the ScanArray 5000 microarray analysis system (Packard BioChip Technologies, Massachusetts). Normally, 95 to 100% laser power and 70 to 80% photomultiplier tube efficiency were selected for scanning.
Data normalization and analysis.
To determine signal intensities for each spot, 16-bit TIFF scanned images were analyzed using the software ImaGene 5.5 (Biodiscovery Inc., California). Prior to normalization, negative spots, empty spots, and poor spots flagged by ImaGene were removed in Excel (Microsoft). Any spots with values that were >2 standard deviations above the local background level in both channels were rejected. The resulting data files were loaded onto GeneSpring, version 5.1 (Silicon Genetics, Redwood, CA), and channel normalization was accomplished by the software with the Lowess method. The Cy5/Cy3 ratio was transformed to a logarithmic (base 2) value, and a gene was considered differentially expressed if the logarithmic value was greater than 1 (up-regulated) or less than −1 (down-regulated). The signal-to-noise ratio (SNR) was also computed for each spot to discriminate true signals from noise. The SNR ratio was calculated as follows: SNR = (signal mean − background mean)/(background standard deviation). A commonly accepted criterion for the minimum signal (threshold) that can be accurately quantified is an SNR of >3.0 (31).
RESULTS
Effects of hybridization temperature on signal intensity and number of genes detected at an SNR of >3.0.
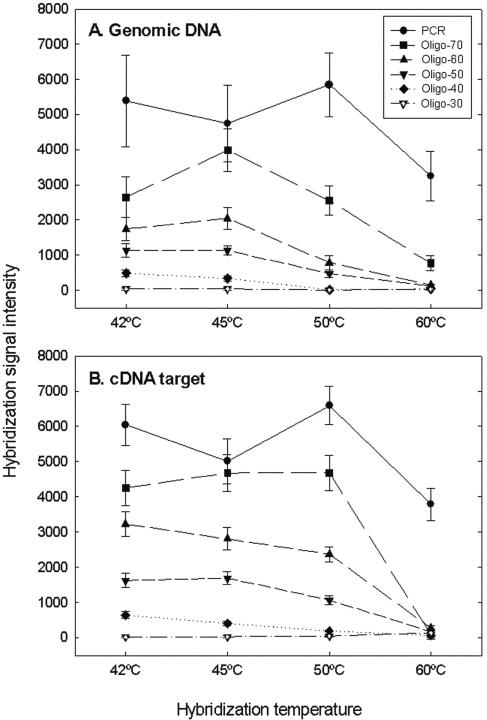
The PCR-oligonucleotide microarray was used to examine the effects of hybridization temperature on signal intensity detected at an SNR of >3.0 when labeled gDNA or labeled cDNA templates of S. oneidensis MR-1 were used as the target. PCR amplicons had the most increased signals at all temperatures tested (42°C, 45°C, 50°C, and 60°C) when the labeled gDNA target (Fig. 1A) or cDNA target was hybridized with the array (Fig. 1B). For example, on average (pooled signal from 96 ORFs), PCR probes had 1.4-, 2.8-, and 6.2-fold-higher signal intensities than 70-mer, 60-mer, and 50-mer oligonucleotides, respectively, at 50°C when they were hybridized with the cDNA target (Fig. 1B). For oligonucleotide probes, the signal intensity increased as the length increased. The 70-mer oligonucleotides produced signal intensities that were 1.7-fold and 2.7-fold higher than those produced by the 60-mer and 50-mer oligonucleotides, respectively, at 45°C when they were hybridized with cDNA target (Fig. 1B). The signal intensities for the 40-mer and 30-mer oligonucleotides were only about 1% of the signal intensities for the 70-mer probes at 45°C (Fig. 1B). In addition, the 70-mer probes had the highest signal intensity at 45°C, and this signal intensity was close to that obtained with PCR probes (Fig. 1). These results indicated that the 70-mer probes had the best sensitivity among the oligonucleotide probes and that this sensitivity was the most comparable to that of the PCR probes.
FIG. 1.
Relationships between hybridization temperature and average hybridization signal intensity of 96 ORFs selected from S. oneidensis MR-1. Five hundred nanograms of genomic DNA (A) or 10 μg of cDNA target (B) was labeled and hybridized with the PCR-oligonucleotide microarray at 42°C, 45°C, 50°C, and 60°C in the presence of 50% formamide for 16 h. The data are averages and standard deviations for three slides and a total of six possible spots.
