Abstract
Fungi are an important and diverse component of soil communities, but these communities have proven difficult to study in conventional biotic surveys. We evaluated soil fungal diversity at two sites in a temperate forest using direct isolation of small-subunit and internal transcribed spacer (ITS) rRNA genes by PCR and high-throughput sequencing of cloned fragments. We identified 412 sequence types from 863 fungal ITS sequences, as well as 112 ITS sequences from other eukaryotic microorganisms. Equal proportions of Basidiomycota and Ascomycota sequences were present in both the ITS and small-subunit libraries, while members of other fungal phyla were recovered at much lower frequencies. Many sequences closely matched sequences from mycorrhizal, plant-pathogenic, and saprophytic fungi. Compositional differences were observed among samples from different soil depths, with mycorrhizal species predominating deeper in the soil profile and saprophytic species predominating in the litter layer. Richness was consistently lowest in the deepest soil horizon samples. Comparable levels of fungal richness have been observed following traditional specimen-based collecting and culturing surveys, but only after much more extensive sampling. The high rate at which new sequence types were recovered even after sampling 863 fungal ITS sequences and the dominance of fungi in our libraries relative to other eukaryotes suggest that the abundance and diversity of fungi in forest soils may be much higher than previously hypothesized.
Fungi are a diverse component of soil microbial communities, in which they function as decomposers, mycorrhizal mutualists, and pathogens. Studying the ecological interactions of these organisms is challenging because of the extremely high diversity of soil fungi, the complexity of the substrate, and the difficulty of direct observation of these communities (4). Mycologists have traditionally examined soil fungal communities by collecting macroscopic sexual reproductive structures (fruiting bodies) that are produced above ground or by using isolation techniques to obtain pure cultures of fungal strains directly from soil (29). However, these approaches have several limitations. Conventional culturing methods are heavily biased towards fast-growing species, and many fungi have specialized growth requirements, so this approach recovers only a small subset of the community. Similarly, fruiting body collection is limited to the detection of species that frequently reproduce sexually unless long-term studies are conducted (33). Furthermore, since fungi vary dramatically in their resource allocation, there is often a lack of correspondence between above-ground and below-ground views of fungal community structure (15). Finally, many fungi are nonculturable, produce inconspicuous or hypogeous fruiting bodies, or do not reproduce sexually at all and thus are overlooked entirely by these methods.
Sequence-based studies of DNA obtained directly from environmental samples are revolutionizing our view of microbial diversity (18). This technique involves using PCR to amplify target regions of DNA extracted directly from environmental samples. These amplicons are then cloned to isolate individual fragments and sequenced. The application of this approach to a wide range of substrates has resulted in the identification of a large number of new bacterial species that cannot be cultivated, many of which constitute novel lineages or are dominant components of microbial communities (11). This technique has become an indispensable tool in bacterial soil ecology, but its use in studies of eukaryotic microorganisms has been limited. Sequence-based studies have been carried out on fungi from grass leaves (6), plant roots (10, 35, 38), and soil (30), but most studies have been restricted to the use of methods based on fragment length polymorphisms (13, 39) or denaturing gradient gel electrophoresis (2, 37). These methods collectively have the potential to improve our understanding of fungal biodiversity and facilitate ecological studies of soil communities.
Most recent studies of uncultured microbial diversity have sequenced genes coding for the small-subunit (SSU) rRNA. Sequencing of SSU rRNA is highly effective in bacteria, but this region does not evolve rapidly enough in eukaryotes for use in more than higher level taxonomic assignments (38). The internal transcribed spacers (ITS) that separate the SSU and large-subunit (LSU) rRNAs in eukaryotes evolve at a much faster rate and can be used to identify fungi to genus and often to species level. This region has become a critical marker for species identification from fungal vegetative structures such as mycorrhizae (14, 35). Due in large part to the use of ITS sequence data for fungal molecular systematic studies (5), a large database of publicly available ITS sequences from diverse fungal taxa now exists (40). If the taxonomic coverage of this database is sufficient such that all sequences from environmental samples will have a close match in the database, the ITS rRNA will provide high-level resolution among different members of fungal communities while simultaneously providing information on taxonomic composition.
