Abstract
The Ets-1 transcription factor plays a critical role in cell growth and development, but the means by which it activates transcription are still unclear (J. C. Bories, D. M. Willerford, D. Grevin, L. Davidson, A. Camus, P. Martin, D. Stehelin, F. W. Alt, and J. C. Borles, Nature 377:635–638, 1995; N. Muthusamy, K. Barton, and J. M. Leiden, Nature 377:639–642, 1995). Here we show that Ets-1 binds the transcriptional coactivators CREB binding protein (CBP) and the related p300 protein (together referred to as CBP/p300) and that this interaction is required for specific Ets-1 transactivation functions. The Ets-1- and c-Myb-dependent aminopeptidase N (CD13/APN) promoter and an Ets-1-dependent artificial promoter were repressed by adenovirus E1A, a CBP/p300-specific inhibitor. Furthermore, Ets-1 activity was potentiated by CBP and p300 overexpression. The transactivation function of Ets-1 correlated with its ability to bind an N-terminal cysteine- and histidine-rich region spanning CBP residues 313 to 452. Ets-1 also bound a second cysteine- and histidine-rich region of CBP, between residues 1449 and 1892. Both Ets-1 and CBP/p300 formed a stable immunoprecipitable nuclear complex, independent of DNA binding. This Ets-1–CBP/p300 immunocomplex possessed histone acetyltransferase activity, consistent with previous findings that CBP/p300 is associated with such enzyme activity. Our results indicate that CBP/p300 may mediate antagonistic and synergistic interactions between Ets-1 and other transcription factors that use CBP/p300 as a coactivator, including c-Myb and AP-1.
The c-Ets-1 (Ets-1) transcription factor is the cellular counterpart of the v-ets proto-oncogene product originally described as part of the tripartite Gag-Myb-Ets fusion protein from the E26 avian leukemia virus (45, 73). Ets-1 is expressed predominantly in B and T cells of adult mice, where it is critical for T- and B-cell function and development (12, 50). Ets-1 often cooperates with other transcription factors, including AP-1 (74, 78) and c-Myb (21, 66), and can be inhibited by MafB (67); however, its mode of transactivation remains unclear.
The Ets family of transcription factors consists of about 30 members characterized by the highly conserved Ets DNA binding domain (73). Outside of this domain, Ets proteins are more diverse, with the exception of the Ets-1 and Ets-2 subfamily, for example (76). Ets-1 can occur in two alternatively spliced variants, p54 (54 kDa) and p68 (68 kDa), that differ in their N termini (73). p68 Ets-1 is present only in birds and reptiles, while p54 Ets-1 is more widely distributed among vertebrates and is the form expressed in mammals (2, 3). In addition to the Ets domain, Ets-1 and Ets-2 have similarity in the Pointed domain, so named for the Drosophila Ets protein Pointed, which cooperates with c-Jun and Ras in Drosophila eye development (18, 57, 72). The Pointed domain spans about 100 amino acids (aa) in the N-terminal half of Ets-1 and lacks transactivation function when fused to a heterologous DNA binding domain, but it is important for synergistic activity with AP-1 and Ras in mammalian cells (38, 73–75, 78). Deletion analysis indicates that Ets-1 contains an activation domain between the Pointed domain and the Ets domain at the C terminus (73). Moreover, p68 Ets-1 and Ets-2 compete for a limiting factor in transcription activation experiments, suggesting that they have a common coactivator (73), although it is still unclear whether p54 Ets-1 uses the same coactivator as p68 Ets-1 or Ets-2.
A growing number of transcription factors, including c-Myb and the AP-1 components Fos and Jun, use the CREB binding protein (CBP) and the related p300 protein (together referred to as CBP/p300) to mediate the transactivation of RNA polymerase II (34). CBP/p300 may also act as a common mediator of synergistic and antagonistic interactions between these factors and others that bind CBP/p300 (36, 51, 55). Physical contact between the transactivation domains and CBP/p300 appears to be necessary, but not always sufficient, to stimulate transcription (70). Although it is unclear how these protein-protein interactions lead to transactivation, one suggestion is that CBP/p300 acts an adaptor between the activation domain and general transcription initiation factors such as TFIID and TFIIB, or possibly RNA polymerase II (1, 37, 39). Alternatively, the recruitment of CBP/p300 itself may be responsible for transactivation (56). Indeed, CBP/p300 has intrinsic histone acetyltransferase (HAT) activity that could potentially activate chromatin-repressed promoters and enhancers by acetylation of histone N-terminal lysine residues or other proteins involved in transcription (11, 56). The importance of correctly regulated CBP-associated HAT activity in tissue-specific transcription is underscored by the t(8;16)(p11;p13) translocation in acute myeloid leukemias, which fuses a putative acetyltransferase to the N terminus of CBP, presumably leading to deregulation of CBP-associated HAT (13).
Here we show that Ets-1 binds CBP and the related p300 and that this association mediates Ets-1 transactivation potential. Because Ets-1 often requires other CBP/p300 binding transcription factors to transactivate target genes, these coactivators may also be critical for mediating Ets-1-dependent transcriptional synergism.
MATERIALS AND METHODS
Antibodies.
Specific antisera were purchased from Santa Cruz Biotechnology. The CBP/p300 cocktail consisted of equal parts of the following antisera: CBP (A-22), CBP (C-20), and CBP (451) [CBP (451) also recognizes p300]. A-22 was used for the CBP N-terminus-specific antiserum. The p300-specific cocktail consisted of equal parts of p300 (N-15) and p300 (C-20) antisera. The 5614 and 5729 antisera were described previously (37) and were raised against glutathione S-transferase (GST)-CBP fusion proteins containing CBP residues 455 to 679 and 1 to 117, respectively (a gift from M. Montminy). The Ets-1-specific antisera, anti-Ets-1(N) and anti-Ets-1(C), are Ets-1 (N-276) and Ets-1 (C-20), respectively. The monoclonal antibodies that recognize the Gal4 DNA binding domain (DBD) and E1A were RK5C1 and M73, respectively. Typically, 1 μg of each antiserum was used for each immunoprecipitation. The normal rabbit serum (NRS; Sigma) control was used at 5 μl per immunoprecipitation (1 μl for coimmunoprecipitations used in the HAT assays). Preblocked antisera were produced by incubating antisera with a 10-fold mass excess of antigenic peptide (Santa Cruz Biotechnology) for 3 h at room temperature or overnight at 4°C. Immunoblots were developed by enhanced chemiluminescence (Amersham).
Plasmids and transient-transfection assays.
KG1a myeloblastic cells (ATCC CCL 246.1) were grown and electroporated with 5 μg of reporter plasmid, 5 μg of Gal fusion protein expression plasmid, 25 ng of cytomegalovirus (CMV) E1A vectors (where applicable), and 4 μg of Rous sarcoma virus (RSV) β-galactosidase (β-gal) or MAP1-SEAP internal control reporter plasmids, as previously described (66). Larger amounts of CMV E1A sometimes led to a general repression of reporters and expression vectors. The cells were harvested after about 16 h, and enzyme assays were performed to assess reporter gene expression. Reporter gene-derived chloramphenicol acetyltransferase (CAT) or luciferase activity was normalized to β-gal activity derived from the RSV β-gal or Renilla luciferase derived from pRL-TK (Promega) or to secreted alkaline phosphatase from MAP1-SEAP internal transfection control reporter plasmids. F9 cells were transfected in 35-mm wells with Superfect (Qiagen) or Lipofectamine (Gibco BRL), with 1 μg of reporter, 1 μg of pEVRFO-Ets-1 or pEVRFO, 8 μg of CMVp300 or 1 μg of pRC/RSVmCBP HA-RK (RSV CBP) or 1 μg of RSV CBP 1–1285, and 50 ng of pRL-TK. Equal molar amounts of a CMVβ plasmid religated without the NotI-HindIII insert were used as controls in the p300 experiments, with the total amount of DNA balanced with pBluescript SKII (Stratagene). F9 luciferase assays were normalized to Renilla luciferase derived from pRL-TK.