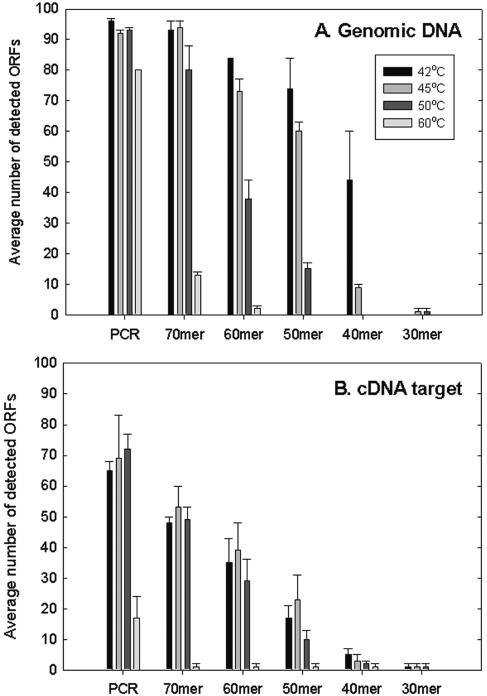
To compare gene coverage for the PCR and oligonucleotide probes, the numbers of detected ORFs out of 96 at an SNR of >3.0 were determined. A total of 95 to 100% of the genes could be detected with the PCR probes when gDNA was used as the target and hybridized at 42°C to 50°C (Fig. 2A). Almost 85% of the ORFs were still detected at 60°C (Fig. 2A). No substantial differences in gene coverage were observed between the 70-mer oligonucleotide probes and the PCR probes at 42°C and 45°C. However, only 85% and 15% of the genes could be detected with the 70-mer probes at 50°C and 60°C, respectively. The 60-mer and 50-mer oligonucleotide probes detected only 88% and 77% of the ORFs at 42°C and 76% and 63% of the ORFs at 45°C, respectively. The 40-mer and 30-mer probes detected less than 50% of the ORFs even at 42°C (Fig. 2A). These results suggested that the 70-mer probes were most comparable to the PCR probes in terms of gene coverage under the hybridization conditions tested (45°C and 50% formamide).
FIG. 2.
Average number of genes detected among 96 ORFs selected from S. oneidensis MR-1 at an SNR of >3.0. Five hundred nanograms of genomic DNA (A) or 10 μg of cDNA target RNA (B) was labeled with a Cy dye and hybridized with the array. The data are averages and standard deviations for three slides (six spots).
Eight ORFs (SO0876, SO0991, SO1193, SO1277, SO1387, SO2320, SO2697, and SO3207) were detected by the 70-mer, 60-mer, and 50-mer probes at 45°C but were not detected at 50°C or higher temperatures. Some of these probes had low G+C contents and Tm values. For example, the G+C contents of the 70-mer probes for SO1193 and SO2697 were 30% and 31%, respectively, and the Tm values for these ORFs were 68°C and 69°C, respectively; these values were lower than the average values (45% and 75°C) for the other probes. These results suggested that the G+C content and/or Tm of an oligonucleotide probe could be partially responsible for the observed temperature-dependent hybridization behavior.
As expected, the number of spots detected decreased when labeled cDNA was used as the target to hybridize with the array (Fig. 2B) because not all ORFs were expressed. At 42°C, 45°C, 50°C, and 60°C, the PCR probes detected 65, 69, 72, and 17 ORFs, respectively, and the 70-mer probes detected 48, 53, 49, and 1 ORFs, respectively. A total of 29 to 39 ORFs were detected by the 60-mer probes and 10 to 23 ORFs were detected by the 50-mer probes at temperatures from 42°C to 50°C. The number of ORFs detected was not dramatically affected by hybridization temperatures ranging from 42°C to 50°C, but only a few ORFs (17 ORFs for PCR probes and 0 to 2 ORFs for oligonucleotide probes) could be detected at 60°C (Fig. 2B). The 40-mer and 30-mer probes did not have substantial signal intensities (Fig. 1) or gene coverage (Fig. 2). The results suggested that the optimal temperatures were 45°C for oligonucleotide probes longer than 50-mer and 50°C for PCR probes, although hybridization differences between 45°C and 50°C were not observed in each case.
Specificity.
In order to determine whether the signals detected were specific at 45°C, PCR-amplified targets or synthesized oligonucleotide targets of four loci (SO1679, SO1744, SO2680, and SO0848) were used for hybridization. Five picograms of PCR target per locus was hybridized with the array at 45°C in the presence of 50% formamide. All four targets were specifically detected by the PCR probes corresponding to the four loci, and no substantial signals were observed with other PCR probes at an SNR of >3.0 (data not shown).
To further evaluate the specificity of oligonucleotide probes, artificial targets of the 70-mer probes were synthesized. Three targets (T1-SO1679, T2-SO1744, and T3-SO2680) were end labeled with Cy5, and one target (T4-SO0848) was end labeled with Cy3. Each target (2.0 pg) was mixed and hybridized with the array at 45°C in the presence of 50% formamide. The results showed that all four targets specifically hybridized with the corresponding oligonucleotide probes (40-mer to 70-mer). The 30-mer oligonucleotide probes detected only two of the four targets at an SNR of >3.0. The SNR values for all other loci varied from 0 to 1.0 (data not shown). The data indicated that all signals detected by the PCR or oligonucleotide probes were specific under the conditions tested (45°C and 50% formamide).
Sensitivity of oligonucleotide arrays compared to sensitivity of the DNA array.