Our objectives in this study were (i) to determine the feasibility of using sequence-based biotic inventories for soil fungi to quantify species richness and to detect correlations between the taxonomic composition of soil communities and environmental variables and (ii) to compare the utility of the ITS and SSU rRNA regions for characterizing soil fungi. We hypothesized that a sequence-based approach would detect higher levels of fungal diversity in the soil, representing a greater taxonomic breadth than is usually detected with traditional survey methods, and that distinct fungal communities could be detected at different depths in the soil profile. We expected the ITS region to provide finer-scale taxonomic resolution of community composition than the SSU while still permitting taxonomic placement of sequences because of the extensive sequence database available. If this approach is successful, it can be used to greatly increase our understanding of the dynamics of soil fungal communities through rapid and unbiased inventories of soil fungi.
MATERIALS AND METHODS
Sampling.
Two plots in Duke Forest, Durham, N.C., were sampled: an early successional well-drained, upland stand dominated by loblolly pine (Pinus taeda) containing a few hardwood species regenerating in the understory and a thermic Ultic Hapludalf soil in the Enon series; and a late successional, low-lying, mature, mixed hardwood stand with an alluvial Fluvaquentic Dystrudept soil in the Chewacla soil series. Each plot was 0.1 ha; the two plots are approximately 6.5 km apart and are two of approximately 250 plots originally established by Peet and Christensen in the 1970s to study forest succession and community diversity (8).
At each site, 1-m2 quadrats were sampled at 5-m intervals along transects spaced approximately 5 m apart, for a total of 50 soil samples per plot. For each quadrat, all of the litter was collected and one soil core 2.5 cm in diameter and 20 cm deep was taken. In each soil core distinct zonation corresponding to O (organic material and detritus), A (organic matter-rich mineral soil), and B (clay-rich mineral soil) horizons could be visually detected. Each soil core was subdivided by zone, and all samples within a soil horizon type were composited to overcome any within-plot spatial heterogeneity. Litter samples were air dried for 24 h and homogenized by grinding in a Wiley mill (Thomas Scientific, Swedesboro, NJ). Organic and mineral soil samples were sieved using a 2-mm sieve to remove large rocks and roots. Samples were stored at 4°C for 1 week or less before processing. Subsamples (50 ml) were finely ground using a sterile mortar and pestle and liquid nitrogen and stored at −20°C until DNA was extracted. Samples were designated according to plot (P, pine; MH, mixed hardwood) and horizon (L, litter; O, organic; A, A horizon; and B, B horizon).
Molecular methods.
DNA was isolated from 0.5 g of the finely ground subsamples using the Mo Bio Ultraclean soil DNA extraction kit (Mo Bio Laboratories, Solana Beach, CA) according to the manufacturer's instructions. The ITS and SSU rRNA regions were amplified from these extracts using universal primers (ITS1, ITS4, NS1, and NS2) (42) as well as taxon-specific primers ITS1F (fungus-specific) and ITS4B (Basidiomycota specific) (16). PCR was performed using the following protocol: initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, 50°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 7 min. A modified PCR protocol consisting of an initial denaturation at 94°C for 5 min, followed by 20 cycles of 94°C for 1 min, 50°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 7 min was used for some PCRs to assess the effect of cycle number and extension time on the generation of chimeric sequences and other PCR artifacts (Table 1). Each 25-μl PCR consisted of 25 μg bovine serum albumin, 0.625 U Taq DNA polymerase (Abgene, Rochester, NY), 1.5 mM MgCl2, deoxynucleoside triphosphates (0.2 mM each), primers (0.5 μM each), and PCR buffer.
TABLE 1.
