Abstract
A gene cluster responsible for the biosynthesis of validamycin, an aminocyclitol antibiotic widely used as a control agent for sheath blight disease of rice plants, was identified from Streptomyces hygroscopicus subsp. jinggangensis 5008 using heterologous probe acbC, a gene involved in the cyclization of d-sedoheptulose 7-phosphate to 2-epi-5-epi-valiolone of the acarbose biosynthetic gene cluster originated from Actinoplanes sp. strain SE50/110. Deletion of a 30-kb DNA fragment from this cluster in the chromosome resulted in loss of validamycin production, confirming a direct involvement of the gene cluster in the biosynthesis of this important plant protectant. A sequenced 6-kb fragment contained valA (an acbC homologue encoding a putative cyclase) as well as two additional complete open reading frames (valB and valC, encoding a putative adenyltransferase and a kinase, respectively), which are organized as an operon. The function of ValA was genetically demonstrated to be essential for validamycin production and biochemically shown to be responsible specifically for the cyclization of d-sedoheptulose 7-phosphate to 2-epi-5-epi-valiolone in vitro using the ValA protein heterologously overexpressed in E. coli. The information obtained should pave the way for further detailed analysis of the complete biosynthetic pathway, which would lead to a complete understanding of validamycin biosynthesis.
Filamentous gram-positive Streptomyces strains produce myriads of secondary metabolites with diversified structures and biological activities, including antibacterial, antifungal, antitumor, immunosuppressant, pesticidal, and herbicidal effects (5). The recently published genome sequences of Streptomyces coelicolor A3(2) (2) and Streptomyces avermitilis (20) further verified their position as a most abundant source of biosynthetic gene clusters encoding various bioactive compounds.
Streptomyces hygroscopicus subsp. jinggangensis 5008 (S. hygroscopicus 5008 or strain 5008 hereafter), isolated from the Jinggang Mountain area of China in 1974 (27), produces at least two antibiotics of agricultural importance. Jingangmycin, a weakly basic water-soluble aminocyclitol antibiotic, which was later proven to be identical to validamycin A (Fig. 1) produced by S. hygroscopicus var. limoneus IFO 12703 (13), has been widely used as a prime control reagent against sheath blight disease of rice plants and dumping-off of cucumber seedlings in China and many other eastern Asian countries. Upon treatment with validamycin, normal extension of the main hyphae is switched to an abnormal branching at the tips and further development of the growing fungi is severely repressed (23). Furthermore, voglibose, a valiolamine derivative produced from validamycin by bioconversion and chemical modifications (10), is widely used for the treatment of diabetes. The other antibiotic, jingsimycin, is an acidic polypeptide similar to saramycetin and has activity against various fungi.
FIG. 1.
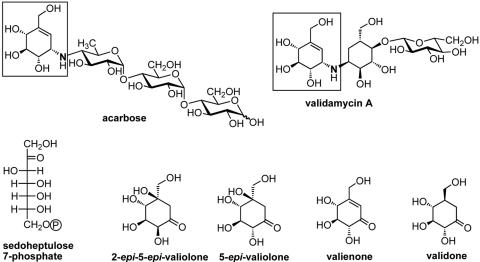
Chemical structures of acarbose and validamycin A (top), and the initial intermediates proposed to be involved in validamycin A biosynthesis (bottom) (7). Boxed regions show the valienamine moiety shared by both compounds.
A structural comparison between validamycin A and acarbose (Fig. 1), a compound used for the treatment of type II insulin-independent diabetes, revealed that they contain an identical C7N aminocyclitol moiety (16), valienamine, whose structure and stereochemistry resemble that of d-glucose and which is responsible for the strong inhibitory activity of acarbose against α-glucosidases. Based on feeding experiments with isotopically labeled precursors, 2-epi-5-epi-valiolone, 5-epi-valiolone, valienone, and validone were suggested to be the intermediates of the validamycin A biosynthetic pathway by Floss and coworkers (7) (Fig. 1).
The incorporation patterns of various putative intermediates seemed to be different between the validamycin-producing S. hygroscopicus var. limoneus and the acarbose-producing Actinoplanes sp. strain SE50/110, but 2-epi-5-epi-valionone was found to be efficiently incorporated into both compounds, suggesting that both biosynthetic pathways share the same initial cyclization reaction catalyzed by common enzymes with similar activities. AcbC, an enzyme closely related to 3-dehydroquinate synthetases (AroB proteins), was proven to be involved in the cyclization of d-sedoheptulose 7-phosphate to 2-epi-5-epi-valionone in acarbose biosynthesis (25). The encoding gene (acbC) was thus used as a heterologous probe for the cloning of the validamycin biosynthetic genes from S. hygroscopicus 5008, which is reported in this paper. The direct involvement of valA, an acbC homologue, in validamycin biosynthesis was confirmed in vivo by gene inactivation as well as in vitro by biochemical characterization of the reaction catalyzed by the ValA protein heterologously overexpressed in E. coli.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and cosmids.
Bacterial strains, phage, plasmids, and cosmids are described in Table 1.
TABLE 1.