G5B CAT was described previously (46). CMV expression vectors for 12S E1A and Δ2–36 E1A (69) were gifts from Bob Rooney. RSV CBP 1–1285 is pRC/RSV-CBP (39) with CBP codon 1286 mutated from GAG to TAG. CMVp300 was constructed by inserting the HindIII-NotI fragment from pCMVβp300 (22) into pCMVβ (Clontech). RSV CBP2 was constructed by inserting a HindIII-XbaI fragment containing mouse CBP sequences that lacked aa 739 to 2394 into the RSV expression vector pGR. Gal-Ets 2–440 was constructed by inserting a BamHI-SpeI fragment encoding mouse p54 Ets-1 aa 2 to 440 from pEVRFO-Ets1 into the Gal4 fusion vector pM2 cut with BamHI and XbaI (53, 64). Gal-Ets 2–165 was made by cutting pEVRFO-Ets1 with SphI, blunting with T4 DNA polymerase, and isolating the Ets-1 fragment released after BamHI digestion. This fragment was ligated into pM2, which had been cut with HindIII, blunted with Klenow fragment, and digested with BamHI. Gal-Ets 2–129 was constructed by inserting the BamHI-XbaI fragment from pEVRFO-Ets-1 into pM2. Gal-Ets 2–155, 2–177, 2–194, and 2–210 were constructed by inserting Ets-1 PCR fragments into pM2. Gal-Ets Δ166–194, Δ177–194, and Δ178–210 were made by PCR site-directed mutagenesis (32). GST-CBP fusion proteins were produced from the following pGEX-based vectors (Pharmacia): GST-CBP 553–679 (GST-KIX S/B) was a gift from M. Montminy (58); pGEX-2T-CBP 1–1891 has a mouse CBP BamHI-SmaI fragment from pRC/RSV-CBP cloned into pGEX-2T cut with BamHI and SmaI (39); pGEX-3X-CBP 1891–2441 contains a SmaI-EcoRI CBP fragment from pRC/RSV-CBP cloned into pGEX-3X cut with SmaI and EcoRI; pGEX-4T-2-CBP 1–117 was made by cutting pRC/RSV-CBP with NcoI, blunting with Klenow fragment, digesting with BamHI, and ligating into pGEX-4T-2 digested with BamHI and SmaI; pGEX-4T-2-CBP 1–141 was made by digesting pRC/RSV-CBP with ApaI, blunting with T4 DNA polymerase, digesting with BamHI, and ligating into pGEX-4T-2 cut with BamHI with SmaI; pGEX-4T-2-CBP 1–270 was made by digesting pRC/RSV-CBP with KpnI, blunting with T4 DNA polymerase, cutting with BamHI, and ligating into pGEX-4T-2 cut with BamHI and SmaI; pGEX-4T-2-CBP 1–312 was made by digesting pRC/RSV-CBP with EcoRV and BamHI and cloning into pGEX-4T-2 cut with BamHI and SmaI; pGEX-2T-CBP 1–452 was made by digesting pGEX-2T-CBP 1–1892 with EcoRI and religating; pGEX-3X-CBP 313–452 was constructed by isolating the EcoRV-EcoRI fragment from pRC/RSV-CBP and ligating it into pGEX-3X cut with SmaI and EcoRI; and pGEX-3X-CBP 357–452 was made by isolating the PvuII-EcoRI fragment from CMV CBP2 and ligating it into pGEX-3X cut with SmaI and EcoRI (17).
Coimmunoprecipitations.
Jurkat or KG1a cells (0.8 × 108 to 1 × 108 cells) were harvested, washed once with short-term labeling medium (phosphate- or methionine-free RPMI 1640, 5% dialyzed fetal bovine serum), resuspended at 5 × 106 cells/ml in this medium, and incubated for 20 min at 37°C to deplete intracellular pools of methionine or phosphate. The cells were then incubated in 20 ml of fresh short-term labeling medium containing 0.18 mCi of [35S]methionine per ml (for 3 h) or 0.15 mCi of [32P]orthophosphate per ml (for 2 h). The cells were harvested and washed twice with 20 ml of ice-cold phosphate-buffered saline, and nuclear extracts were prepared as previously described (65), except that the buffers also contained 0.5 μg of leupeptin per ml (and sometimes 10 ng of calyculin per ml), and used for coimmunoprecipitation. The first immunoprecipitation was performed by adding antiserum and 50 μl of protein A-Sepharose (50% slurry) to about 80 to 100 μl of nuclear extract diluted with 3 volumes of ice-cold PC+100 buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% Nonidet P-40, 20% glycerol, 0.01% bovine serum albumin [BSA], 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml) followed by rotation at 4°C for 3 h (58). The first immunoprecipitation pellet was washed twice with 0.5 ml of ice-cold PC+100 buffer, and antigens were released by boiling for 2 min in 100 μl of boiling buffer (20 mM Tris-HCl [pH 8.0], 0.5% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol). For experiments showing a primary immunoprecipitate of Ets-1 and CBP/p300, PC+100 containing 400 mM KCl (PC+400) and 1% BSA was used in the immunoprecipitation, and for the four subsequent 1-ml washes, PC+400 containing 0.01% BSA was used. After centrifugation, the supernatant was removed, diluted with 4 volumes of RIPA buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate), and used in a second immunoprecipitation with 50 μl of antibody-protein A-Sepharose (50% slurry) at 4°C for 3 h or overnight. The immunocomplex was washed three times with 0.5 ml of RIPA buffer (containing 0.1% SDS), and the pellet was boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. Samples were sometimes normalized for counts per minute before SDS-PAGE (large format for examining primary immunoprecipitates, which were not normalized) and fluorography of the dried gel.
Coimmunoprecipitation HAT assays.
Nuclear extracts from nonlabeled KG1a or Jurkat cells were prepared as described above. Extracts were precleared by adding protein A-Sepharose and NRS, and the supernatants were used for immunoprecipitations, as detailed above. Each immunoprecipitation experiment was performed in quadruplicate, with each immunocomplex washed three times with ice-cold PC+100 buffer. HAT assays were performed as previously described by Bannister and Kouzarides (11), except that phosphocellulose filters were washed with the buffer used by Ogryzko et al. (56). HAT assays based on histones or BSA control were performed in duplicate with four separate immunoprecipitations for each antiserum. Background counts per minute were determined from HAT assays performed with two NRS immunoprecipitations without additional protein substrates. The average background counts per minute (usually about 100) was subtracted from the experimental values.
Coimmunoprecipitation of GST–Ets-1 and CBP 1–714.
35S-labeled CBP 1–714 protein was produced with a coupled reticulocyte lysate system (Promega), using CMV CBP2 plasmid cut with SphI (17). GST–Ets-1 fusion protein (GST-Ets 2–440) was eluted from glutathione-agarose (GSH beads; Sigma) after expression in Escherichia coli. Coimmunoprecipitation was carried out by incubating about 0.5 μg of GST-Ets 2–440 and 1 μl of CBP 1–714 in 30 μl of PC+100 buffer for 20 min at room temperature. After the addition of antiserum, the immunocomplexes were washed and analyzed by SDS-PAGE followed by fluorography.
Coimmunoprecipitation of Ets-1 and CBP Δ739–2394.
Jurkat cells (45 × 106 to 50 × 106 cells) were electroporated with 50 μg of the RSV CBP2 plasmid at 960 μF and 250 V. Then 180 × 106 to 200 × 106 electroporated cells were pooled and labeled overnight in 60 ml of methionine-free RPMI 1640 supplemented with 10% regular RPMI 1640, 10% fetal bovine serum, and 0.12 mCi of [35S]methionine per ml, before preparation of nuclear extracts and coimmunoprecipitation.
GST pull-down assays.
GST fusion proteins were purified from E. coli, and GST pull-down assays were performed as previously described (58). GSH beads were preblocked for nonspecific protein interactions by using NRS and two washes with PC+100 buffer (58). In vitro-transcribed and -translated [35S]methionine-labeled protein (2 to 4 μl) was added to the GSH beads, and the mixture was stirred for 30 min. The GSH beads were washed three to four times and boiled in 20 μl of 2× SDS gel sample buffer before SDS-PAGE was performed. Quantitation was performed with Molecular Dynamics Storm.
RESULTS
Transactivation of the Ets-1- and c-Myb-dependent CD13/APN promoter requires CBP/p300.
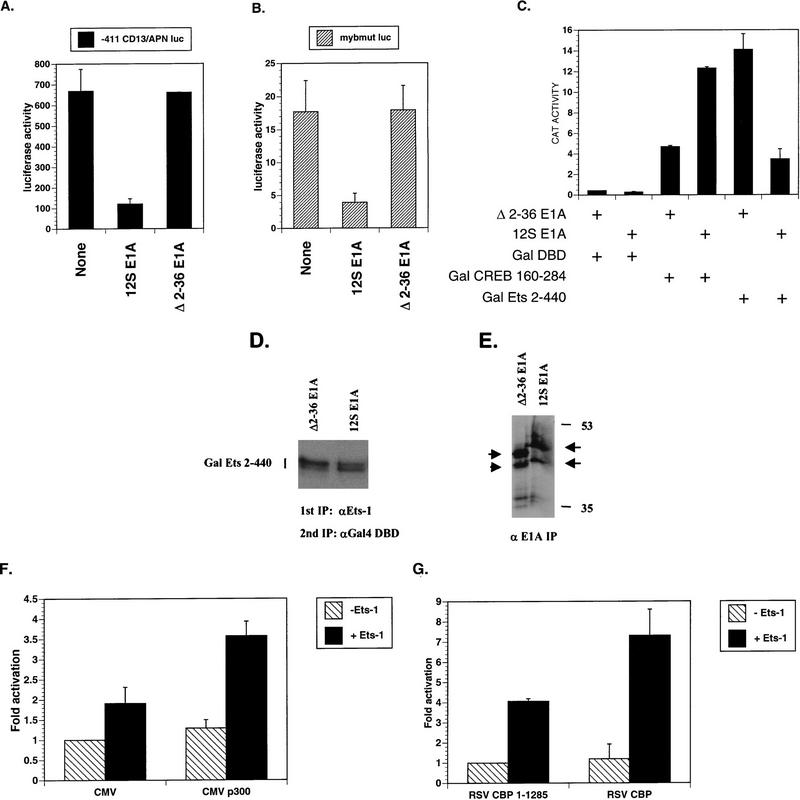
Transcriptional activation of the CD13/APN gene in hematopoietic cells of the myeloid lineage depends upon c-Myb and Ets-1 binding sites in the promoter (66). Since c-Myb has been shown to use CBP as a coactivator, we sought to determine if CBP/p300 was necessary for c-Myb/Ets-1 synergism on the CD13/APN promoter (20, 55). To this end, we used the adenovirus 12S E1A protein, which specifically binds the CH3 region of CBP/p300 and inhibits associated transactivator function (5, 44). 12S E1A repressed CD13/APN promoter activity about sixfold in transiently transfected myeloblastic KG1a cells (Fig. 1A). By contrast, an E1A mutant incapable of binding CBP/p300 (Δ2–36 E1A) had no effect on CD13/APN promoter activity (Fig. 1A) (69). Thus, CBP/p300 appears to be required for activation of the CD13/APN promoter in myeloid cells. To ascertain if E1A was targeting the Ets-1 component of CD13/APN promoter activity, we tested this promoter when the c-Myb binding site was mutated (66). Although mutation of the c-Myb binding site reduced CD13/APN activity about 40-fold, the residual Ets-1-dependent activity was further repressed about 5-fold by 12S E1A (Fig. 1B), suggesting that E1A indeed targets Ets-1 transactivation function.