Number of sequences and OTUs recovered from each library
| Library | Amt of template DNA (μg) | No. of sequences | No. of OTUs | Ha | No. of chimeras |
|---|---|---|---|---|---|
| MH-L-1 | 30 | 43 | 18 | 3.98 | 2 |
| MH-L-2b | 240 | 101 | 67 | 5.60 | 0 |
| MH-O-1 | 3 | 25 | 23 | 4.45 | 0 |
| MH-O-2 | 3 | 50 | 39 | 5.15 | 0 |
| MH-O-3 | 3 | 18 | 16 | 4.04 | 1 |
| MH-O-4 | 150 | 72 | 53 | 5.57 | 3 |
| MH-O-5c | 3 | 29 | 25 | 4.56 | 0 |
| MH-O-6c | 3 | 23 | 16 | 3.58 | 0 |
| MH-A-1b | 240d | 101 | 39 | 3.62 | 13 |
| MH-A-2b | 240d | 87 | 62 | 5.60 | 0 |
| MH-B-1b | 240d | 70 | 33 | 4.33 | 2 |
| P-L-1b | 240d | 68 | 48 | 5.34 | 9 |
| P-O-1b | 240d | 62 | 43 | 5.14 | 4 |
| P-A-1 | 30 | 103 | 62 | 5.61 | 3 |
| P-B-1 | 30 | 123 | 61 | 5.44 | 3 |
Shannon-Weiner diversity index.
Library generated from two pooled PCR products amplified using the modified “soil PCR” thermal cycler profile (see Materials and Methods).
PCR primers ITS1F (MH-O-5 and -6) and ITS4B (MH-O-6 only) were used in place of ITS1 and ITS4.
Template consisted of three pooled Mo Bio DNA preps.
Fragments were cloned into Escherichia coli plasmids using the Topo-TA 5-minute PCR cloning kit (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The maximum recommended incubation times were used for each step with 4 μl of PCR product. For SSU fragments, the L, O, A, and B PCR products were pooled and used to generate a single library.
White colonies were screened for inserts by adding whole cells directly to PCRs and amplifying the inserts using M13 F and R primers (Invitrogen, Carlsbad, CA) and a “colony PCR” protocol: initial denaturation of 94°C for 10 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 10 min. PCR products containing the insert were purified by affinity chromatography using Qiaquick spin columns (QIAGEN, Valencia, CA). Cycle sequencing was performed with Big Dye chemistry version 3.1 (Applied Biosystems, Foster City, CA) and sequences were determined with an ABI 3700 (Applied Biosystems). All PCR and sequencing reactions were run on a Primus 96 Plus thermal cycler (MWG AG Biotech, High Point, NC).
Bioinformatics and phylogenetic analyses.
Forward and reverse sequence reads were assembled using Sequencher 4.0 (Gene Codes, Ann Arbor, MI). Conserved motifs were identified in the SSU and LSU regions flanking each ITS sequence and the sequences were trimmed to these motifs to ensure that all sequences had the same start and end point. Pairwise alignments were generated among all ITS sequences using the Needleman-Wunsch global alignment algorithm (26) as implemented in the program Needle in the EMBOSS software suite (28). This program calculates pairwise similarity values by dividing the number of matching nucleotides by the total length of the alignment.
Operational taxonomic units (OTUs) were defined as a group of sequences sharing at least 97% pairwise similarity, which is within the range of intraspecific ITS sequence divergence. A Perl script (written by J. E. Stajich using the Bioperl Toolkit [32] and available from the authors upon request) was used to cluster sequences using single-linkage clustering. Since terminal gaps were treated as mismatches in these analyses, pairwise similarity was calculated separately for sequences where the terminal motifs could not be identified by using only homologous parts of the sequences to avoid underestimating their similarity to the other full-length sequences.
Representative sequences for each ITS OTU were queried against GenBank using BLAST (1). All sequences for which the ITS1 and ITS2 regions matched unrelated sequences (i.e., different genera) from the database with an alignment score ≥200 were assumed to be chimeric sequences resulting from PCR recombination and were excluded from subsequent analyses. Each sequence was identified to the lowest taxonomic rank common to all of the top BLAST hits using the taxonomy of Kirk et al. (21). Due to the high degree of sequence conservation in the 5.8S rRNA (located between the ITS1 and ITS2 regions), all sequences had moderate to high BLAST scores for this portion of the sequence. Thus, a match was required in at least half of the ITS1 or ITS2 region to confidently identify sequences below kingdom or phylum. Most sequences with an alignment score over 600 could be identified to the level of genus, but this was not possible for some taxonomically problematic groups. We did not attempt to identify OTUs to the level of species. The taxonomic placement of sequences with high similarity to anamorphic fungi were determined either by looking at the best BLAST match for a teleomorph or by consulting phylogenetic studies which included that species (23, 31).