Strains, phage, plasmids, and cosmids used in this study
| Strain, phage, or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| S. hygroscopicus strains | ||
| 5008 | Wild-type producer of validamycin | 27 |
| YU-1 | Nonproducer of validamycin generated by gene replacement of a ca. 30-kb region including acbC homolog with aac(3)IV | This work (Fig. 3) |
| JXH-1 | As above, but replacing a 563-bp DNA fragment internal to valA | This work (Fig. 5) |
| S. lividans 66 ZX1 | rec-46 str-6 pro-2 | 30 |
| E. coli strains | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 D(ara, leu)1697 galU galK λ−rpsL nupG | GIBCO-BRL |
| ET12567(pUZ8002) | dam dcm hsdS/pUZ8002 | 21 |
| LE392 | supE44 supF58 hsdR514, used for infection with in vitro-packaged cosmids | 3 |
| BL21Gold(DE3)/pLysS | F−ompT hsdSB (rB− mB−) gal dcm(DE3) pLysS (Cmr) | 18 |
| XL-1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac | Stratagene |
| Pellicularia sasakii | Fungal indicator strain sensitive to validamycin A | SJUSC |
| Plasmids | ||
| pAS8/7 | pIJ6021 with a 1,264-bp NdeI-EcoRI fragment containing acbC in the corresponding sites | 25 |
| pBlueScript II SK(+) | bla lacZ orif1 | Stratagene |
| pHZ1358 | Cosmid used for the construction of 5008 genomic library; pIJ101 derivative; tsr Ltz−sti+oriT | 26 |
| pHZ2229 | pBlueScript II SK(+) carrying a ca. 6-kb sequenced BamHI fragment (Fig. 2) from cosmid 3G8 | This study |
| pHZ2234 | Construct after religation of BamHI-digested 3G8 | This study |
| pHZ2236 | Insertion in BamHI site of pHZ2234 of an aac(3)IV gene sandwiched between the two 2.1-kb genomic DNA fragments | This study |
| pIJ2925 | bla lacZ | 12 |
| pJTU504 | Insertion of a 1.33-kb XbaI-EcoRI fragment from pJTU690 in pBlueScript II SK(+) | This study |
| pJTU514 | pJTU504 derivative with the insertion of aac(3)IV in EcoRI site | This study |
| pJTU515 | Insertion of 1.5-kb PvuII-HindIII fragment from pJTU696 into EcoRV-HindIII sites of pJTU514 | This study |
| pJTU516 | Insertion of a 4.23-kb BglII fragment from pJTU515, | This study |
| which contains a linked 1.33-kb (left arm)-1.4-kb aac(3)IV-2.1-kb (right arm) for valA inactivation, in BamHI site of pHZ1358 | ||
| pJTU690 | Part of the 6-kb sequenced BamHI fragment (Fig. 2) including the 1.33-kb XbaI-EcoRI fragment inserted into pBlueScript II SK(+) | This study |
| pJTU696 | Part of the 6-kb sequenced BamHI fragment (Fig. 2) including the 1.5-kb PvuII-HindIII fragment inserted into pBlueScript II SK(+) | This study |
| pKMW5 | pUC18 derivative carrying the strD and strE genes of the streptomycin pathway | 24 |
| pRSET-B | bla reppUC, T7 promoter | Invitrogen |
| pUC18 | bla lacZ | 28 |
| Phage | ||
| φC31 | Wild type; c+attP+int+ | 31 |
Ltz (lethal zygosis), pock formation caused by plasmid transfer; mel, tyrosinase gene for melanin production; oriT, origin of transfer of plasmid RK2; tsr, thiostrepton resistance gene; aac(3)IV, apramycin resistance gene; sti, origin for second-strand synthesis of the multicopy plasmid pIJ101 (6); vph, viomycin resistance gene; xylE, catechol 2,3-dioxygenase from Pseudomonas putida; Cmr, chloramphenicol resistance gene.
Culture techniques, transformation, and conjugation.
S. hygroscopicus 5008 and its derivatives were grown on SFM agar plates or in TSB liquid medium supplemented with 10.3% (wt/vol) sucrose and 1% (wt/vol) yeast extract (15) at 28°C for growth of mycelia, isolation of total DNA, or conjugation. FM-II liquid medium (containing [per liter] soluble starch, 60 g; sucrose, 30 g; beef extract, 35 g; NaCl, 0.75 g; MgSO4 2 g; CaCO3 8 g; initial pH, 7.8.) was used for fermentation. Protoplast preparation and transformation were performed according to Hopwood et al. (9). Escherichia coli strains were cultured according to Sambrook et al. (22). Cosmid clones were selected after infection of E. coli LE392 on LB agar containing 100 μg ampicillin ml−1 or 30 μg apramycin ml−1. For selection of Streptomyces transformants, apramycin and thiostrepton were both used at 30 μg ml−1 in SFM agar medium and at 5 μg ml−1 in liquid media.
Cloning techniques.
Plasmid and total DNA was isolated from Streptomyces strains according to Kieser et al. (15). Restriction enzymes, T4 DNA ligase, Taq polymerase, and alkaline phosphatase were purchased from various companies (New England Biolabs, Takara, and MBI Fermentas). The gel recovery kit (Shanghai Watson and QIAGEN) was used for DNA recovery from agarose gels. For the generation of cosmid libraries, total DNA samples were partially digested with MboI, dephosphorylated with calf intestinal alkaline phosphatase, and size-fractionated by sedimentation analysis using a sucrose gradient (9). DNA fragments between 30 and 40 kb were mixed in a 1:1 molar ratio with BamHI-digested cosmid vector pHZ1358 and ligated at ca. 200 μg ml−1 DNA. Packaging was done with λ packaging mixes prepared according to Sambrook et al. (22).
DNA probes, PCR primers, and Southern hybridization.
An NdeI-EcoRI fragment carrying the acbC gene from Actinoplanes sp. strain SE50/110 producing acarbose was excised from plasmid pAS8/7 (25) and used as a heterologous probe. The two oligonucleotide primers used for PCR amplification were ValA-F (5′-GGATCCACATATGACCATGACCAAG-3′) and ValA-R (5′-GAATTCACACCCCCATGTCC-3′). For Southern hybridization experiments, S. hygroscopicus 5008 genomic DNA was cleaved with restriction enzymes, separated on 0.8% agarose gels, and transferred onto Hybond-N+ nylon membrane (Amersham-Pharmacia). α-[32P]dCTP-labeled radioactive probes using a random priming kit (Roche) were used for both Southern blots and in situ colony hybridization.
Construction of pHZ2236 for targeted deletion of 30-kb region within the ca. 70-kb contig.
Complete digestion of cosmid 3G8 DNA by BamHI and religation resulted in the construction of pHZ2234, in which an internal ca. 30 kb (marked by a solid bar in Fig. 2) of 3G8 was found to be deleted and the two 2.1-kb flanking fragments were connected. Then a 1.4-kb BamHI fragment carrying aac(3)IV (apramycin resistance gene) was inserted between the two 2.1-kb fragments with the same transcriptional direction as valA, which generated pHZ2236 for subsequent conjugation.