FIG. 1.
Ets-1 transactivation function requires CBP/p300. (A) The Ets-1- and c-Myb-responsive CD13/APN promoter is repressed by adenovirus 12S E1A protein, an inhibitor of CBP/p300. KG1a myeloblastic cells were transiently cotransfected with the CD13/APN promoter luciferase reporter (CD13/APN Luciferase), a secreted alkaline phosphatase gene reporter (MAP1-SEAP), and either expression plasmids for 12S E1A (12S E1A), the Δ2–36 12S E1A mutant incapable of binding CBP/p300 (Δ2–36 E1A), or empty expression vector (None). Luciferase values were normalized to SEAP activity derived from the internal transfection control reporter. 12S E1A has little effect on the MAP1-SEAP internal control reporter (data not shown). (B) E1A represses Ets-1-dependent transcription in KG1a cells from a CD13/APN promoter lacking c-Myb binding sites (mybmut luc). Mybmut luc reporter activity was assayed as in panel A. (C) Gal-Ets 2–440 is inhibited by 12S E1A in KG1a cells. KG1a cells were transiently transfected with G5B CAT reporter plasmid, RSV β-gal, and expression vectors for Gal4 DBD (Gal DBD), Gal4 DBD fused to the CBP/p300-independent-glutamine-rich activator from CREB aa 160 to 284 (Gal CREB 160–284), Gal DBD fused to murine Ets-1 aa 2 to 440 (Gal Ets 2–440), 12S E1A, or Δ2–36 E1A. CAT activity was normalized to β-gal activity derived from the internal transfection control RSV β-gal reporter plasmid. Empty expression vector gave similar results to those of the mutant Δ2–36 12S E1A (data not shown). (D) 12S E1A does not inhibit the expression of Gal-Ets 2–440 in KG1a cells. The cells were transiently transfected with the indicated expression vectors before undergoing metabolic labeling with [35S]methionine; this was followed by whole-cell extract preparation and two sequential immunoprecipitations (IP) with anti-Ets-1(N- and C-terminal-specific) and anti-Gal4 DBD antibodies. (E) 12S E1A and Δ2–36 E1A are expressed at comparable levels in transiently transfected KG1a cells. The cells were labeled with [35S]methionine before whole-cell extract preparation and immunoprecipitation with anti-E1A antibody. Arrows point to the respective E1A proteins. Molecular size markers are indicated (in kilodaltons). (F) p300 potentiates Ets-1 activity in transiently transfected F9 cells. The CD13/APN luciferase reporter was cotransfected with expression vectors for Ets-1 (+Ets-1) or empty vector (−Ets-1), p300 (CMVp300), or empty vector (CMV). Luciferase activity derived from the CD13/APN reporter was normalized to Renilla luciferase derived from the internal control reporter pRL-TK. (G) Full-length CBP potentiates Ets-1 activity in F9 cells. Transfections were performed as in panel F, except that full-length CBP (RSV CBP) was compared to a CBP expression vector (RSV CBP 1–1285) containing a nonsense mutation at CBP codon 1286. Results are means and standard errors (n ≥ 2).
CBP/p300 is necessary for Ets-1-mediated transactivation.
12S E1A-mediated repression of the CD13/APN promoter lacking c-Myb binding sites suggested that Ets-1 also uses CBP/p300 as a coactivator. To more rigorously test this prediction, we assessed the effects of 12S E1A on the transactivation potential of a Gal-Ets 2–440 fusion protein containing the Gal4 DNA binding domain (Gal DBD) fused to mouse Ets-1 residues 2 to 440 (wild-type Ets-1 is 1 to 440); this strategy allows one to determine Ets-1 transactivation function without interference from endogenous Ets proteins. Gal-Ets 2–440 transactivated a reporter gene containing five Gal4 binding sites (G5B CAT) about 33-fold more efficiently than did Gal DBD in the presence of Δ2–36 E1A (Fig. 1C) or empty expression vector (data not shown). However, Gal-Ets 2–440 was inhibited approximately fourfold by 12S E1A, indicating that Ets-1 requires CBP/p300 as a coactivator in KG1a cells. To control for the specificity of 12S E1A-dependent inhibition of Gal-Ets 2–440, we also used a CBP-independent activator, Gal-CREB 160–284, which fuses the Gal DBD to the glutamine-rich activation domain of CREB, termed Q2 (Q2 binds the TFIID component dTAF110 [16, 25, 61, 77]). Gal-CREB 160–284 stimulated the reporter about 12-fold compared to Gal DBD in KG1a cells, but, in contrast to Gal-Ets 2–440, its activity was stimulated about 2.5-fold by 12S E1A, demonstrating the specificity of 12S E1A repression on Gal-Ets 2–440 activity (Fig. 1C). The divergent effects of 12S E1A on Gal-Ets 2–440 and Gal CREB 160–284 suggested that E1A was not repressing Gal-Ets 2–440 by lowering its expression (both Gal fusion proteins are expressed from similar simian virus 40 early-promoter/ori-driven expression vectors). We confirmed this by comparing Gal-Ets 2–440 expression in KG1a cells cotransfected with 12S E1A or Δ2–36 E1A (Fig. 1D). Furthermore, the Δ2–36 E1A protein was expressed comparably to 12S E1A in KG1a cells (Fig. 1E), consistent with the notion that the ability of E1A to inhibit Ets-1 is dependent on E1A N-terminal residues implicated in binding CBP/p300 (69).
To determine if CBP/p300 could potentiate Ets-1 activity, we expressed p300 in F9 mouse teratocarcinoma cells. When cotransfected with CD13/APN promoter reporter and Ets-1 expression vector, Ets-1 modestly activated the reporter about twofold, with addition of p300 potentiating Ets-1 activity an additional 50 to 100% (Fig. 1F). p300 alone had little effect. This result was reinforced when we compared full-length CBP (aa 1 to 2441) to a mutant truncated CBP (CBP 1–1285) containing a termination triplet at codon 1286 (Fig. 1G). CBP 1–1285 is missing one of the Ets-1 binding regions (Fig. 5C) and a C-terminal transactivation domain (71), suggesting that the N-terminal 1,285 aa of CBP is not sufficient to fully cooperate with Ets-1. Differences in the absolute levels of Ets-1 activity observed in these experiments could be due to the different combinations of expression vectors used in the two systems.
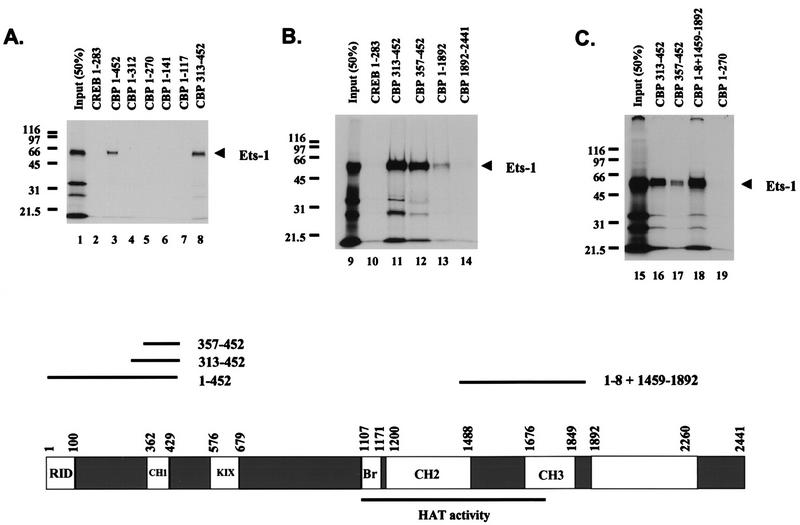
FIG. 5.