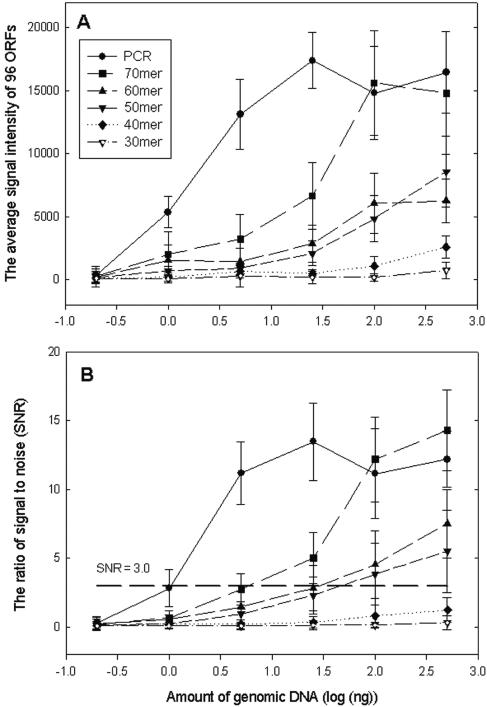
To assess sensitivity, different amounts (0.2, 1.0, 5.0, 25, 100, and 500 ng) of gDNA from S. oneidensis MR-1 were labeled with the Cy3 dye and hybridized with the array at 45°C in the presence of 50% formamide. The signal intensities detected by the PCR probes increased as the gDNA concentration increased (0.2 to 5 ng) and did not change dramatically from 25 to 500 ng (Fig. 3A). The 70-mer probes gave signal intensities similar to those observed with the PCR probes at relatively high concentrations of gDNA (100 and 500 ng), but decreased signals were detected at low target concentrations (0.2 to 25 ng). The 70-mer oligonucleotides had two- to threefold-stronger signals than the 60-mer and 50-mer oligonucleotide probes (Fig. 3A).
FIG. 3.
Relationships between the amount of genomic DNA (0.2, 1, 5, 25, 100, and 500 ng) and the average signal intensity (A) or the average ratio of signal to noise (B) for 96 ORFs selected from S. oneidensis MR-1. The data are averages and standard deviations for three slides. For each point there were 576 possible spots on the array.
The amount of gDNA was also plotted against the average SNRs of the genes detected (Fig. 3B). As expected, the results showed that the PCR probes had the highest sensitivity. Some genes could be detected at 1 ng of gDNA (SNR, 2.84 ± 1.32), and most genes were detected with 5.0 ng (SNR, 11.19 ± 2.26). The 70-mer probes detected some genes at 5 ng (SNR, 2.74 ± 1.12) and most of the genes at 25 ng (SNR, 5.02 ± 1.87). The 50-mer and 60-mer probes had detection limits between 25 and 100 ng. No substantial signals were detected with the 40-mer and 30-mer probes at the gDNA concentrations tested.
For simplicity and quantitative comparison, the sensitivity of the PCR-oligonucleotide microarray was evaluated by measuring the minimum amount of targets (gDNA or PCR products) required for detecting more than 50% of the ORFs with an SNR of >3.0. Based on this working definition, the numbers of ORFs detected (of a total of 96 ORFs) at an SNR of >3.0 are shown in Table 1 for different probes hybridized with different concentrations of gDNA. The sensitivity of the PCR probes was fivefold higher than that of the 70-mer probes, which was fourfold higher than that of the 60-mer and 50-mer oligonucleotide probes.
TABLE 1.
Numbers of detected ORFs out of 96 at an SNR of >3.0 by PCR and oligonucleotide probes when different amounts of genomic DNA from S. oneidensis MR-1 were labeled and hybridized with the PCR-oligonucleotide mixed array
| gDNA (ng) | pg/gene | No. of ORFs detecteda
|
|||||
|---|---|---|---|---|---|---|---|
| PCR | 70-mer | 60-mer | 50-mer | 40-mer | 30-mer | ||
| 0.2 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.0 | 0.2 | 41 ± 3.3 | 0 | 0 | 0 | 0 | 0 |
| 5.0 | 1.0 | 95 ± 0.7 | 38 ± 4.7 | 8 ± 3.2 | 2.3 ± 1.3 | 0 | 0 |
| 25 | 5 | 95 ± 0.7 | 56 ± 4.3 | 14 ± 5.3 | 7.3 ± 3.4 | 0 | 0 |
| 100 | 20 | 95 ± 0.8 | 94 ± 0.7 | 66 ± 6.7 | 57 ± 4.5 | 0 | 0 |
| 500 | 100 | 96 ± 0.0 | 95 ± 0.4 | 86 ± 6.0 | 66.7 ± 6.7 | 2.3 ± 1.3 | 0 |
The data are averages ± standard deviations for three slides with a total of six spots.
Differential gene expression under pH stress conditions.