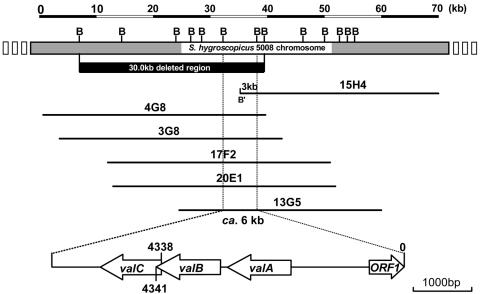
FIG. 2.
Overlapping cosmids covering genes for validamycin biosynthesis and gene organization of the 6-kb sequenced region. Top: BamHI restriction map of the ca. 70-kb contig for validamycin biosynthesis. The solid bar indicates the 30-kb deleted region in Fig. 3, which abolished validamycin production. Bottom: 6-kb sequenced region including the acbC homologue (valA). B: BamHI site. B′: regenerated BamHI site from BamHI (vector) and MboI (insert) sites.
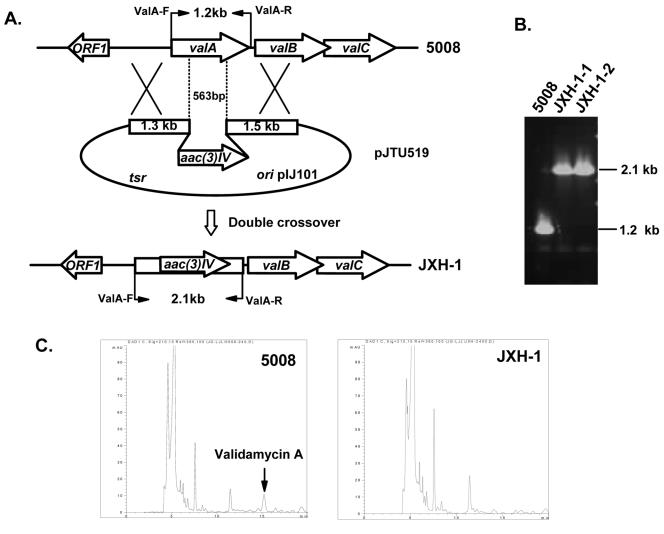
Construction of pJTU519 for targeted deletion of 563 bp of DNA internal to valA.
A 1.33-kb XbaI-EcoRI fragment overlapping one side of the valA gene on the ca. 6-kb sequenced BamHI fragment (Fig. 2) in pHZ2229 was excised from pJTU690 and inserted into the corresponding site of pBluescript II SK(+), resulting in pJTU504. A 1.4-kb EcoRI fragment carrying the apramycin resistance gene aac(3)IV was inserted into the EcoRI site of pJTU504 to generate pJTU514, which was subsequently digested with EcoRV and HindIII to accept a 1.5-kb PvuII-HindIII fragment of pJTU696 overlapping the other end of the valA on the ca. 6-kb sequenced BamHI fragment (Fig. 2) to generate pJTU515. A total of 4.23 kb of DNA linking 1.33 kb (left arm)-1.4 kb aac(3)IV-1.5 kb (right arm), was transferred as an XbaI fragment to the corresponding site of pIJ2925 to construct pJTU516, for the final excision as a BglII fragment to be inserted into the unique BamHI site of pHZ1358, resulting in pJTU519 as a final construct for the targeted replacement of a 563-bp DNA fragment internal to valA by the 1.4-kb aac(3)IV, in the wild-type strain 5008.
Antibiotic assay.
Production of validamycin by S. hygroscopicus 5008 and its derivatives was detected using a bioassay and high performance liquid chromatography (HPLC). For the bioassay, 1 milliliter of fermentation supernatant extracted with chloroform was mixed with 14 ml of melted agar (8 g of agar, 1 liter of water). An agar plug with Pellicularia sasakii (Shanghai Jiaotong University Stock Collection, Shanghai Jiaotong University, Shanghai), which is sensitive to validamycin, was transferred to the center of the agar plate and after 24 h incubation at 30°C the diameter of the colony was measured, which is inversely related to the inhibitory potency. For HPLC analysis, the strains were cultured in 40 ml of FM-II fermentation medium in 250-ml baffled flasks at 37°C and 220 rpm for 6 days. The fermentation broth was centrifuged at 12,000 rpm for 5 min followed by chloroform extraction. The extracted supernatant was directly loaded onto a Nucleosil C18 column (250 mm by 4.6 mm, Sigma-Aldrich) for HPLC analysis (Waters 220). The mobile phase (0.005 M sodium phosphate buffer-acetone, 97:3) was applied with the flow rate of 1 ml min−1 at room temperature. The elute was monitored at 210 nm with a Waters 996 photodiode array detector and the data were analyzed with a Waters Millennium Chromatography Manager.
Sequence analysis.
DNA sequencing was done at Shanghai Sangon Ltd. using pUC18 as the vector. Sequencing reactions were carried out using the Amersham Thermosequenase sequencing kit containing fluorescent dye terminators and an Applied Biosystems model 377 automated DNA sequencer. Sequence analysis was performed with the Lasergene DNA analysis tools (DNASTAR) (Madison, MI). Open reading frames and ribosome binding sites were predicted with FramePlot (11). Nucleotide and amino acid sequence comparisons against public databases were done using the BLAST program (1).
Cloning and heterologous overexpression of recombinant His6-tagged ValA.
The valA gene was amplified by PCR with Platinum Pfx DNA polymerase (Invitrogen) using the cosmid clone 3G8 as the template and primers ValA-F3, 5′-GAAGATCTGCATATGACCAAGCAGAGTTCCTTATCC-3′ (BglII and NdeI), and ValA-R2, 5′-GGAATTCTCACACCCCCATGTCCACGGCACCG-3′ (EcoRI). PCR amplification was done in a Thermocycler (Eppendorf, Mastercycler gradient) under the following conditions: 33 cycles of 90 s at 95°C, 45 s at 60°C, and 45 s at 72°C. The PCR products were digested with BglII and EcoRI, and subsequently ligated into BamHI- and EcoRI-digested pRSET-B. The constructs were transformed into E. coli XL-1-Blue and plated on LB agar plates containing 100 μg ml−1 ampicillin.