Ets-1 binds at least two separate regions of CBP in vitro. Shown are the results of three separate experiments demonstrating specific Ets-1 binding sites on CBP. GST-CBP pull-down experiments were performed with [35S]methionine-labeled Ets-1 produced in vitro and GST-CBP fusion proteins purified from E. coli. Molecular mass markers are indicated (in kilodaltons). Input (50%) shows 50% of the Ets-1 used in each pull-down assay. GST CREB 1–283 (CREB 1–283) is GST fused to CREB aa 1 to 283 and acts a negative control for Ets-1 binding. Comparable amounts of GST fusion proteins were used in each assay. A scale drawing depicting the CBP domain structure, along with approximate domain boundaries in residues from the N terminus (position 1) to the C terminus (position 2441) is shown at the bottom. The receptor interaction domain (RID), the three cysteine-plus histidine-rich domains (CH1, CH2, and CH3), the CREB binding KIX domain (KIX), and the bromodomain (Br) are shown (6, 19, 22, 36, 58). CBP residues 1099 to 1758 sufficient for HAT activity are underlined (11). Also shown are scale-drawn lines corresponding to CBP fragments with inclusive endpoints sufficient to bind Ets-1 when fused to GST (1 to 452, 313 to 452, 357 to 452, 1 to 8 plus 1459 to 1892) and their relative positions in CBP. This figure was produced using Macintosh versions of Adobe Photoshop and Microsoft PowerPoint. The results are representative of at least two independent experiments.
CBP/p300 and Ets-1 associate in nuclear extracts.
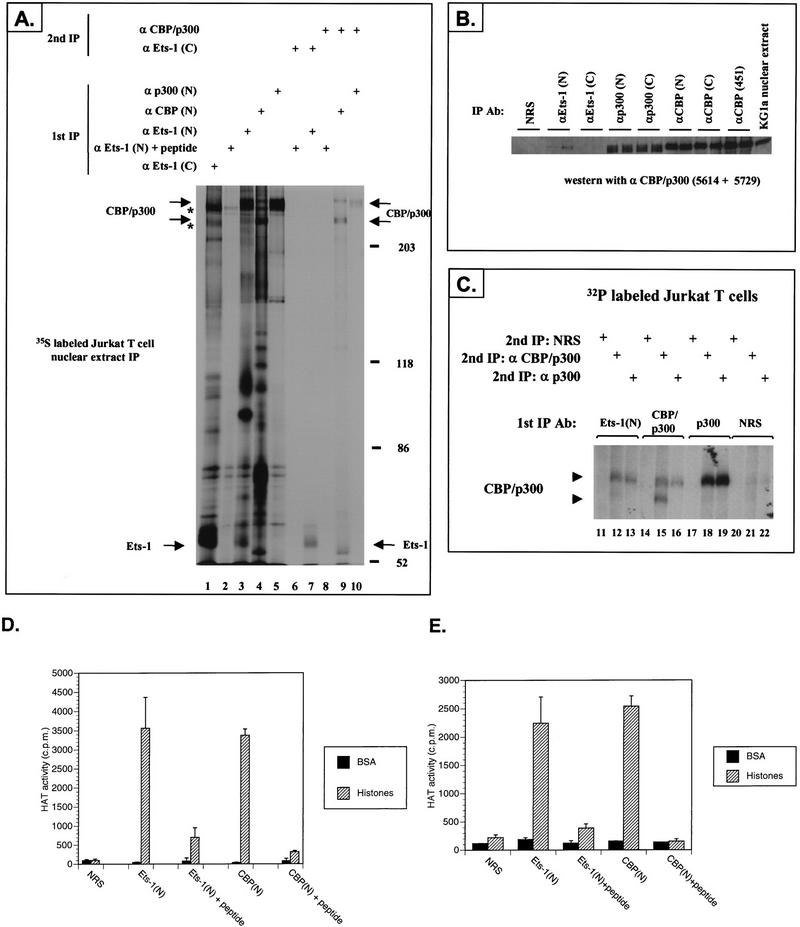
Inhibition of Gal-Ets 2–440 activity by 12S E1A, and synergism between Ets-1 and CBP/p300, indicated that Ets-1 may interact with CBP/p300 in the nucleus. We tested this possibility by coimmunoprecipitating Ets-1 and CBP/p300 with an Ets-1 N-terminus-specific antiserum [anti-Ets-1(N)] from nuclear extracts prepared from Jurkat T cells labeled with [35S]methionine (Fig. 2A). To confirm the SDS-PAGE positions of Ets-1 and CBP/p300 following a primary immunoprecipitation we performed in parallel two sequential immunoprecipitations, the first under mild buffer conditions followed by antigen release and the second under more stringent conditions (Fig. 2A, lane 7 for Ets-1 and lanes 9 and 10 for CBP/p300 [note the broad p300 band]). Bands corresponding to CBP/p300 and Ets-1 were present in the primary immunoprecipitation with anti-Ets-1(N) (lane 3), but not when this antiserum was preblocked with an excess of peptide antigen (lane 2). Interestingly, an Ets-1 C-terminus-specific antiserum [anti-Ets-1(C)] that was more efficient at immunoprecipitating Ets-1 was noticeably less proficient at coimmunoprecipitating CBP/p300 (lane 1). This suggests that in nuclear extracts the anti-Ets-1(C) antiserum disrupts an Ets-1–CBP/p300 complex or preferentially immunoprecipitates Ets-1 that is not complexed with CBP/p300. Although anti-Ets-1(N) and a p300 N-terminus-specific antiserum [anti-p300(N)] appear to immunoprecipitate similar amounts of p300 (compare lanes 3 and 5), this is probably due to the inability of the commercial antisera to immunoprecipitate more than a small fraction of CBP/p300 in extracts (15a). The presence of CBP/p300 in the Ets-1 immunoprecipitate was confirmed by immunoprecipitation of KG1a nuclear extracts followed by immunoblotting with the CBP/p300-specific antisera 5614 and 5729 (Fig. 2B). We also observed Ets-1 in immunoprecipitates with antisera 5614 and 5729 (but not NRS), suggesting that CBP/p300 antisera could also coimmunoprecipitate Ets-1 and CBP/p300 (data not shown). Thus, anti-Ets-1(N) specifically recognized a complex containing Ets-1 and CBP/p300 in KG1a myeloblastic and Jurkat T-cell nuclear extracts. Together, these results suggest that the amount of Ets-1 complexed with CBP/p300 is variable, depending on the cell type and probably other factors.
FIG. 2.
CBP/p300 coimmunoprecipitates with Ets-1 from nuclear extracts. (A) Jurkat T cells were labeled for 3 h with [35S]methionine. Nuclear extracts were then prepared, and either single or two sequential immunoprecipitations were performed as described in Materials and Methods (1st IP, 2nd IP). The first immunoprecipitation was performed with the indicated antisera specific for the following proteins: Ets-1 N terminus or C terminus [α Ets-1 (N), α Ets-1 (C)], Ets-1 N-terminus antiserum preblocked with a 10-fold mass excess of antigenic peptide [α Ets-1 (N) + peptide], CBP or p300 N terminus [α CBP (N), α p300 (N)], both CBP and p300 (α CBP/p300). Second immunoprecipitations were performed with the following antisera: α Ets-1 (C) or α CBP/p300. The positions of CBP/p300, Ets-1, and molecular mass markers (in kilodaltons) are indicated. Asterisks indicate the positions of nonspecific high-molecular-mass bands present in the primary immunoprecipitates. Note that CBP can appear as two bands and p300 appears as a single broad band. (B) CBP/p300 immunoprecipitates with Ets-1 N-terminal-specific antiserum from KG1a cell nuclear extracts. Duplicate immunoprecipitations were performed with the indicated antisera. NRS was used as the control. CBP/p300 present in the immunoprecipitations was detected by immunoblotting with a cocktail of 5614 and 5729 CBP/p300-specific antisera. KG1a nuclear extract starting material is indicated. (C) 32P-labeled CBP/p300 coimmunoprecipitates with endogenous Ets-1 from Jurkat nuclear extracts. The cells were labeled with [32P]orthophosphate for 2 h. Two sequential immunoprecipitations were performed on nuclear extracts as in panel A. α p300 is the antiserum specific for p300. (D and E) Ets-1 coimmunoprecipitates with HAT activity from both KG1a nuclear extracts (D) and Jurkat nuclear extracts (E). HAT activity with BSA or histones used as substrates was measured following immunoprecipitation with the indicated antisera. CBP was immunoprecipitated with the CBP N-terminus-specific antiserum A22 [CBP(N)]. To control for the specificity of the Ets-1 N-terminus-specific and CBP antisera, we preblocked the antiserum with an excess of antigenic peptide [Ets-1(N) + peptide, CBP(N) + peptide]. NRS also served as a negative control. HAT activity is reported in counts per minute derived from [3H]acetyl groups transferred to substrate proteins. The means and standard errors of two assays performed in a single representative experiment are shown. This figure was produced with Macintosh versions of Adobe Photoshop and Microsoft PowerPoint. The results represent at least two independent experiments.