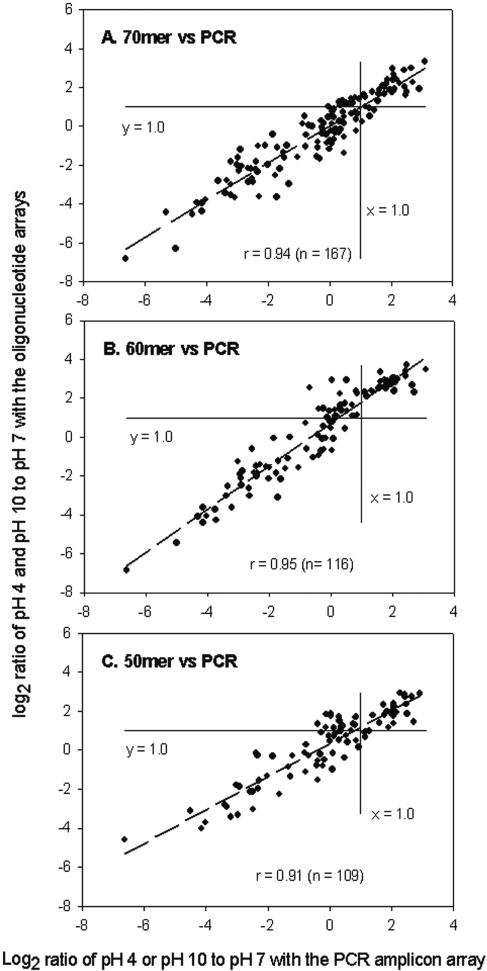
To determine whether oligonucleotide probes had similar power for differentiating gene expression patterns compared to PCR probes, S. oneidensis MR-1 cells were exposed to low-pH (pH 4.0) and high-pH (pH 10.0) conditions for 30 and 60 min, and the results were compared to the results obtained at pH 7.0. Total RNA was isolated, reverse transcribed, labeled, and hybridized with the array. Detailed microarray data are shown in Table S3 in the supplemental material. Significant correlations for the fold changes in gene expression detected under the conditions examined were obtained between PCR probes and 70-mer probes (Fig. 4A), 60-mer probes (Fig. 4B), or 50-mer probes (Fig. 4C). Based on the numbers of detections (167, 116, and 109 for the 70-mer, 60-mer, and 50-mer probes, respectively) and the correlations (0.94, 0.95, and 0.91 for the 70-mer, 60-mer, and 50-mer probes, respectively) between the oligonucleotide probes and the PCR amplicons, the results indicated that the 70-mer probes performed most like the PCR probes.
FIG. 4.
Relationships of logarithmic ratios in gene expression between the PCR probes and the 70-mer (A), 60-mer (B), and 50-mer (C) oligonucleotide probes.
We compared the significantly up-regulated and down-regulated genes detected by using PCR amplicon and oligonucleotide arrays (Table 2). At pH 4.0 (30 min), 95, 89, 68, and 69 ORFs were detected by the PCR, 70-mer, 60-mer, and 50-mer probes, respectively, and 83, 48, 36, and 25 ORFs were detected after exposure to pH 10.0 (30 min) (Table 2). The number of ORFs detected did not change significantly at pH 4.0 after 60 min of exposure, but at pH 10.0 the number increased from 30 min of exposure to 60 min of exposure. The numbers of up-regulated or down-regulated ORFs detected by different probes were very similar at pH 4.0. For example, the PCR, 70-mer, 60-mer, and 50-mer probes detected 35, 38, 39 and 38 ORFs that were induced, respectively, and 5, 6, 3, and 3 ORFs that were repressed, respectively, when S. oneidensis MR-1 cells were exposed to pH 4.0 for 30 min. After 60 min of exposure at pH 4.0, fewer ORFs were observed to be up-regulated and the number of down-regulated ORFs did not change compared to the number observed with 30 min of exposure (Table 2). After exposure to pH 10.0 for 30 min, most of the ORFs were down-regulated, and few ORFs were up-regulated. For example, with pH 10.0 treatment for 30 min, 2, 2, 0, and 1 ORFs were found to be up-regulated and 76, 42, 33, and 20 ORFs were found to be down-regulated by the PCR, 70-mer, 60-mer, and 50-mer probes, respectively. The patterns of up- or down-regulated ORFs changed from 30 min of exposure to 60 min of exposure at pH 10.0, and more ORFs were induced and fewer ORFs were repressed (Table 2). The 30-mer and 40-mer probes detected less than 50% of the selected ORFs, and the data were not compared to the data for the other probes.
TABLE 2.
Numbers of genes that were detected (SNR, >3.0), induced (fold change, >2.0), and repressed (fold change, <0.5) when S. oneidensis MR-1 cells were exposed to pH 4.0 and pH 10.0 for 30 and 60 mina
| Probe | Parameter | pH 4
|
pH 10
|
||
|---|---|---|---|---|---|
| 30 min | 60 min | 30 min | 60 min | ||
| PCR | Detected gene no. | 95 ± 0.7 | 92 ± 2.3 | 83 ± 2.5 | 89 ± 3.2 |
| Induced gene no. | 35 ± 1.8 | 25 ± 2.7 | 2 ± 0.0 | 7 ± 01.2 | |
| Repressed gene no. | 5 ± 0.5 | 5 ± 0.6 | 76 ± 4.9 | 32 ± 3.7 | |
| 70-mer | Detected gene no. | 89 ± 5.3 | 79 ± 4.3 | 48 ± 2.7 | 66 ± 3.1 |
| Induced gene no. | 38 ± 3.2 | 21 ± 2.5 | 2 ± 0.3 | 8 ± 1.6 | |
| Repressed gene no. | 6 ± 0.4 | 5 ± 0.6 | 42 ± 3.8 | 20 ± 2.1 | |
| 60-mer | Detected gene no. | 68 ± 5.4 | 65 ± 2.5 | 36 ± 3.0 | 52 ± 4.1 |
| Induced gene no. | 39 ± 1.9 | 22 ± 3.7 | 0 | 10 ± 2.2 | |
| Repressed gene no. | 3 ± 0.5 | 3 ± 0.5 | 33 ± 3.8 | 13 ± 2.5 | |
| 50-mer | Detected gene no. | 69 ± 3.3 | 54 ± 5.0 | 25 ± 3.2 | 38 ± 4.1 |
| Induced gene no. | 38 ± 2.1 | 21 ± 2.8 | 1 ± 0.0 | 5 ± 1.6 | |
| Repressed gene no. | 3 ± 0.5 | 2 ± 0.3 | 20 ± 2.3 | 13 ± 1.8 | |
The data were determined by using PCR amplicon or oligonucleotide arrays and are averages ± standard deviations for three slides with a total of six spots.