The plasmid DNA was isolated and introduced by heat-pulse transformation into E. coli BL21Gold(DE3)/pLysS (Stratagene), which was then plated onto LB agar plates containing 100 μg ml−1 ampicillin and 25 μg ml−1 chloramphenicol. The transformants were grown in 20 ml LB medium containing ampicillin and chloramphenicol at 37°C to an optical density at 600 nm of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM and the incubation was continued at 28°C for 24 h. The cells were harvested by centrifugation at 3,500 rpm for 15 min and stored frozen at −80°C until further use.
Preparation of cell extracts and purification of His6-tagged ValA.
Cells were thawed and resuspended in disruption buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). The suspension was sonicated three times for 25 s each and cell debris was removed by centrifugation at 10,000 rpm for 10 min. The protein solution was applied to an Ni-nitrilotriacetic acid spin column (QIAGEN) and centrifuged at 2,000 rpm for 2 min. The column was washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0, and 50 mM NaH2PO4, 300 mM NaCl, 100 mM imidazole, pH 8.0). The His6-tagged protein was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 8.0) and dialyzed for 24 h against 1 liter of dialysis buffer (20 mM potassium phosphate [pH 7.4], 0.05 mM CoCl2, 2 mM KF and 0.5 mM dithiothreitol). Protein concentration was measured by the Bradford protein microassay with bovine serum albumin as the standard.
Enzyme assay.
The enzyme assay was carried out at 30°C for 3 to 12 h in a 100-μl volume of 20 mM potassium phosphate (pH 7.4), 0.05 mM CoCl2, 2 mM KF, 1 mM NAD+, 5 mM sedoheptulose 7-phosphate, and 50 μl of protein solution (2.4 mg/ml protein). The reaction progress was monitored by thin-layer chromatography analysis. The reaction mixture was lyophilized and the reaction products were extracted with methanol. This extract was then dried and a few drops of SIGMA-SIL-A (SIGMA) were added. The solvent was removed in a flow of argon gas and the products were reextracted with n-hexane and injected for gas chromatography-mass spectroscopy (GC-MS) (Hewlett Packard 5890 series II gas chromatograph).
Nucleotide sequence accession number.
The DNA and deduced protein sequences reported in this paper have been deposited in GenBank under accession number AY753181.
RESULTS
S. hygroscopicus 5008 contains a homologue of the acbC gene required for acarbose biosynthesis in Actinoplanes.
Probing a Southern transfer of BamHI-digested genomic DNA of 5008 with the labeled acbC gene (a 1,264-bp NdeI-EcoRI fragment carrying a gene involved in the cyclization of d-sedoheptulose 7-phosphate to 2-epi-5-epi-valiolone of the acarbose biosynthetic pathway) of Actinoplanes sp. strain SE50/110 gave a weak signal at ca. 6 kb at low stringency in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 65°C (data not shown). To isolate the hybridizing fragment, further hybridization with the same probe in a pHZ1358 library was carried out with the same stringency. Six of the 39 initial isolates were confirmed positive by successive hybridization experiments. Five (4G8, 3G8, 17F2, 20E1, and 13G5, Fig. 2) out of the six positive overlapping cosmids shared a common 6-kb fragment hybridizing to the above-mentioned probe, while the sixth cosmid (15H4) gave a hybridization signal to a fragment of only about 3 kb (Fig. 2).
The BamHI digestion patterns of the six cosmids resulted in a contig spanning ca. 70 kb, without apparent genomic rearrangement. The different hybridization signal originating from BamHI-digested 15H4 was found to have resulted from the regeneration of a BamHI (designated B′ in Fig. 2) site between the cloning site (BamHI) of the vector and an accidental MboI site of the insert during library construction, as this fragment was found to lie at the very end of the cloned fragment in cosmid 15H4.
Cosmid 3G8 contains genes involved in validamycin biosynthesis.
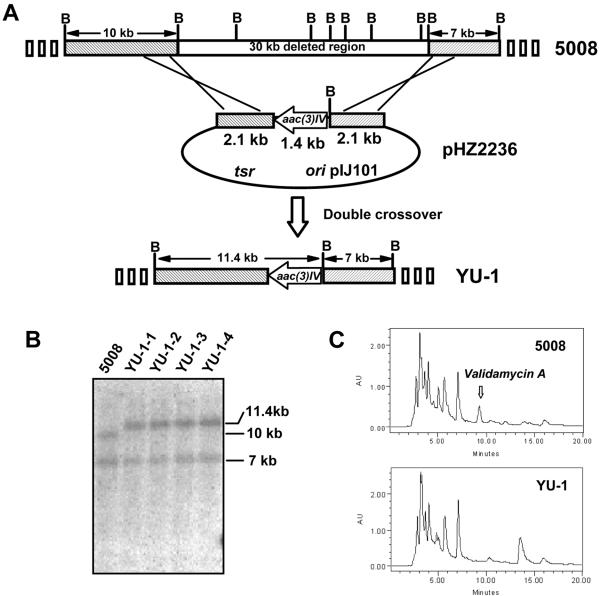
One of the six pHZ1358-derived (26) bifunctional cosmids, 3G8 (Fig. 2), carrying a tsr gene suitable for selection in Streptomyces and a 34.2-kb insert under the control of pIJ101 origin of replication (6), was proven to be extremely unstable (ca. 98% loss after one round of nonselective growth on SFM medium) in strain 5008. For targeted gene replacement, ca. 30 kb of the strain 5008 DNA insert containing the acbC homologue in cosmid 3G8 was replaced by a 1.4-kb apramycin resistance (Aprr) determinant, aac(3)IV, which resulted in pHZ2236 for subsequent conjugation.
pHZ2236 was transferred by conjugation from E. coli ET12567(pUZ8002) into strain 5008. About 10−8 exconjugants per donor were obtained which were initially selected to be Aprr. Surprisingly, all four randomly selected exconjugants were found to be sensitive to thiostrepton. A Southern transfer of total DNA (Fig. 3B) from the four Aprr Thios exconjugants (YU-1-1 to YU-1-4) together with wild-type 5008 was probed with the labeled 5.6-kb insert from pHZ2236 (Fig. 3A). As expected, the ca. 30-kb fragment in the 5008 chromosome was found to be replaced by a 1.4-kb fragment in the YU-1 mutants, resulting in a fusion of the 10-kb leftward BamHI fragment with the BglII end of the 1.4-kb aac(3)IV to form a new 11.4-kb BamHI fragment (Fig. 3A), which is distinguishable from the 10-kb BamHI fragment of the wild-type 5008 in Fig. 3B.