Since CBP and p300 are phosphoproteins, we addressed whether the high-molecular-weight proteins that coimmunoprecipitated with Ets-1 were phospho-CBP and phospho-p300. Similar to 35S-labeled cells, endogenous Ets-1 and CBP/p300 were coimmunoprecipitated from Jurkat cells labeled with [32P]orthophosphate (Fig. 2C, lanes 12 and 13). p300-specific antisera also immunoprecipitated p300 after the initial Ets-1 immunoprecipitation (lane 13), confirming that p300 is present in an immunocomplex with Ets-1. Only background bands were detected with NRS as a nonspecific antiserum control (Fig. 2C).
Ets-1–CBP/p300 immunocomplexes have HAT activity.
Recently, CBP and p300 have been shown to have intrinsic HAT activity and to bind the HAT protein PCAF, suggesting that HAT activity should be present in the Ets-1–CBP/p300 immunocomplex (11, 56, 79). To confirm this prediction, we performed immunoprecipitations with KG1a (Fig. 2D) and Jurkat (Fig. 2E) nuclear extracts with anti-Ets-1(N) and CBP N-terminus-specific antiserum [anti-CBP(N)], followed by HAT assays of the immunocomplexes with [3H]acetyl coenzyme A and BSA or histones as protein substrates. The anti-Ets-1(N) and anti-CBP(N) immunocomplexes contained acetyltransferase activities that were specific for histones, while the NRS control showed little activity. Similar results were obtained with p300-specific antiserum (data not shown). Moreover, preblocking anti-Ets-1(N) and anti-CBP(N) with their respective antigenic peptides significantly reduced HAT activity, showing that the presence of HAT correlates with the presence of an Ets-1–CBP/p300 complex and CBP in the respective immunocomplexes (Fig. 2D and 2E). Intriguingly, the similarity in HAT activity between the Ets-1 and CBP/p300 immunoprecipitations could be due to intrinsic differences in HAT function between an Ets-1–CBP/p300 complex and a CBP or p300 complex without Ets-1. While other factors such as inefficient immunoprecipitation or HAT inhibition by CBP or p300 antibodies may also account for this observation, we are investigating if Ets-1 modulates HAT activity as part of its transactivation functions.
Ets-1–CBP/p300 association is independent of DNA binding.
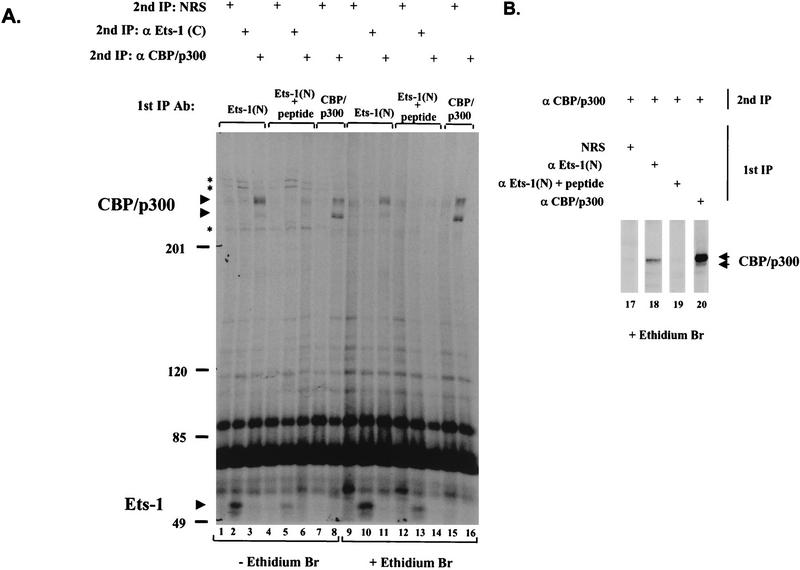
Our coimmunoprecipitation data suggested that Ets-1 binds CBP/p300 by protein-protein interactions. To rule out an indirect association between Ets-1 and CBP/p300 through a contaminating nucleic acid “link” between the Ets-1 DBD domain and a DNA binding CBP/p300 complex, we used ethidium bromide to disrupt protein-DNA interactions (30, 40). Immunoprecipitations with Ets-1 antiserum (and control antisera and washes) were performed with buffer containing 50 μg of ethidium bromide per ml, a concentration that preferentially inhibits DNA-protein interactions (30, 40). The Ets-1–CBP/p300 complex coimmunoprecipitated from Jurkat nuclear extracts was insensitive to ethidium bromide (Fig. 3A, compare lanes 3 and 11), suggesting that Ets-1 and CBP/p300 interact through protein-protein contacts. Similar results were obtained with KG1a cell nuclear extracts (Fig. 3B). Although the Ets-1–CBP/p300 coimmunoprecipitation was unaffected by ethidium bromide, several other bands were lost (Fig. 3A, compare lanes 1 to 6 with lanes 9 to 14), showing that ethidium bromide did affect other proteins associated with the immune complex. Consistent with the results in Fig. 2, addition of anti-Ets-1(N) antigenic peptide blocked coimmunoprecipitation of Ets-1 and CBP/p300 (Fig. 3, compare lanes 2 and 5 and lanes 10 and 13 for Ets-1; compare lanes 3 and 6, lanes 11 and 14, and lanes 18 and 19 for CBP/p300). Thus, the physical association of Ets-1 with CBP/p300 in nuclear extracts appears to be independent of DNA binding.
FIG. 3.
The Ets-1–CBP/p300 immunocomplex from Jurkat and KG1a nuclear extracts is resistant to ethidium bromide, a disrupter of protein-DNA interactions. (A) Jurkat cells were transiently transfected with an Ets-1 expression vector and labeled overnight with [35S]methionine. Nuclear extracts were prepared, and two sequential immunoprecipitations (1st IP and 2nd IP) were performed with the indicated antisera as in Fig. 2. The first immunoprecipitation and washes that contained 50 μg of ethidium bromide per ml are indicated (+ Ethidium Br). Asterisks indicate proteins lost from the immunocomplex when ethidium bromide was used. Molecular mass markers (in kilodaltons) are indicated, as are CBP and p300 (CBP/p300) and Ets-1. In lanes 9, 12, and 15, ethidium bromide caused the appearance of a nonspecific band migrating near CBP/p300 when NRS was used in the second immunoprecipitation. (B) Nontransfected KG1a cells were labeled overnight with [35S]methionine, and nuclear extracts were prepared and analyzed by immunoprecipitation as for panel A. This figure was produced with Macintosh versions of Adobe Photoshop and Microsoft PowerPoint. The results represent at least two independent experiments.
Ets-1 associates with the N terminus of CBP.
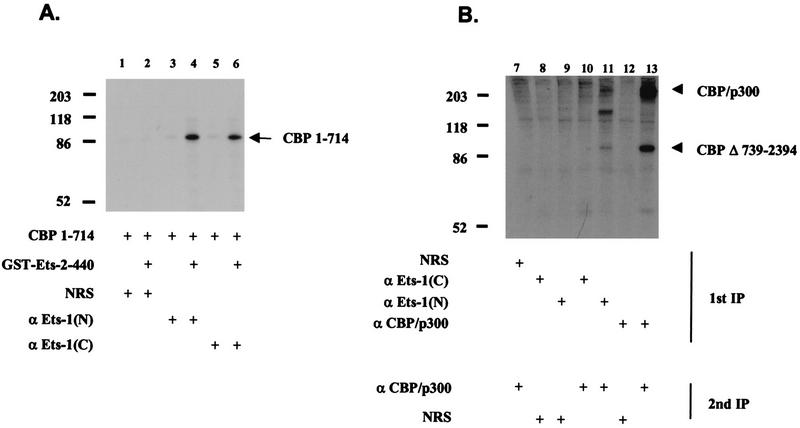
To define regions of CBP necessary for Ets-1 interactions, we performed coimmunoprecipitations in vitro with GST-Ets 2–440 fusion protein purified from E. coli and in vitro-translated CBP containing residues 1 to 714. CBP 1–714 was specifically coimmunoprecipitated with two different Ets-1-specific antisera only in the presence of GST-Ets 2–440 (Fig. 4A). This suggested that CBP residues 1 to 714 were sufficient to bind GST-Ets 2–440 and that other cellular factors, unless present in the reticulocyte lysate, were not necessary to mediate the Ets-1–CBP interaction. To confirm this result by using nuclear extracts, we transiently transfected Jurkat cells with a plasmid expressing a mutated CBP gene lacking residues 739 to 2394 (CBP Δ739–2394; wild-type CBP is 1 to 2441) (17). Consistent with the in vitro data, coimmunoprecipitation with a nuclear extract from these cells showed that CBP Δ739–2394 and endogenous CBP/p300 coimmunoprecipitated with anti-Ets-1(N) (Fig. 4B, lanes 11 and 13). However, anti-Ets-1(C) was much less efficient at coimmunoprecipitating CBP/p300 and CBP Δ739–2394 from Jurkat nuclear extracts (Fig. 4B, lane 10), even though it could coimmunoprecipitate CBP 1–714 in vitro (Fig. 4A, lane 6). Therefore, there are differences in CBP–Ets-1 interactions in nuclear extracts from those in more purified systems.