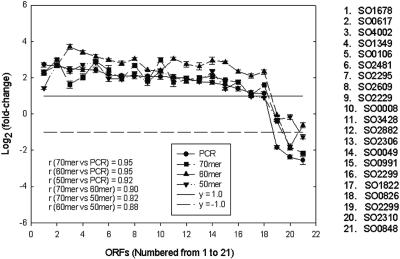
When the data from the 30-min pH 4.0 exposure were compared for the different probes, 21 ORFs were consistently detected by the PCR probes and the 50-mer to 70-mer oligonucleotides (Fig. 5). In this analysis gene expression with a fold change of 2.0 or more was considered up-regulated, gene expression with a fold change of 0.5 or less was considered down-regulated, and gene expression with a fold change between 0.5 and 2.0 was considered no change. Of the 21 ORFs, 18 were detected as up-regulated genes by the PCR amplicon array and the 70-mer, 60-mer, and 50-mer oligonucleotide arrays. The PCR amplicon array detected three genes that were down-regulated, and two, one, and one genes were detected by the 70-mer, 60-mer, and 50-mer oligonucleotide arrays, respectively (Fig. 5). The correlation coefficients between PCR probes and 70-mer, 60-mer, and 50-mer oligonucleotide probes were 0.95, 0.95, and 0.92, respectively, for the 21 genes selected. In addition, the correlation coefficients were 0.90 for the 70-mer and 60-mer probes, 0.92 for the 70-mer and 50-mer probes, and 0.88 for the 60-mer and 50-mer probes (Fig. 5).
FIG. 5.
Comparison of fold changes for 21 ORFs consistently detected by PCR amplicon, 70-mer, 60-mer, and 50-mer arrays when S. oneidensis MR-1 cells were exposed to pH 4 for 30 min. ORFs were up-regulated above the line at y = 1.0 (solid line), ORFs were down-regulated below the line at y = −1.0 (dashed line), and ORFs were not changed between the two lines.
However, some differences in the expression patterns were observed for the different probes. For example, the PCR probe indicated that SO2320 was up-regulated 4.2-fold, but a change in expression was not detected by the 50-mer, 60-mer, and 70-mer oligonucleotide probes. SO1157 was detected as an up-regulated locus by the PCR probe (2.2-fold) and the 70-mer probe (2.8-fold), but no changes were detected by the 50-mer probe (1.6-fold) or the 60-mer probe based on our criteria (Table 3). Possible reasons for the observed differences in detection are discussed below. It should be noted that spots with low signal intensities or low quality (0 to 3%) were flagged and manually or automatically removed during data analysis.
TABLE 3.
ORFs and average fold changes in gene expression determined with PCR or oligonucleotide probes when S. oneidensis MR-1 cells were exposed to pH 4.0 for 30 mina
| Locus | Fold change
|
|||
|---|---|---|---|---|
| PCR | 70-mer | 60-mer | 50-mer | |
| SO0105 | 8.6 ± 1.77b | 10.0 ± 2.01 | 11.0 ± 1.96 | NAc |
| SO0017 | 7.5 ± 1.05 | 3.8 ± 0.32 | NA | 2.7 ± 0.30 |
| SO1383 | 5.7 ± 0.63 | 3.4 ± 0.29 | NA | 3.6 ± 0.42 |
| SO2743 | 4.8 ± 0.50 | 4.6 ± 0.67 | NA | 7.6 ± 0.68 |
| SO4180 | 4.4 ± 0.39 | 5.5 ± 0.73 | NA | NA |
| SO2320 | 4.2 ± 0.26 | NA | NA | NA |
| SO3051 | 4.2 ± 0.34 | 5.2 ± 0.45 | NA | 2.6 ± 0.31 |
| SO2697 | 3.7 ± 0.41 | 5.2 ± 0.61 | NA | 2.2 ± 0.29 |
| SO4661 | 3.6 ± 0.45 | 4.5 ± 0.51 | 7.5 ± 0.65 | NA |
| SO1679 | 3.6 ± 0.51 | 4.6 ± 0.37 | NA | 4.0 ± 0.49 |
| SO2389 | 3.5 ± 0.30 | 3.2 ± 0.36 | 5.7 ± 0.46 | NA |
| SO2953 | 3.5 ± 0.42 | 3.1 ± 0.37 | 5.8 ± 0.49 | NA |
| SO2113 | 3.0 ± 0.25 | 3.1 ± 0.34 | 5.9 ± 0.68 | NA |
| SO2654 | 2.5 ± 0.22 | 1.4 ± 0.21 | 5.2 ± 0.55 | NA |
| SO1157 | 2.2 ± 0.18 | 2.8 ± 0.25 | NA | 1.6 ± 0.27 |
| SO2930 | 2.1 ± 0.19 | 1.2 ± 0.35 | NA | NA |
| SO1277 | 0.4 ± 0.08 | NA | 1.0 ± 0.09 | 0.6 ± 0.05 |
| SO2886 | 0.2 ± 0.07 | 0.5 ± 0.08 | NA | 0.8 ± 0.22 |
All 18 ORFs were detected by the PCR probes, but at least one oligonucleotide probe failed to detect one of the genes at an SNR of >3.0.