FIG. 3.
Replacement of a 30-kb region containing the acbC homologue by aac(3)IV in strain 5008. (A) Schematic representation of the replacement of the 30 kb of DNA mediated by 2.1-kb genomic fragments flanking both sides of the 1.4-kb aac(3)IV apramycin resistance determinant in pHZ2236. The mutants generated by double crossover (YU-1-1 to YU-1-4) had a fusion of the 10-kb leftward BamHI fragment with the BglII end of the 1.4-kb aac(3)IV to form an 11.4-kb new BamHI fragment, but keep the rightward 7-kb fragment unchanged. (B) Southern hybridization using the α-[32P]dCTP-labeled 5.6 (2.1 + 1.4 + 2.1)-kb insert from pHZ2236 as the probe after genomic DNAs of the wild-type 5008 and its derivatives (YU-1-1 to YU-1-4) were digested with BamHI. (C) HPLC analysis demonstrating that validamycin A is produced by wild-type strain 5008, but not by YU-1.
All four mutants were tested for their ability to inhibit the growth of Pellicularia sasakii, a plant pathogen causing rice sheath blight disease. While P. sasakii stopped growing on agar plates containing the fermentation supernatant of strain 5008 (as a positive control), the fungus continued to grow on plates containing the fermentation supernatants of each of the four candidate strains (YU-1-1 to YU-1-4). HPLC analysis revealed that the mutant candidates did not produce validamycin like the wild-type strain. Unlike with the wild-type strain 5008, no peak corresponding to validamycin A (retention time of 9.5 min) was found in samples obtained from the individual mutant candidates (Fig. 3C). This is consistent with the presumption that genes essential for validamycin biosynthesis including the acbC homologue have been deleted.
Sequencing analysis of a DNA fragment including the acbC homologue.
The common 6-kb BamHI fragment shared by the five cosmids (4G8, 3G8, 17F2, 20E1, and 13G5, Fig. 2) including the acbC homologue was cloned into pBluescript II SK(+) and sequenced. The overall G+C content of the sequenced region was 67.9%, lower than that of S. coelicolor A3(2), which showed an overall G+C content of 72.1%. FramePlot (11) analysis revealed three open reading frames (valA, valB, and valC) transcribed in the same direction, with valA separated from valB by 5 bp, while valB and valC overlap by 3 bp, suggesting that they are transcribed into one polycistronic mRNA (Fig. 2).
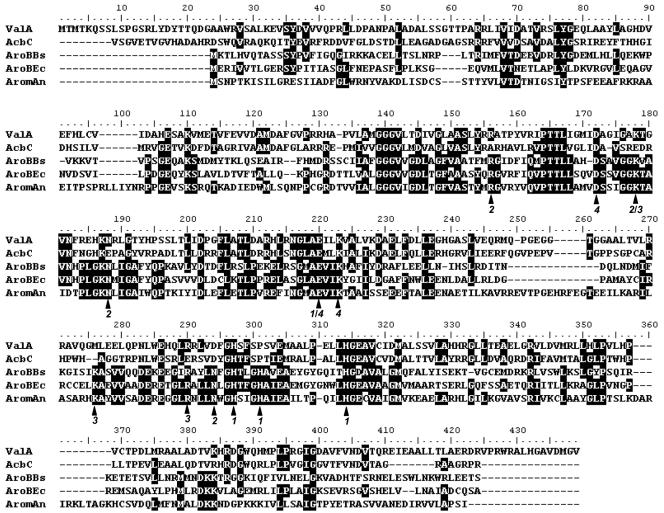
The valA gene seems to encode a polypeptide of 412 amino acids in length, with a putative ribosome binding site (GTGA) 10 bp preceding the putative translational start codon (ATG). The nucleotide sequence of valA has 59.5% identity with the Actinoplanes acbC gene, while the deduced ValA protein has 48% identity with the AcbC protein (25) (Fig. 4). The ValA protein also showed significant similarity (37% identity) to the AroB protein from Emericella nidulans (Fig. 4), which is known to be responsible for the cyclization of 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) to dehydroquinate (4).
FIG. 4.
Alignment of ValA with AcbC and three AroB proteins. Deduced amino acid sequences are from the following organisms: AcbC, Actinoplanes sp. (Y18523.3); AroBBs, Bacillus subtilis (M80245); AroBEc, E. coli (X03867); and AromAn, Emericella (formerly Aspergillus) nidulans (395-amino-acid DHQS domain at the N terminus of the pentafunctional AROM protein; X05204). A functional attribution of the amino acid residues indicated with black arrows, based on the analysis of the three-dimensional structure of the dehydroquinate synthase (DHQS) domain of E. nidulans (4), is given below the alignment by the following code: 1 = Co2+ binding (Zn2+ in the fungal protein instead); 2 = heptulose phosphate group binding; 3 = C-1 hydroxyl fixation; and 4 = C-4 hydroxyl fixation.
The deduced amino acid sequence (373 amino acids) of ValB shows 33% identity with GlgC, a glucose-1-phosphate adenylyltransferase isolated from Bacillus halodurans C-125 (8). ValC (351 amino acids) shows 30% identity to AcbM, the 2-epi-5-epi-valiolone 7-kinase from the acarbose biosynthetic gene cluster, whose demonstrated function is to phosphorylate 2-epi-5-epi-valiolone to form 2-epi-5-epi-valiolone 7-phosphate. Additionally, ValC also shows weaker homology with glucokinase from a Bacillus species (29% identity and 43% similarity).
There is an incomplete open reading frame (tentatively designated ORF1) at the 5′ end of the fragment, which is transcribed in the opposite direction and shows 31% identity to the N-terminal amino acids of dTDP-4-dehydrorhamnose reductase, RmlD, whose known function is to reduce dTDP-6-deoxy-l-xylo-4-hexulose to dTDP- l-rhamnose in Geobacillus stearothermophilus (19).