FIG. 4.
Coimmunoprecipitation of Ets-1 and CBP N-terminal polypeptides in vitro and in vivo. (A) Immunoprecipitations with purified GST-Ets 2–440, in vitro-translated CBP 1–714 labeled with [35S]methionine, control antisera (NRS), and anti-Ets-1(N) and anti-Ets-1(C) [α Ets-1(N) and α Ets-1(C)] are shown. CBP 1–714, encompassing the first 714 residues of CBP, is indicated, as are molecular mass markers (in kilodaltons). (B) Coimmunoprecipitation from nuclear extract of Ets-1, endogenous CBP/p300, and CBP Δ739–2394 expressed in transiently transfected Jurkat cells labeled with [35S]methionine. Two sequential immunoprecipitations were performed as in Fig. 2 (1st IP, 2nd IP) with the indicated antisera. This figure was produced with Macintosh versions of Adobe Photoshop and Microsoft PowerPoint. The results represent at least two independent experiments.
Ets-1 binds two regions of CBP.
To determine the Ets-1 binding region within the first 714 residues of CBP, we performed GST ‘pull-down’ assays with in vitro-translated Ets-1 and GST-CBP fusion proteins purified from E. coli. About 25% of the input Ets-1 was retained on GST-CBP 1–452 or GST-CBP 313–452 prebound to GSH beads (Fig. 5A), localizing the interaction domain to CBP 313–452. By contrast, Ets-1 did not bind other GST fusion proteins containing the CBP N-terminal 312 residues (Fig. 5A). Nor did GST-CBP 1892–2441 (Fig. 5B) or GST-CBP 553–679 (data not shown) bind Ets-1, in contrast to GST-CBP 357–452, further localizing an Ets-1 interaction domain on CBP to residues 357 to 452 (Fig. 5B, lane 12). CBP 357–452 sequences include the cysteine- and histidine-rich CH1 region, which has been proposed to contain the TAZ motif (transcriptional adaptor putative zinc fingers; Cys-X4-Cys-X8-His-X3-Cys) (60). The ability of Ets-1 to bind the CBP CH1 region indicated that it may also bind to another CBP cysteine- and histidine-rich region with TAZ motifs (CH3) (60). Since CH3 approximately spans CBP residues 1676 to 1849, we tested whether Ets-1 would bind a GST-CBP protein containing CBP residues 1 to 8 fused to residues 1459 to 1892. Figure 5C shows that Ets-1 was bound by GST-CBP 1–8+1459–1892 to approximately the same extent as by GST CBP 313–452 and GST CBP 357–452. Thus, Ets-1 can bind at least two separate regions of CBP that are rich in cysteines and histidines and have TAZ motifs.
Ets-1 transactivation function correlates with binding to CBP residues 1 to 714.
To more precisely correlate Ets-1 sequences that bind CBP 1–714 and also activate transcription, we performed pull-down assays with GST–Ets-1 fusion proteins and transactivation assays with the cognate Gal–Ets-1 fusion genes in KG1a cells. Figure 6A and B shows the results of a typical pull-down assay; the results of four independent experiments are summarized in Fig. 6C. Figure 6C also summarizes the relative activities of the Gal–Ets-1 fusions from three experiments. GST–Ets-1 proteins fell into three classes based on their ability to bind CBP 1–714 (Fig. 6C): weak binders bound less than 6% of that seen with GST-Ets 2–440 (GST, GST-Ets 2–129, and GST-Ets 2–153); moderate binders bound about 30 to 60% of that seen with GST-Ets 2–440 (GST-Ets 2–177, GST-Ets Δ166–194, GST-Ets Δ178–194, and GST-Ets Δ178–210); and a strong binder bound about 150% of that seen with GST-Ets 2–440 (GST Ets 2–210). This grouping correlated well with the activities of the Gal–Ets-1 fusions (Fig. 6C): poor activators had less than about 2.5% of the activity of Gal-Ets 2–440 (Gal DBD, Gal-Ets 2–129, and Gal-Ets 2–153); moderate activators had between 9 and 17% of Gal-Ets 2–440 (Gal-Ets 2–165, Gal-Ets 2–177, Gal-Ets Δ166–194, Gal-Ets Δ178–194, and Gal-Ets Δ178–210); and strong activators had about 130 to 150% the activity of Gal-Ets 2–440 (Gal-Ets 2–194 and Gal-Ets 2–210). Similar amounts of GST–Ets-1 fusion proteins were used (Fig. 6B), and the Gal–Ets-1 fusions were expressed comparably (Fig. 6D and E). Thus, it appears that Ets-1 residues between 153 and 210 are necessary for full activity in KG1a cells and efficient binding to CBP 1–714 in vitro.
FIG. 6.

Gal–Ets-1 activity correlates with binding to CBP 1–714. (A) Ets-1 residues between 153 and 210 are necessary for efficient binding to CBP 1–714 in vitro. GST pull-down assays were performed with the indicated GST–Ets-1 fusion proteins and [35S]CBP 1–714. The input lane has 5% of the material used in each assay. Molecular mass markers (in kilodaltons) are indicated. The arrow points to CBP 1–714 detected by fluorography. The results are representative of four experiments. (B) Equivalent amounts of GST–Ets-1 proteins were used in the pull-down assays (as in panel A). Proteins were detected by Coomassie brilliant blue staining. Residual BSA from the wash buffer is indicated. Note the distortion of the CBP 1–714 band by the GST-Ets Δ178–194 protein. The results are representative of four experiments. (C) Ets-1 residues between 153 and 210 are necessary for transactivation and efficient binding to CBP 1–714. The relative activities of Gal–Ets-1 fusion proteins in transiently transfected KG1a cells were measured. The means and standard errors derived from three experiments are shown. Gal-Ets 2–440 is set at 100%. Quantitation of CBP 1–714 binding to cognate GST–Ets-1 fusion proteins as in panel A is shown as means and standard errors derived from four pull-down experiments. GST-Ets 2–440 is set at 100%. n.d. indicates not determined due to degradation of the GST-Ets fusion proteins purified from E. coli. Ets-1 residues present or deleted in the fusion proteins are indicated at the left of the scale drawing, showing the position of the mutations relative to the Pointed and Ets domains. The Ets-1 region necessary for binding CBP 1–714 is indicated. (D and E) Gal–Ets-1 fusion proteins are expressed comparably in transiently transfected Cos-7 cells. Nuclear extracts were prepared, and the indicated Gal–Ets-1 proteins were detected by immunoblotting with anti-Gal4 DBD antibody. The Control lane contains an extract without a Gal–Ets-1 fusion protein. Molecular size markers are indicated in kilodaltons. This figure was produced with Macintosh versions of Adobe Photoshop and Microsoft PowerPoint. The results represent at least two independent experiments.
DISCUSSION
Our findings indicate that Ets-1 physically interacts with two distinct but similar regions of CBP, CH1 (aa 357 to 452) and CH2/CH3 (aa 1459 to 1892). Mapping the sequences of Ets-1 that bind the CH1 region revealed that the amino-terminal half of Ets-1 (aa 1 to 210), including the Pointed domain (aa 33 to 130) and a portion of the activation domain (aa 131 to 270) (75), is sufficient for strong interaction and that residues between 153 and 210 are necessary for optimal Ets-1–CH1 binding. The Pointed domain is not sufficient, and it is unclear if it is necessary, for binding. Moreover, the binding efficiency of Ets-1 to the CH1 region qualitatively correlated with its ability to transactivate Gal4 promoter constructs (Fig. 6), demonstrating that the Ets-1–CBP/p300 interactions are functionally significant. Testing of Ets-1 internal deletions and truncations in this region suggests that there is not a singular compact CBP N-terminus-binding motif but that multiple residues may contribute to binding and transactivation. Whether Ets-1 transactivation functions also correlate with binding to the CBP carboxyl-terminal CH2/CH3 region remains to be shown, but the presence of the TAZ motif in both the CH1 and CH3 regions suggests that Ets-1 interacts similarly with both of these regions (60). We do not know if Ets-1 affects the binding of the coactivator accessory factors RNA helicase A, TFIIB, or PCAF to the CH3 region. It is also possible that the Ets-1–CH2/CH3 interaction will show a stronger quantitative correlation between binding and transactivation than that seen with Ets-1–CH1, although in the latter instance the lack of a strict quantitative linear relationship may be due to comparisons of protein–protein interactions in vivo and in vitro. It is also conceivable that binding of CBP/p300 by Ets-1 is not sufficient for transactivation, which would be consistent with an exacting quantitative comparison of the transactivation and Ets-1–CBP 1–714 binding assays.