The data are averages ± standard deviations for three slides with a total of six spots.
NA, data are not available because of empty spots, poor spots, or manually removed bad spots.
A pH shock was used to test the applicability of the designed probes under experimental conditions. For example, the product of SO0024 is predicted to be a potassium uptake protein, TrkH, and previous studies have suggested that a high K+ level may be necessary in Escherichia coli under acidic conditions (30). The results indicated that the SO0024 level did not increase dramatically (∼1.2-fold) when cells were exposed to pH 4.0 for 30 min, but it did significantly increase approximately threefold after the 60-min treatment. SO2743, one of the most significantly up-regulated loci during the pH 4.0 exposure, is predicted to encode an acetyl-coenzyme A synthetase (EC 6.2.1.1), which plays an important role in primary metabolism. SO0848 is predicted to encode a periplasmic nitrate reductase (EC 1.7.99.4), and it was significantly down-regulated at low or high pH.
DISCUSSION
The length of oligonucleotide probes is a concern not only because of the higher cost of longer oligonucleotide probes but also due to the effects on sensitivity and specificity, as well as the design criteria for unique probes. The alternative to oligonucleotide probes, PCR-generated probes, can provide more sensitivity, but they can be more labor- and cost-intensive at the front end of microarray construction. The use of oligonucleotide probes can overcome many of the caveats for PCR-generated probes, and different studies have used various sizes of oligonucleotides. However, a systematic comparison of probe length is not available in the literature.
Previous experiments of Relógio et al. (20) indicated that 60-mer oligonucleotide probes provided significantly improved (10-fold) sensitivity compared with 25-mer oligonucleotide probes, but the specificity was significantly lower (∼fourfold) than that obtained with 25-mer oligonucleotide probes. In this study, the signal intensities detected by 70-mer probes were significantly higher than those detected by 50-mer probes when labeled gDNA or cDNA templates were used as targets. The results suggested that the 70-mer oligonucleotide probes achieved sensitivity that was closest to that of the PCR probes and that the 50-mer and 60-mer oligonucleotide probes were able to provide reasonably good sensitivity (20-fold lower) compared to the sensitivity of the PCR probes. The data also indicated that 40-mer or 30-mer probes were not suitable for construction of oligonucleotide arrays using the methods described here when they were compared to 50-mer, 60-mer, or 70-mer probes.
Sensitivity is a critical parameter, particularly for environmental studies, when biomass can be low, or for low-level molecules in individual cells. When PCR-generated functional gene arrays were used, the detection limit for nirS (nitrite reductase) genes was approximately 1 ng of pure gDNA and 25 ng of soil community DNA (34). A previous study demonstrated that when oligonucleotide arrays with capture and detector probes were used, the detection sensitivity for Geobacter chapellei small-subunit rRNA gene sequences in soil extracts was approximately 500 ng of total RNA (26). Recently, studies with 50-mer functional gene arrays showed that the detection limit for some functional genes could be 5 to 10 ng of pure gDNA and 50 to 100 ng in a mixture of gDNA from different organisms (21, 29).
As expected, the PCR-generated probes had the best sensitivity, but the 70-mer probes had fourfold-greater sensitivity than the 60-mer and 50-mer probes (25 ng versus 100 ng). These results were in general agreement with the previous observations (21, 26, 29, 34) when the types of probes and samples were considered.
In order to determine the sensitivity of PCR probes for individual targets, the same array was used to hybridize with a mixture of four PCR targets generated from four loci (SO1679, SO1744, SO2680, and SO0848). The results showed that three loci (SO1744, SO2680, and SO0848) could be detected at a level of 0.05 pg of PCR target per locus and one locus (SO1679) could be detected at a level of 1.0 pg by the four PCR-generated probes, suggesting that the sensitivities of individual genes can differ to a large degree (data not shown). This gene-dependent phenomenon has been observed previously with 50-mer oligonucleotide arrays in bioremediation studies (21), but it has not been understood. Due to the gene-dependent nature, it is helpful to distinguish between microarray sensitivity and probe sensitivity. We define probe sensitivity as the minimum amount of target required for individual probes to obtain reproducible signals at a defined SNR threshold (e.g., 3.0). In the example described above, the probe sensitivities for SO1744 and SO1679 were 0.05 and 1.0 pg of PCR products, which are equivalent to 0.5 and 10 ng of gDNA, respectively. This may be a very important consideration for the assessment of individual genes in environmental studies. We define microarray sensitivity as the minimum amount of target required for a defined number of genes (e.g., 50%) to give reproducible signals at a defined SNR (e.g., 3.0). By this definition, the PCR, 70-mer, 60-mer, and 50-mer probes had sensitivities of 5.0, 25, 100, and 100 ng of gDNA, respectively. Microarray sensitivity may be more applicable to whole-genome arrays because the number of genes is generally known and most changes in transcript levels are expected to be detected.