All unique SSU sequences were queried against GenBank using BLAST (1). Chimeric sequences were identified using the program Chimera Check from the Ribosomal Database Project II (25) and eliminated from subsequent analyses. The remaining sequences were subjected to automated multiple alignment using ClustalW (36) and the resulting alignment was edited by eye using MacClade 4.0 (24). Ambiguously aligned regions were excluded from the alignment. Phylogenetic analysis was conducted with the neighbor joining method using PAUP*4.0b10 (34).
Diversity analyses.
All diversity calculations were carried out using the program EstimateS 6.0b (9). Shannon-Weiner diversity indices were calculated for each clone library using OTUs as a proxy for species. Species-effort curves were used to determine how sensitive the observed richness was to the number of clones sampled. These curves were constructed by randomly resampling different numbers of sequences from the data set without replacement, and determining the number of OTUs in each sample. This resampling was repeated 1000 times for each sample size and the mean and variance of these richness estimators were calculated. Species-effort curves also were constructed for estimated richness using the ACE nonparametric richness estimator (7). Differences in community composition among soil horizons within and between plots were investigated by plotting relative abundances of different taxonomic groups at hierarchically nested taxonomic scales for each library.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank, with accession numbers AY969316 to AY970290 for the ITS sequences and AY969135 to AY969315 for the SSU sequences. Tables listing the BLAST identity of each OTU and their distribution across libraries can be found in the supplemental material.
RESULTS
Description of libraries.
Fifteen ITS clone libraries were generated for this study: six from MH-O (designated MH-O-1 through MH-O-6), two each from MH-L and MH-A (designated MH-L-1, MH-L-2, MH-A-1, and MH-A-2), and one each from MH-B, P-L, P-O, P-A, and P-B. All libraries were generated using the primers ITS1 and ITS4 for initial PCR amplification, except for MH-O-5 and MH-O-6, for which ITS1F/ITS4 (specific to fungi) and ITS1F/ITS4B (specific to basidiomycete fungi) were used, respectively (Table 1). The number of colonies sequenced per library varied from 18 to 123, with a minimum of 62 sequences (P-O) and a maximum of 217 sequences (MH-O) per soil sample (Table 1). In total, 1,015 sequences were obtained, 40 of which were determined to be chimeric and removed from the data set.
The 975 nonchimeric sequences comprised 454 unique OTUs, 301 of which were singletons occurring only once in the entire data set. The remaining 153 OTUs ranged in abundance from 2 to 51 sequences (0.2% and 5.2% of the data set, respectively). Since primer specificity affects the breadth of phylogenetic diversity recovered, only libraries generated with the universal primer pair ITS1 and ITS4 were considered when comparing diversity and community composition among soil horizons and between plots.
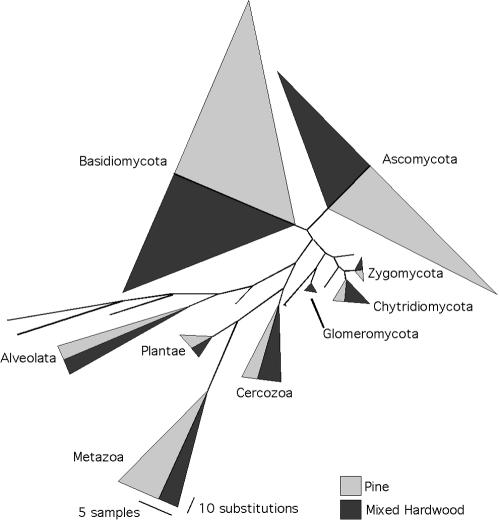
One SSU library was generated for each plot, and 100 and 92 clones were sequenced for the P and MH plots, respectively. Eleven of these sequences were determined to be chimeric and removed from subsequent analyses. A total of 153 unique sequence types were recovered, with each occurring between 1 and 12 times in the data set. The phylogenetic analysis of SSU sequences confirmed the results of BLAST searches as all Ascomycota and Basidiomycota sequences formed monophyletic groups (Fig. 1). Four sequences that matched members of the Chytridiomycota and Zygomycota fell outside their corresponding clusters, but all fungal sequences grouped together.