Functional synergism between Ets-1 and c-Myb was first noted with the tripartite Gag-Myb-Ets fusion protein isolated from the E26 avian leukemia virus, where expression of the fusion protein or coexpression of individual c-Myb and Ets-1 proteins results in higher rates of transformation than those seen upon expression of either protein alone (26, 48). In addition, c-Myb and Ets-1 can act synergistically to transactivate the myeloid cell-specific CD13/APN promoter (66), and Gag-Myb-Ets activates many promoters, including mim-1 (21). These examples of functional synergy suggest that Ets-1 and c-Myb may also interact physically. However, aside from the Gag-Myb-Ets protein, such interactions have been difficult to demonstrate (21, 28, 47–49), indicating an additional level of complexity and perhaps involving a contribution from accessory or bridging molecules. CBP/p300 fits the requirements for such a bridging molecule. Indeed, preliminary experiments suggest that Ets-1 mutants deficient for binding the CBP CH1 region are also impaired in their ability to synergize with c-Myb on the CD13/APN promoter in C33A cells (data not shown). c-Myb binds to CBP residues 590 to 699 within the KIX domain (20, 55), a region physically distinct from sites that bind Ets-1 (Fig. 5), raising the possibility of a ternary complex of Ets-1–CBP/p300–c-Myb. Transcriptional synergism could therefore result from increases in CBP/p300 binding affinity and concomitant contact with the basal transcriptional machinery or from alteration of CBP/p300-associated enzymatic activities. However, even in the absence of ternary-complex formation, CBP/p300 could mediate c-Myb/Ets-1 cooperation through other mechanisms, such as kinetic synergism, whereby Ets-1 and c-Myb would sequentially act upon CBP/p300 (31). Studies investigating these possibilities are under way.
The concurrent binding of c-Myb and Ets-1 by CBP/p300 may also explain other attributes of the Gag-Myb-Ets fusion protein. Gag-Myb-Ets represses transactivation by the retinoic acid receptor (RAR) and thyroid hormone receptor (TR) (62), which also bind and require CBP/p300 to function (19, 36). Gag-Myb-Ets would be predicted to bind CBP/p300, since the CBP/p300-binding regions of c-Myb and Ets-1 are both present in the oncogene (42, 54), which might sequester CBP/p300 or impede its binding to RAR and TR. In addition, expression of Gag-Myb-Ets overcomes the inhibition of AP-1 activity by RAR and TR (62), and since the AP-1 components Jun, Fos, and JunB also interact with CBP/p300 (10, 41, 57), Gag-Myb-Ets may block RAR and TR binding to CBP/p300, thus allowing AP-1 to act productively. Indeed, competition for limiting CBP has been invoked to explain cross-talk among various signaling pathways as well as the multiple developmental anomalies associated with Rubinstein-Taybi syndrome (33, 36, 51, 59).
Direct demonstration of the functional necessity of CBP/p300 for transactivation is not always straightforward due to a lack of CBP and p300 mutant cell lines. Another option is to test if CBP/p300 can augment transcription factor activity. Indeed, we observed a synergistic effect of CBP and p300 on Ets-1 activity in F9 cells (Fig. 1), the modest nature of which may be due to the low activity of Ets-1 or to the reporter/cell system used. However, this is not uncommon, since several other studies have noticed similarly weak (2.5-fold or less) effects of CBP/p300 on CREB (7, 52), MyoD (23), E47 (23), c-Jun (68), p65 (27), and Sap-1a (35). In other systems, effects larger than fourfold can be observed (9, 29), sometimes after cotransfection of large amounts of CBP or p300 expression vector (39, 44). Alternatively, the ability of the 12S E1A protein to inhibit CBP-dependent transcriptional activity depends on its ability to bind CBP/p300, providing a reliable indicator of CBP/p300-dependent transactivation (5, 44). Consistent with a role for functional CBP/p300 in Ets-1 transactivation function, 12S E1A specifically inhibited both CD13/APN promoter and Gal-Ets 2–440 activity (Fig. 1A to C), while the 12S Δ2–36 E1A mutant was not inhibitory to reporter levels. However, in accord with other systems (7), overexpression of CBP or p300 in KG1a cells does not significantly augment Gal-Ets 2–440 activity or CD13/APN promoter activity (data not shown), possibly because Ets-1 activity in KG1a cells is not limited by CBP/p300 or perhaps requires additional factors. Interestingly, Gal-Ets 2–440, which showed CBP/p300-dependent transcriptional activation in KG1a myeloid cells, was virtually inactive in Jurkat T cells (data not shown), although we could clearly show an Ets-1–CBP/p300 complex in both of these cell types. Thus, Ets-1–CBP/p300 binding appears not to be sufficient for Ets-1 activation. This observation may also explain why Ets-1 and c-Myb transcriptional synergy does not occur on every promoter containing c-Myb and Ets sites (66). Perhaps additional requirements for c-Myb and Ets-1 cooperation are needed, including binding-site geometry and/or other promoter- or cell-type-specific considerations. In this regard, phosphorylation of Ets-1 Thr38 does not appear to be necessary for binding to the CBP CH1 region, even though there are reports that Thr38 is critical for Ets-1 transactivation function (75, 78). One possibility is that Thr38 phosphorylation is required in addition to CBP/p300 binding for transactivation to occur. Indeed, this could explain why we could coimmunoprecipitate Ets-1–CBP/p300 from Jurkat cells where Gal-Ets 2–440 was inactive. Alternatively, Thr38 phosphorylation may not be required for all Ets-1 transactivation functions; in this light, one of us (L.H.S.) has observed that mutation of Thr38 to Ala has no effect on Ets-1 transactivation or cooperation with c-Myb on the CD13/APN promoter (data not shown). The Gal–Ets-1 system will facilitate our analysis of this intriguing component of transcription factor interaction and tissue specific-gene regulation.
We also showed a physical interaction between Ets-1 and CBP/p300 in nuclear extracts and with semipurified proteins, supporting the notion that direct contact between Ets-1 and CBP/p300 is required for transcriptional activity. Previously, CREB and CBP interactions were demonstrated by a mammalian cell two-hybrid protein interaction assay (17). We were unable to demonstrate an Ets-1–CBP interaction by this method for several cell types, including Jurkat and KG1a cells (data not shown), although it is not uncommon for results from two-hybrid assays to disagree with protein binding data derived by other methods (4).
The results from the coimmunoprecipitation assays argue strongly that p300 and probably CBP are complexed with Ets-1 in the nucleus. Because our CBP/p300 antiserum cocktail recognized both CBP and p300, we cannot be certain that CBP is complexed with Ets-1 in these assays. However, the likelihood of such complexes appears high, based on our in vitro CBP–Ets-1 binding data (Fig. 4 and 5). It should also be noted that two protein species were usually present in the CBP/p300 control immunoprecipitations from Jurkat nuclear extracts (Fig. 2C, lane 15), consistent with the multiple CBP and/or p300 species observed by other workers (8, 37). The difference between these species is unclear because the cocktails we used included both N- and C-terminus-specific CBP and p300 antisera, precluding the identification of specific epitopes present in each band. The more slowly migrating species is generally more abundant in the Ets-1 coimmunoprecipitations, perhaps indicating that this protein is preferentially associated with Ets-1 in these extracts. This observation will form the basis for further studies.
In a broader context, our findings suggest that CBP/p300 interactions may play a role in the unique transforming properties of the Gag-Myb-Ets oncoprotein. Furthermore, since CBP and p300 integrate multiple intracellular signals for transcription (19, 33, 34, 36, 51) and since Ets-1 functions primarily in combination with other transcription factors (14, 15, 24, 43, 63, 66, 67, 74, 78), it follows that CBP/p300 is an important mediator of synergistic and antagonistic interactions between Ets-1 and its numerous transcription factor partners. Not surprisingly, some of these Ets-1-cooperating factors also bind CBP/p300; one example is AP-1, which synergizes with Ets-1 and Ras on specific promoters (74, 78). In a similar context, CBP/p300 has recently been implicated in the mediation of Ras-dependent AP-1/Ets-2 synergy (33). It is also intriguing to speculate that CBP/p300 may be the target of the Ets-1 repressor MafB (67).
ACKNOWLEDGMENTS
We thank Xiaoying Wang, Mike Long, and Geli Gao for technical assistance; Marc Montminy for the gift of GST-KIX (S/B) plasmid and CBP antisera; Richard Goodman and Roland Kwok for pRC/RSV mCBP HA-RK; Bob Rooney for the E1A expression vectors; and Barbara Graves for the Ets-1 mutant constructs. We also thank David Shapiro, Barbara Graves, and John Cleveland for helpful comments on the manuscript.