DNA replacement in valA is abolished validamycin biosynthesis.
Direct evidence for the involvement of valA in the biosynthesis of validamycin came from the replacement of a 563-bp DNA fragment internal to valA with aac(3)IV (Fig. 5A). This was performed by using a pHZ1358-derived plasmid (pJTU519, detailed in Materials and Methods), in which aac(3)IV (apramycin resistance gene) was sandwiched between sequences of 1.33 kb flanking to the left, and 1.5 kb flanking to the right of the 563-bp DNA to be deleted (Fig. 5A). JXH-1, a 5008 derivative, was obtained after introduction of pJTU519 into wild-type strain 5008 by conjugation from E. coli ET12567(pUZ8002), initial selection by thiostrepton, and further screening for the Thios Aprr phenotype. Total DNA from the two independent Thios Aprr exconjugants (JXH-1-1 and JXH-1-2) and from the wild-type strain 5008 was used as template for PCR amplification using two oligonucleotide primers (ValA-F and ValA-R) (Fig. 5B).
FIG. 5.
Inactivation of valA of the validamycin biosynthetic gene cluster. (A) Schematic representation of the replacement of a 563-bp internal fragment of valA with the 1.4-kb aac(3)IV. In shuttle plasmid pJTU519, aac(3)IV was inserted between 1.3-kb and 1.5-kb genomic fragments originally flanking the deleted 563-bp region. While wild-type 5008 should give a 1.2-kb PCR-amplified product, mutant JXH-1 should yield a 2.1-kb product using a pair of primers (ValA-F and ValA-R) designed for amplification of full-length valA. (B) PCR analysis of wild-type 5008 and mutant JXH-1. (C) HPLC comparison between 5008 and JXH-1. The peak corresponding to validamycin A is absent from mutant JXH-1.
The 5008 DNA gave a 1.2-kb PCR product, while both JXH-1-1 and JXH-1-2 gave an expected 2.1-kb PCR product (Fig. 5B), which confirmed that a 563-bp DNA fragment internal to valA (Fig. 5A) has been removed from the chromosome of these mutants. No inhibition of Pellicularia sasakii could be detected in the bioassay, as in the case of YU-1 reported above, and no peak corresponding to validamycin A could be detected in the JXH-1-1 sample by HPLC analysis (Fig. 5C) as opposed to that of 5008, confirming the complete loss of validamycin production in JXH-1-1.
Characterization of ValA activity using heterologously overexpressed protein.
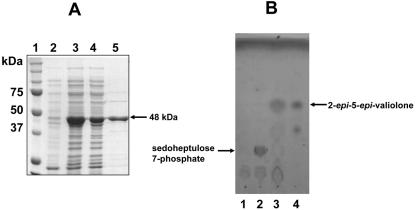
In order to confirm the function of its gene product as a 2-epi-5-epi-valiolone synthase, valA was heterologously expressed in E. coli. A 1.24-kb DNA fragment containing the valA gene was amplified from cosmid 3G8 by PCR, introducing BglII and NdeI restriction sites with the forward primer and an EcoRI site with the reverse primer. The PCR product was subcloned into the expression vector pRSET-B as a BglII/EcoRI fragment, transformed into E. coli XL-1-Blue. The correct plasmids were subsequently transformed into E. coli BL21Gold(DE3)pLysS. Expression of valA under the control of the T7 promoter was induced by isopropyl-β-d-thiogalactopyranoside (IPTG), which gave rise to a 48-kDa soluble polyhistidine-tagged protein. Affinity purification on a Ni-nitrilotriacetic acid spin column (QIAGEN) or a BD TALON column gave a protein that was >80% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 6A).
FIG. 6.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the His6-tagged ValA protein. Lanes: 1, molecular weight markers; 2, soluble protein of cell extract of E. coli BL21Gold(DE3)pLysS/valA before induction with IPTG; 3, total protein of cell extract of E. coli BL21Gold(DE3)pLysS/valA after induction with IPTG; 4, soluble protein of cell extract of E. coli BL21Gold(DE3)pLysS/valA after induction with IPTG; 5, purified His6-tagged ValA. (B) Thin-layer chromatography analyses of ValA assay (solvent system, n-butanol:ethanol:H2O, 9:7:4). Lane 1, ValA without substrate; lane 2, boiled ValA with sedoheptulose 7-phosphate; lane 3, ValA with sedoheptulose 7-phosphate; lane 4, 2-epi-5-epi-valiolone.
To test the catalytic activity of ValA, the enzymatic reaction was carried out using sedoheptulose 7-phosphate as the substrate under reaction conditions used previously for the AcbC protein of acarbose biosynthesis (25). The reaction extracted with methanol gave rise to a product which has the same Rf value as authentic 2-epi-5-epi-valiolone on thin-layer chromatography (Fig. 6B). On the other hand, incubation with cell extracts of E. coli harboring the empty pRSET-B vector gave no product.
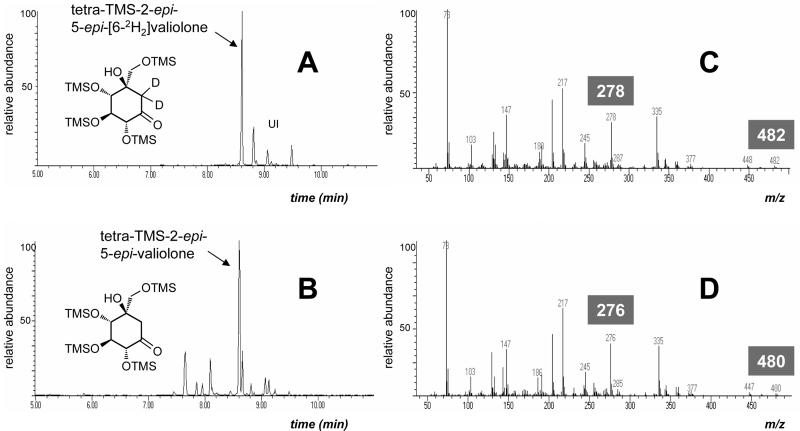
The product was extracted from the lyophilized reaction mixture by methanol, converted to its trimethylsilylated derivative, and analyzed by GC-MS (Fig. 7). An isotopically labeled authentic sample, 2-epi-5-epi-[6-2H2]valiolone, chemically synthesized from d-mannose (25) was used for comparison (Fig. 7A). The enzyme product was detected as a tetratrimethylsilyl derivative of 2-epi-5-epi-valiolone [m/z 480 (M+)] with major fragment ions at m/z 335, 276, 217, 147, and 73 (Fig. 7D). This fragmentation pattern is consistent with that of the authentic sample, which showed m/z 482 (M+), 335, 278, 217, 147, and 73 (Fig. 7C).