FIG. 1.
Proportions of SSU sequences belonging to different fungal phyla and other eukaryotic kingdoms. The width of triangles corresponds to the number of sequences in each, and the height corresponds to the maximum amount of sequence divergence within clades (see scale bars).
Community composition.
For all but one ITS library, the proportion of sequences determined to be of fungal origin ranged from 84% to 100% (Fig. 2A). The only exception was MH-A-1, which possessed 65 nonfungal sequences out of 101, 49 of which belonged to a single OTU. Because this OTU was not found at high frequency in MH-A-2, this result was considered an artifact of library construction and MH-A-1 was excluded from diversity and community composition comparisons. Seventy-eight percent of SSU sequences were of fungal origin, while a slightly higher percentage (88%) of fungal ITS sequences could be unambiguously identified to a fungal phylum by BLAST.
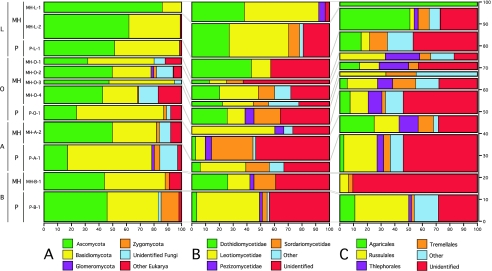
FIG. 2.
Proportional distribution of different taxonomic groups in ITS clone libraries. The x axis indicates the proportion of sequences assigned to each taxonomic group, and bar width corresponds to the proportion of all sequences belonging to each library. A. Proportion of all ITS sequences belonging to each fungal kingdom. B. Proportion of Ascomycota ITS sequences belonging to each subclass. C. Proportion of Basidiomycota ITS sequences belonging to each order.
The majority of fungal sequences recovered in this study belonged to the Ascomycota and Basidiomycota, with a slightly greater proportion of Ascomycota sequences (Ascomycota relative frequency = 46% ITS, 35% SSU; Basidiomycota relative frequency = 41% ITS, 34% SSU). However, the relative abundance of Ascomycota and Basidiomycota ITS sequences varied greatly among libraries, ranging from 18% Ascomycota and 60% Basidomycota for P-A to 69% Ascomycota and 30% Basidiomycota for MH-L (Fig. 2A). There were no apparent trends in relative abundance of these phyla with plot type or soil horizon; Ascomycota sequences were most abundant in MH-O, MH-A, MH-L, P-L, and P-B, while Basidiomycota sequences were most abundant in MH-B, P-A, and P-O. Zygomycota and Glomeromycota sequences were present in both SSU and ITS libraries, but were rarer in ITS libraries (Zygomycota relative frequency = 1.5% ITS, 4% SSU; Glomeromycota relative frequency = 0.5% ITS, 1% SSU). Four percent of SSU sequences matched the Chytridiomycota, but no ITS sequences matched members of this phylum.
Composition of Ascomycota sequences.
Sixty-six percent of all Ascomycota ITS sequences recovered in this study could be identified to subclass or lower. The most abundant subclasses were the Leotiomycetidae and Dothidomycetidae. Members of the Sordariomycetidae, Chaetothyriomycetidae, Pezizomycetidae, and Eurotiomycetidae also were found in many samples. There were no clear compositional differences with depth at this taxonomic scale, but Dothidiomycetidae sequences were most abundant in the mixed hardwood plot, while Leotiomycetidae sequences dominated in the pine plot (Fig. 2B). Twenty-five percent of the Ascomycota ITS sequences could be identified to genus, with the most abundant genera including Phialophora (Leotiomycetidae), Hormonema (Dothidiomycetidae), and Alternaria (Dothidiomycetidae) (Table 2).
TABLE 2.