This work was supported by NIH grants CA70909 (to L.H.S.) and RO1 CA76385 (to P.K.B.), National Cancer Institute Cancer Center Support (CORE) grant P30 CA21765, and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
REFERENCES
- 1.Abraham S E, Lobo S, Yaciuk P, Wang H G, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 2.Albagli O, Flourens A, Crepieux P, Begue A, Stehelin D, Leprince D. Phylogeny of the p68c-ets-1 amino-terminal transactivating domain reveals some highly conserved structural features. Oncogene. 1992;7:1435–1439. [PubMed] [Google Scholar]
- 3.Albagli O, Soudant N, Ferreira E, Dhordain P, Dewitte F, Begue A, Flourens A, Stehelin D, Leprince D. A model for gene evolution of the ets-1/ets-2 transcription factors based on structural and functional homologies. Oncogene. 1994;9:3259–3271. [PubMed] [Google Scholar]
- 4.Allen J B, Walberg M W, Edwards M C, Elledge S J. Finding prospective partners in the library: the two-hybrid system and phage display find a match. Trends Biochem Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 5.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 6.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 7.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 8.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A S. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional coactivator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 9.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 10.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 12.Bories J C, Willerford D M, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt F W, Borles J C. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 13.Borrow J, Stanton V P, Jr, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Volinia S, Watmore A E, Housman D E. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 14.Bradford A P, Conrad K E, Tran P H, Ostrowski M C, Gutierrez-Hartmann A. GHF-1/Pit-1 functions as a cell-specific integrator of Ras signaling by targeting the Ras pathway to a composite Ets-1/GHF-1 response element. J Biol Chem. 1996;271:24639–24648. doi: 10.1074/jbc.271.40.24639. [DOI] [PubMed] [Google Scholar]
- 15.Bradford A P, Conrad K E, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Functional interaction of c-Ets-1 and GHF-1/Pit-1 mediates Ras activation of pituitary-specific gene expression: mapping of the essential c-Ets-1 domain. Mol Cell Biol. 1995;15:2849–2857. doi: 10.1128/mcb.15.5.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Brindle, P. Unpublished data.
- 16.Brindle P, Linke S, Montminy M. Protein-kinase-A-dependent activator in transcription factor CREB reveals new role for CREM repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 17.Brindle P, Nakajima T, Montminy M. Multiple PK-A regulated events are required for transcriptional induction by cAMP. Proc Natl Acad Sci USA. 1995;92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- 19.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 20.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 21.Dudek H, Tantravahi R V, Rao V N, Reddy E S, Reddy E P. Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc Natl Acad Sci USA. 1992;89:1291–1295. doi: 10.1073/pnas.89.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 23.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 24.Espinas M L, Roux J, Ghysdael J, Pictet R, Grange T. Participation of Ets transcription factors in the glucocorticoid response of the rat tyrosine aminotransferase gene. Mol Cell Biol. 1994;14:4116–4125. doi: 10.1128/mcb.14.6.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frampton J, Kouzarides T, Doderlein G, Graf T, Weston K. Influence of the v-Myb transactivation domain on the oncoprotein’s transformation specificity. EMBO J. 1993;12:1333–1341. doi: 10.1002/j.1460-2075.1993.tb05778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graf T. Myb: a transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr Opin Genet Dev. 1992;2:249–255. doi: 10.1016/s0959-437x(05)80281-3. [DOI] [PubMed] [Google Scholar]
- 29.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 30.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 31.Herschlag D, Johnson F B. Synergism in transcriptional activation: a kinetic view. Genes Dev. 1993;7:173–179. doi: 10.1101/gad.7.2.173. [DOI] [PubMed] [Google Scholar]
- 32.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 33.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 35.Janknecht R, Nordheim A. Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene. 1996;12:1961–1969. [PubMed] [Google Scholar]
- 36.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 37.Kee B L, Arias J, Montminy M R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 38.Klambt C. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development. 1993;117:163–176. doi: 10.1242/dev.117.1.163. [DOI] [PubMed] [Google Scholar]
- 39.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 40.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J S, See R H, Deng T, Shi Y. Adenovirus E1A downregulates c-Jun- and JunB-mediated transcription by targeting their coactivator p300. Mol Cell Biol. 1996;16:4312–4326. doi: 10.1128/mcb.16.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leprince D, Gegonne A, Coll J, de Taisne C, Schneeberger A, Lagrou C, Stehelin D. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983;306:395–397. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- 43.Logan S K, Garabedian M J, Campbell C E, Werb Z. Synergistic transcriptional activation of the tissue inhibitor of metalloproteinase-1 promoter via functional interaction of AP-1 and Ets-1 transcription factors. J Biol Chem. 1996;271:774–782. doi: 10.1074/jbc.271.2.774. [DOI] [PubMed] [Google Scholar]
- 44.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 45.Macleod K, Leprince D, Stehelin D. The ets gene family. Trends Biochem Sci. 1992;17:251–256. doi: 10.1016/0968-0004(92)90404-w. [DOI] [PubMed] [Google Scholar]
- 46.Martin K J, Lillie J W, Green M R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990;346:147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- 47.Melotti P, Calabretta B. Ets-2 and c-Myb act independently in regulating expression of the hematopoietic stem cell antigen CD34. J Biol Chem. 1994;269:25303–25309. [PubMed] [Google Scholar]
- 48.Metz T, Graf T. v-myb and v-ets transform chicken erythroid cells and cooperate both in trans and in cis to induce distinct differentiation phenotypes. Genes Dev. 1991;5:369–380. doi: 10.1101/gad.5.3.369. [DOI] [PubMed] [Google Scholar]
- 49.Metz T, Graf T. Fusion of the nuclear oncoproteins v-Myb and v-Ets is required for the leukemogenicity of E26 virus. Cell. 1991;66:95–105. doi: 10.1016/0092-8674(91)90142-l. [DOI] [PubMed] [Google Scholar]
- 50.Muthusamy N, Barton K, Leiden J M. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 51.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 53.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 54.Nunn M F, Seeburg P H, Moscovici C, Duesberg P H. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306:391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- 55.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 56.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 57.O’Neill E M, Rebay I, Tjian R, Rubin G M. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 58.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C M, Masuno M, Tommerup N, van Ommen G-J B, Goodman R H, Peters D J M, Breuning M H. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 60.Ponting C P, Blake D J, Davies K E, Kendrick-Jones J, Winder S J. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 61.Quinn P G. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 62.Rascle A, Ferrand N, Gandrillon O, Samarut J. Myb-Ets fusion oncoprotein inhibits thyroid hormone receptor/c-ErbA and retinoic acid receptor functions: a novel mechanism of action for leukemogenic transformation by E26 avian retrovirus. Mol Cell Biol. 1996;16:6338–6351. doi: 10.1128/mcb.16.11.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddy M A, Yang B S, Yue X, Barnett C J, Ross I L, Sweet M J, Hume D A, Ostrowski M C. Opposing actions of c-ets/PU.1 and c-myb protooncogene products in regulating the macrophage-specific promoters of the human and mouse colony-stimulating factor-1 receptor (c-fms) genes. J Exp Med. 1994;180:2309–2319. doi: 10.1084/jem.180.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 65.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shapiro L H. Myb and Ets proteins cooperate to transactivate an early myeloid gene. J Biol Chem. 1995;270:8763–8771. doi: 10.1074/jbc.270.15.8763. [DOI] [PubMed] [Google Scholar]
- 67.Sieweke M H, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 68.Smits P H, de Wit L, van der Eb A J, Zantema A. The adenovirus E1A-associated 300 kDa adaptor protein counteracts the inhibition of the collagenase promoter by E1A and represses transformation. Oncogene. 1996;12:1529–1535. [PubMed] [Google Scholar]
- 69.Stein R W, Corrigan M, Yaciuk P, Whelean J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun P, Maurer R A. An inactivating point mutation demonstrates that interaction of cAMP response element binding protein (CREB) with the CREB binding protein is not sufficient for transcriptional activation. J Biol Chem. 1995;270:7041–7044. doi: 10.1074/jbc.270.13.7041. [DOI] [PubMed] [Google Scholar]
- 71.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 72.Treier M, Bohmann D, Mlodzik M. JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell. 1995;83:753–760. doi: 10.1016/0092-8674(95)90188-4. [DOI] [PubMed] [Google Scholar]
- 73.Wasylyk B, Hahn S L, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 74.Wasylyk B, Wasylyk C, Flores P, Begue A, Leprince D, Stehelin D. The c-ets proto-oncogenes encode transcription factors that cooperate with c-Fos and c-Jun for transcriptional activation. Nature. 1990;346:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 75.Wasylyk C, Bradford A P, Gutierrez-Hartmann A, Wasylyk B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- 76.Watson D K, McWilliams M J, Lapis P, Lautenberger J A, Schweinfest C W, Papas T S. Mammalian ets-1 and ets-2 genes encode highly conserved proteins. Proc Natl Acad Sci USA. 1988;85:7862–7866. doi: 10.1073/pnas.85.21.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xing L, Gopal V K, Quinn P G. cAMP response element-binding protein (CREB) interacts with transcription factors IIB and IID. J Biol Chem. 1995;270:17488–17493. doi: 10.1074/jbc.270.29.17488. [DOI] [PubMed] [Google Scholar]
- 78.Yang B S, Hauser C A, Henkel G, Colman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]