FIG. 7.
GC-MS analysis of the trimethylsilylated derivatives of authentic 2-epi-5-epi-[6-2H2]valiolone and the ValA reaction product. A, total ion chromatogram of the authentic tetratrimethylsilyl-2-epi-5-epi-[6-2H2]valiolone (Rt = 8.60 min); B, TIC chromatogram of ValA reaction product, tetratrimethylsilyl-2-epi-5-epi-valiolone (Rt = 8.63 min); C, electron ionization mass spectrometry fragmentation pattern of tetratrimethylsilyl-2-epi-5-epi-[6-2H2]valiolone; D, EI-MS fragmentation pattern of ValA reaction product tetratrimethylsilyl-2-epi-5-epi-valiolone. UI, unidentified impurities.
DISCUSSION
Considerable effort has been made in our laboratories during the last few years to clone biosynthetic genes for the aminocyclitol antibiotic validamycin, which is commercially produced on a large scale in China and some other eastern Asian countries and is widely used for the protection of rice seedlings against fungal sheath blight. Genetic manipulation of the structural and/or regulatory genes is expected to be of great help for strain improvement, e.g., to increase yield or the relative abundance of the validamycin A component in the fermentation broth, and possibly to make new validamycin derivatives.
A first effort by heterologous expression in Streptomyces lividans 66, based on the proposed biosynthetic pathway (7) that implied that the validamycin gene cluster may possess a limited number of genes, failed in an initial trial. A second attempt using radioactively labeled strD encoding dTDP-glucose synthase of the streptomycin biosynthetic gene cluster of Streptomyces griseus as a probe also yielded no signal, which suggests that the activation of glucose in validamycin biosynthesis is catalyzed by an enzyme different from StrD, which seems to be specific for 6-deoxyhexose pathways.
The success of using the acbC gene from the acarbose cluster of Actinoplanes to probe for the validamycin biosynthetic gene cluster was not surprising, as both compounds contain the same valienamine moiety in their structures, whose initial biosynthetic step involves cyclization of d-sedoheptulose 7-phosphate to form 2-epi-5-epi-valionone. The significant sequence homologies between ValA and ValC of the validamycin pathway and AcbC and AcbM (40) of the acarbose pathway strongly support the idea that they have similar functions. This suggests that 2-epi-5-epi-valiolone in validamycin biosynthesis has the same fate of being phosphorylated (by ValC) as was demonstrated in acarbose biosynthesis (29).
It was proposed that all of the intermediates from 2-epi-5-epi-valiolone to valienone in the acarbose biosynthetic pathway are phosphorylated (29), which explains the lack of incorporation of the nonphosphorylated putative intermediates, such as 5-epi-valiolone and valienone. On the contrary, nonphosphorylated 5-epi-valiolone, valienone, and validone were found to be efficiently incorporated into validamycin A (7). If ValC has a role in phosphorylating the intermediates in the validamycin pathway similar to that of AcbM in acarbose biosynthesis, its substrate specificity or catalytic ability to phosphorylate all of the cyclitol intermediates, including 2-epi-5-epi-valiolone, remains to be explored.
Both ValA and AcbC show significant similarities to AroB-related DHQS proteins from diverse organisms (4, 8, 17), whose catalytic function is known to be cyclization of DAHP to dehydroquinate. Based on the analysis of the three-dimensional structure of the DHQS domain of the functional AroM protein of the filamentous fungus Emericella nidulans, a total of 13 amino acid residues were identified to be important for catalysis and as being involved in Zn2+ binding (Co2+ in bacteria instead; indicated as 1 in Fig. 4), heptulose phosphate group binding (indicated as 2 in Fig. 4), C-1 hydroxyl fixation (indicated as 3 in Fig. 4), or C-4 hydroxyl fixation (indicated as 4 in Fig. 4) (4). In ValA, three of the four identified residues for Co2+ binding (1), three of the four for heptulose phosphate group binding (2), two of the three for C-1 hydroxyl fixation (3), and all three for C-4 hydroxyl fixation (4), are conserved. Therefore, ValA seems to be more related to AroB proteins than AcbC.
Preceding valC is a cotranscribed gene, valB, encoding a putative adenyltransferase (ValB). Conceivably, this activity could be involved in the conversion of glucose 1-phosphate, possibly from primary metabolism, into dTDP-glucose. Incorporation of activated glucose has been proven through feeding experiments with validoxylamine A to be the final step in validamycin biosynthesis (14). Alternatively, ValB may be involved in the activation of one of the cyclitol intermediates, e.g., 1-epi-valienol 1-phosphate, to its nucleotide derivative, setting the stage for the coupling reaction that leads to a pseudodisaccharide intermediate.
The combination of the results of the previous feeding experiments (7) with the genetic and biochemical information obtained, especially the results of the in vivo gene inactivation and in vitro enzymatic characterization of ValA, strongly suggests that the identified genes are involved in validamycin biosynthesis. From the overlapping cosmid contig we know that the flanking DNA to the left and right of the 6-kb sequenced region covering the complete valA, valB, and valC genes extends to 29 kb and 35 kb, respectively, likely to cover the complete set of genes necessary for validamycin formation. We can thus expect a detailed understanding of the biosynthesis of validamycin, which is critical for more targeted strain improvement or generating novel validamycin derivatives by combined genetic and biochemical approaches. This will become possible when the entire gene cluster has been completely sequenced and the genes have been individually characterized, work that is now in progress.