Microarray hybridization and washing conditions greatly affect microarray sensitivity and specificity, so experimental conditions need to be optimized. For example, sensitivity can be increased by destroying stable secondary structures of probes and enhancing duplex formation (25). Specificity may be increased by preventing interactions between probes and noncomplementary targets and/or preventing nonspecific binding (25). Increased hybridization temperatures may improve the specificity of oligonucleotide probes, but the sensitivity may decrease substantially, especially for shorter oligonucleotides. In our experiments, all hybridizations were performed at 45°C in the presence of 50% formamide, and washing was at 37°C for 5 min.
Long oligonucleotide probes (50-mer to 70-mer) showed power for differentiating gene expression patterns under experimental conditions similar to that of PCR probes, but some differences in the expression patterns were also observed for the different probes (Table 3). Differences in sensitivity between PCR and oligonucleotide probes might explain this observation. For example, the mRNA levels of genes that showed different patterns were low and were just above the sensitivity level for PCR probes but below the sensitivity level of 70-mer, 60-mer, and 50-mer probes. The hybridization behavior of some ORFs listed in Table 3, such as SO0105, SO4180, SO2320, SO4661, SO2389, SO2953, SO2113, SO2654 and SO2930, could be explained by sensitivities that decrease in the following order: PCR, 70-mer, 60-mer, and 50-mer probes.
It is still difficult to explain why some ORFs (i.e., SO0017, SO1383, SO2743, SO3051, SO2697, SO1679, SO1157, SO1277, and SO2886) were detected by a shorter oligonucleotide probe instead of a longer oligonucleotide probe. A possible explanation could be related to probe or probe-target interactions on the microarray surface. Differences in signal intensity for individual probes are complicated by many factors, such as steric hindrance on the microarray surface (23) and stable secondary structures (25). To examine if any of the oligonucleotide probes shown in Table 3 have strong secondary structures, the oligonucleotides were checked with the MFOLD software (39). The predictions indicated that significant secondary structures were not present at 60°C (approximately equivalent to the conditions at 45°C with 50% formamide). As mentioned above, the Tm and G+C content may affect hybridization signals to some degree. For the 70-mer oligonucleotides in Table 3, the G+C contents and Tm values for SO0105 (60% and 80°C) and SO0017 (52% and 78°C) are higher and the G+C contents and Tm values for SO4180 (28% and 68°C), SO2697 (31% and 69°C), and SO1383 (35% and 70°C) are lower than the G+C contents and Tm values for other 70-mer probes (averages, 45% and 75°C). When the expression levels across a whole genome are analyzed, some individual probes may have to be reevaluated. Additional work is needed to discern the binding behavior for DNA molecules on glass surfaces and how these parameters can be incorporated into appropriate software algorithms.
Although detecting the responses to pH stress was not the focus of this study, some of the changes detected appear to be biologically meaningful for possible responses to pH stress. For example, in E. coli, the product of SO2743 is thought to be involved in acetate utilization as cells enter the stationary phase of growth (12). Perhaps the induction of a similar protein allows the consumption of acid in response to an acid shock in S. oneidensis MR-1. Another example is SO0848 encoding a periplasmic nitrate reductase, and the enzyme may be repressed by ammonium produced by degradation of some amino acids under pH stress conditions (27). The observations described above indicated that the differential gene expression data were correlated with physiological roles of S. oneidensis cells in response to pH stress and that changes in the expression patterns of the biologically relevant ORFs were detected with both the PCR and longer oligonucleotide probes.
Based on the concentrations of PCR and oligonucleotide probes, the copy number of oligonucleotide probes should be 20 to 500 times higher than that of PCR probes. This allows oligonucleotide probes to have a wider dynamic range than PCR probes so that different levels of RNA can be detected. If the amount of probes binding to the glass slide was saturated under our test conditions, as shown by Relógio (20), the copy number of the oligonucleotide probes was still more than 10 times higher than that of the PCR probes. Owing to the low sensitivity, oligonucleotide arrays may be useful for detecting RNA species at a high levels that may be saturated on PCR amplicon arrays.
The G+C content and Tm of an oligonucleotide are interesting. An oligonucleotide design program should constrict Tm values in a narrow range (e.g., 5°C), and it should also consider the overall G+C content of the whole genome. For example, the S. oneidensis MR-1 genome has a G+C content of 54%, and an oligonucleotide probe should have a G+C content close to the genome G+C content. In this study, the results showed that some oligonucleotides with lower G+C contents and Tm values did have low hybridization signals. Therefore, when probes for a genome with a substantially decreased G+C content are designed, the probe-target relationship may have to be reevaluated if stretches of more evenly distributed G+C content are not available.
Supplementary Material
Acknowledgments
We thank Tingfen Yan for technical assistance.