Number of sequences recovered for the most common genera encountereda
| Genus (order/subclass) | No. of sequences in library:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MH-L | P-L | MH-O | P-O | MH-A | P-A | MH-B | P-B | Total | |
| Russula (Russulales) | 0 | 0 | 4 | 2 | 2 | 12 | 2 | 13 | 35 |
| Mycena (Agaricales) | 11 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 16 |
| Athelia (Polyporales) | 1 | 2 | 4 | 1 | 0 | 0 | 0 | 1 | 9 |
| Phialophora (Leotiomycetidae) | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 8 |
| Tomentella (Thelephorales) | 0 | 0 | 3 | 0 | 2 | 1 | 0 | 1 | 7 |
| Hormonema (Dothidiomycetidae) | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| Alternaria (Dothidiomycetidae) | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Cryptococcus (Tremellales) | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 1 | 5 |
| Hygrocybe (Agaricales) | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 5 |
| Tylospora (Polyporales) | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 5 |
Excluding MH-O-5, MH-O-6, and MH-A-1.
Composition of Basidiomycota sequences.
For Basidiomycota ITS sequences, 64% could be identified to order or lower. Over half of all identifiable sequences matched members of two Basidiomycota orders, the Agaricales and Russulales, with the Thelephorales, Polyporales, and Tremellales constituting an additional 29%. ITS clone libraries from the L horizons were enriched in Agaricales sequences relative to those at greater soil depth, while the proportions of Russulales sequences were much lower in the L horizons of both plots compared to the O, A, and B horizons (Fig. 2C). These differences can be attributed primarily to the distributions of the most abundant genus in these two orders. The genus Russula made up 75% of Russulales sequences in A and B horizon samples and 44% in the O horizon samples, but was absent from the L horizon libraries (Table 2). Similarly, Mycena (Agaricales) was one of the most abundant genera overall, but 11 out of 16 Mycena sequences occurred in the mixed hardwood L horizon.
Richness and diversity estimates.
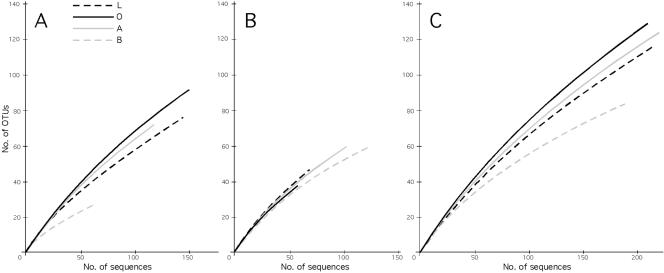
Shannon-Weiner diversity values for clone libraries ranged from 3.58 for MH-O-6 to 5.61 for P-A (Table 1). Overall, diversity was positively correlated with sample size, except for library MH-A-1 (101 sequences, H = 3.62), for which 49% of the sequences belong to a single OTU. Species-effort curves for all samples have a positive slope throughout and show no evidence of approaching saturation, indicating that true richness is much higher than we have captured in our samples (19) (Fig. 3A and B). Similar results were obtained when we examined the data pooled by horizon (Fig. 3C) or plot (Fig. 4).
FIG. 3.
Species-effort curves of richness for fungal OTUs. A. Mixed hardwood plot. B. Pine plot. C. Pooled data from both plots.
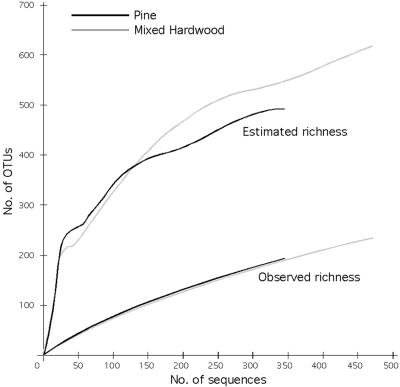
FIG. 4.
Species-effort curves for observed and estimated fungal OTU richness in each plot.
The curves for all samples had similar slopes except for MH-B, which appeared to be more saturated than other samples from the mixed hardwood plot (Fig. 3A). When data were pooled by horizon, O horizon richness was highest, followed by the A and L horizons, with the B horizon having the lowest richness (Fig. 3C). When sequences were pooled by plot, the curves were indistinguishable (Fig. 4). Pine plot richness was estimated to be 491 using the ACE estimator; the estimated richness of the mixed hardwood plot was 616. Predicted richness continued to climb with increasing sample size (Fig. 4), suggesting that these values are likely to be underestimates of true richness.