Acknowledgments
We thank T. Koch and G. Wei for preliminary experiments. We thank David A. Hopwood, Tobias Kieser, Keith F. Chater, Mervyn J. Bibb, Mark Buttner, and Wolfgang Piepersberg for gifts of plasmid, phage, and the strDE probe.
This work received 973 and 863 Funds from the Ministry of Science and Technology, the National Science Foundation of China, the Ph.D. Training Fund from the Ministry of Education, and the Shanghai Municipal Council of Science and Technology. Work at Oregon State University and the University of Washington was supported by NIH grants RAI061528A and AI20264, respectively.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Borck, K., J. D. Beggs, W. J. Brammar, A. S. Hopkins, and N. E. Murray. 1976. The construction in vitro of transducing derivatives of phage lambda. Mol. Gen. Genet. 146:199-207. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter, E. P., A. R. Hawkins, J. W. Frost, and K. A. Brown. 1998. Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. Nature 394:299-302. [DOI] [PubMed] [Google Scholar]
- 5.Demain, A. L. 2000. Microbial biotechnology. Trends Biotechnol. 18:26-31. [DOI] [PubMed] [Google Scholar]
- 6.Deng, Z. X., T. Kieser, and D. A. Hopwood. 1988. “Strong incompatibility” between derivatives of the Streptomyces multicopy plasmid pIJ101. Mol. Gen. Genet. 214:286-294. [DOI] [PubMed] [Google Scholar]
- 7.Dong, H., T. Mahmud, I. Tornus, S. Lee, and H. G. Floss. 2001. Biosynthesis of the validamycins: identification of intermediates in the biosynthesis of validamycin A by Streptomyces hygroscopicus var. limoneus. J. Am. Chem. Soc. 123:2733-2742. [DOI] [PubMed] [Google Scholar]
- 8.Henner, D. J., L. Band, and H. Shimotsu. 1985. Nucleotide sequence of the Bacillus subtilis tryptophan operon. Gene 34:169-177. [DOI] [PubMed] [Google Scholar]
- 9.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces-a laboratory manual. The John Innes Foundation, Norwich, Conn.
- 10.Horii, S., H. Fukase, T. Matsuo, Y. Kameda, N. Asano, and K. Matsui. 1986. Synthesis and alpha-d-glucosidase inhibitory activity of N-substituted valiolamine derivatives as potential oral antidiabetic agents. J. Med. Chem. 29:1038-1046. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 12.Janssen, G. R., and M. J. Bibb. 1993. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124:133-134. [DOI] [PubMed] [Google Scholar]
- 13.Kameda, Y., N. Asano, T. Yamaguchi, K. Matsui, S. Horii, and H. Fukase. 1986. Validamycin G and validoxylamine G, new members of the validamycins. J. Antibiot. (Tokyo) 39:1491-1494. [DOI] [PubMed] [Google Scholar]
- 14.Kameda, Y., S. Horii, and T. Yamano. 1975. Microbial transformation of validamycins. J. Antibiot. (Tokyo) 28:298-306. [DOI] [PubMed] [Google Scholar]
- 15.Kieser, T., M. J. Bibb, K. F. Chater, M. J. Butter, and D. A. Hopwood. 2000. Practical Streptomyces genetics. a laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 16.Mahmud, T. 2003. The C7N aminocyclitol family of natural products. Nat. Prod. Rep. 20:137-166. [DOI] [PubMed] [Google Scholar]
- 17.Millar, G., and J. R. Coggins. 1986. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 200:11-17. [DOI] [PubMed] [Google Scholar]
- 18.Moffatt, B. A., and F. W. Studier. 1987. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell 49:221-227. [DOI] [PubMed] [Google Scholar]
- 19.Novotny, R., C. Schaffer, J. Strauss, and P. Messner. 2004. S-layer glycan-specific loci on the chromosome of Geobacillus stearothermophilus NRS 2004/3a and dTDP-L-rhamnose biosynthesis potential of G. stearothermophilus strains. Microbiology 150:953-965. [DOI] [PubMed] [Google Scholar]
- 20.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Shibata, M., M. Uyeda, and K. Mori. 1981. Stimulation of the extension of validamycin-inhibited hyphae by the hyphal extension factor present in Rhizoctonia solani. J. Antibiot. (Tokyo) 34:447-451. [DOI] [PubMed] [Google Scholar]
- 24.Stockmann, M., and W. Piepersberg. 1992. Gene probes for the detection of 6-deoxyhexose metabolism in secondary metabolite-producing streptomycetes. FEMS Microbiol. Lett. 69:185-189. [DOI] [PubMed] [Google Scholar]
- 25.Stratmann, A., T. Mahmud, S. Lee, J. Distler, H. G. Floss, and W. Piepersberg. 1999. The AcbC protein from Actinoplanes species is a C7-cyclitol synthase related to 3-dehydroquinate synthases and is involved in the biosynthesis of the alpha-glucosidase inhibitor acarbose. J. Biol. Chem. 274:10889-10896. [DOI] [PubMed] [Google Scholar]
- 26.Sun, Y., X. Zhou, J. Liu, K. Bao, G. Zhang, G. Tu, T. Kieser, and Z. Deng. 2002. 'Streptomyces nanchangensis', a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 27.Xia, T. H., and R. S. Jiao. 1986. Studies on glutamine synthetase from Streptomyces hygroscopicus var. jinggangensis. Sci. Sin. B 29:379-388. [PubMed] [Google Scholar]
- 28.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, C. S., A. Stratmann, O. Block, R. Bruckner, M. Podeschwa, H. J. Altenbach, U. F. Wehmeier, and W. Piepersberg. 2002. Biosynthesis of the C(7)-cyclitol moiety of acarbose in Actinoplanes species SE50/110. 7-O-phosphorylation of the initial cyclitol precursor leads to proposal of a new biosynthetic pathway. J. Biol. Chem. 277:22853-22862. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, X., X He, A Li, T. Kieser, and Z. Deng. 2004. The Streptomyces lividans 66 chromosome has a genomic island which is not present in Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 70:7110-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zvenigorodskii, V. I., and N. D. Lomovskaya. 1974. The induction of prophage by a virulent mutant of actinophage φC31 of Streptomyces coelicolor A3(2). Sov. Genet. 8:629-634. [PubMed] [Google Scholar]