This research was supported by the United States Department of Energy under the Genomics:GTL, Microbial Genome, and Natural and Accelerated Bioremediation (NABIR) programs of the Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schäffer, J. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debouck, C., and P. N. Goodfellow. 1999. DNA microarrays in drug discovery and development: progress and potential. Biochem. Pharmacol. 62:1311-1336. [Google Scholar]
- 4.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 5.Gao, H., Y. Wang, X. Liu, T. Yan, L. Wu, E. Alm, A. Arkin, D. K. Thompson, and J. Zhou. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186:7796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golub, T. R., D. K. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek, J. P. Mesirov, H. Coller, M. L. Loh, J. R. Downing, M. A. Caligiuri, C. D. Bloomfield, and E. S. Lander. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531-537. [DOI] [PubMed] [Google Scholar]
- 7.He, Z., L. Wu, X. Li, M. W. Fields, and J. Zhou. 2005. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 71:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 9.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Showmarker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov, I., C. Schaab, S. Planitzer, U. Teichmann, A. Machl, S. Theml, S. Meier-Ewert, B. Seizinger, and H. Loferer. 2000. DNA microarray technology and antimicrobial drug discovery. Pharmacogenomics 1:169-178. [DOI] [PubMed] [Google Scholar]
- 11.Kane, M. D., T. A. Jatkoe, C. R. Stumpf, J. Lu, J. D. Thomas, and J. M. Madore. 2000. Assessment of the specificity and sensitivity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 28:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumari, S., E. J. Simel, and A. J. Wolfe. 2000. σ70 is the principal sigma factor responsible for transcription of acs, which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:551-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lessard, I. A. D., G. J. Domingo, A. Borges, and R. N. Perham. 1998. Expression of genes encoding the E2 and E3 components of the Bacillus stearothermophilus pyruvate dehydrogenase complex and the stoichiometry of subunit interaction in assembly in vitro. Eur. J. Biochem. 258:491-501. [DOI] [PubMed] [Google Scholar]
- 14.Liotta, L., and E. Petricoin. 2000. Molecular profiling of human cancer. Nat. Rev. Genet. 1:48-56. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., J. Zhou, M. Omelchenko, A. Beliaev, A. Venkateswaran, J. Stair, L. Wu, D. K. Thompson D. Xu, I. B. Rogozin, E. K. Gaidamakova, M. Zhai, K. S. Makarova, E. V. Koonin, and M. J. Daly. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 100:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 17.Luo, Z., and D. H. Geschwind. 2001. Microarray application in neuroscience. Neurobiol. Dis. 8:183-193. [DOI] [PubMed] [Google Scholar]
- 18.Ochs, M. F., and A. K. Godwin. 2003. Microarray in cancer: research and application. BioTechniques 34:S4-S15. [Google Scholar]
- 19.Petricoin, E. F., J. L. Hackett, L. J. Lesko, R. K. Puri, S. I. Gutman, K. Chumakov, J. Woodcock, D. W. Feigal, Jr., K. C. Zoon, and F. D. Sistare. 2002. Medical application of microarray technologies: a regulatory science perspective. Nat. Genet. 32:474-479. [DOI] [PubMed] [Google Scholar]
- 20.Relógio, A., C. Schwager, A. Richter, W. Ansorge, and A. Valcarcel. 2002. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 30:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee, S. K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of biodegradation and biotransformation genes in microbial communities using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shchepinov, M. S., S. C. Case-Green, and E. M. Southern. 1997. Steric factors influencing hybridization of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 25:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoemaker, D. D., E. E. Schadt, C. D. Armour, Y. D. He, P. Garrett-Engele, P. D. McDonagh, P. M. Loerch, A. Leonardson, P. Y. Lum, G. Cavet, L. F. Wu, S. J. Altschuler, S. Edwards, J. King, J. S. Tsang, G. Schimmack, J. M. Scheliter, J. Koch, M. Ziman, M. J. Marton, B. Li, P. Cundiff, T. Ward, J. Castle, M. Krolewski, M. R. Meyer, M. Mao, J. Burchard, M. J. Kidd, H. Dai, J. W. Phillips, P. S. Linsley, R. Stoughton, S. Scherler, and M. S. Boguski. 2001. Experimental annotation of the human genome using microarray technology. Nature 409:922-927. [DOI] [PubMed] [Google Scholar]
- 25.Southern, E., K. Mir, and M. Shchepinov. 1999. Molecular interactions on microarrays. Nat. Genet. 21:5-9. [DOI] [PubMed] [Google Scholar]
- 26.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taroncher-Oldedburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-675. [DOI] [PubMed] [Google Scholar]
- 30.Trchounian, A., and H. Kobayashi. 1999. Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Lett. 447:144-148. [DOI] [PubMed] [Google Scholar]
- 31.Verdick, D., S. Handran, and S. Pickett. 2002. Key considerations for accurate microarray scanning and image analysis, p. 83-98. In G. Kamberova (ed.), DNA image analysis: nuts and bolts, DNA Press LLC, Salem, MA.
- 32.Wan, X., N. C. VerBerkmoes, L. A. McCue, D. Stanek, H. Connelly, L. Wu, X. Liu, T. Yan, A. Leaphart, R. L. Hettich, J. Zhou, and D. K. Thompson. 2004. Defining the Shewanella oneidensis FUR regulon: integration of genome-wide expression analysis, proteome characterization, and regulatory motif discovery. J. Bacteriol. 186:8385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 34.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, D., G. Li, L. Wu, J. Zhou, and Y. Xu. 2002. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 18:1432-1437. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, J., and D. K. Thompson. 2002. Challenge in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13:204-207. [DOI] [PubMed] [Google Scholar]
- 39.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.