DISCUSSION
Methodological considerations.
Two difficulties inherent in molecular diversity surveys are determining an appropriate sequence divergence cutoff for defining OTUs that correspond to traditionally recognized species and elimination of PCR artifacts. PCR artifacts encountered in this study included the generation of chimeric sequences due to PCR recombination and biased amplification of a single sequence type. Approximately 5% of the sequences with high BLAST matches in both spacers showed strong differences between ITS1 and ITS2 BLAST matches and were therefore deemed chimeric sequences. Amplification bias in a single PCR could be detected by comparing the frequency of OTUs in independent libraries (e.g., 49 copies of one OTU in MH-A-1 versus 2 copies in MH-A-2). Since adjustments to template concentrations and thermal cycler conditions and pooling of PCR products were unable to completely eliminate these problems, we recommend making multiple independent libraries from separate DNA extracts to accurately evaluate OTU abundances.
The large range of intraspecific ITS variation reported in the literature complicates the determination of an appropriate sequence similarity cutoff. In some cases, different species have been found to have ITS similarity greater than 99% (12, 20) while intraspecific ITS similarity of 90% or lower has been reported for others (22, 27). The degree of ITS sequence variation within and between fungal species is an issue that deserves greater attention if we are to continue to rely on this gene region to delimit species, but clustering sequences with 97% or greater sequence similarity is likely a conservative measure of species richness, especially when sampling from a single locality.
Comparisons of ITS sequencing with traditional fungal survey methods.
Despite methodological limitations, this study demonstrates the potential of large-scale sequencing projects targeting the ITS rRNA for quantifying and characterizing soil fungal diversity. We recovered 412 unique OTUs from a total of 863 fungal sequences. This level of richness is comparable to that found in a 1,500-m2 forest plot in Switzerland, where 408 species were recovered from fruiting body surveys (33), and to richness levels found for endophytic fungi isolated from leaves of two tropical trees, where 418 morphospecies corresponding to an estimated 347 genetic species were recovered (3). However, the study in Switzerland required 21 years of sampling and identification of 71,222 fruiting bodies (33); likewise, the endophyte study involved examining 1,472 cultures that were isolated from 1,992 leaf segments (3).
We captured this level of richness from 863 fungal sequences isolated from only two 2,000-m2 forest plots in a single locale at a single point in time. Given the lack of saturation in our species-effort curves, we would have vastly exceeded their richness estimates if our sampling effort had been comparable to that of these other studies. Using DNA sequences as the primary sampling unit also requires much less processing of each sample than more labor-intensive methods such as fruiting body collection and culture-based census taking and is therefore amenable to automation of data collection and analysis, making large-scale biotic surveys conducted over short time periods feasible.
Comparisons of ITS sequencing with other molecular survey methods.
While sequencing rRNA clones is standard practice in environmental bacteriology, most fungal studies using environmental sampling have employed either fingerprinting methods based on banding patterns obtained from restriction site polymorphisms (13) or denaturing gradient gel electrophoresis profiles (2, 37). These methods can be useful for detecting overall community shifts in response to environmental variables and for tracking the presence or absence of individual OTUs in different samples. However, they are limited in their ability to enumerate species richness in complex communities and do not provide any information about the taxonomic affiliation of the OTUs.
When sequencing efforts have been conducted, most studies have targeted either SSU or LSU rRNA. These regions are useful for phylogenetic characterization of novel lineages (30, 38), but lack sufficient resolution to discriminate among closely related species, resulting in underestimation of diversity. While most fungal ITS sequences were identifiable, 12% could not be assigned to a phylum. Many of these unidentifiable sequences may represent novel lineages with no representatives known from cultures or fruiting bodies, but almost all SSU sequences cluster within known fungal groups in phylogenetic analysis, suggesting that they may represent known fungi which are not yet represented in the ITS database. Since the ITS region allows fine-scale taxonomic resolution but lacks sufficient conservation for phylogenetic analyses of unknown sequences, we recommend that future studies coamplify the ITS with either the SSU or LSU rRNA region as a single fragment.
Taxonomic breadth of libraries and compositional differences with depth in soil profile.
Comparing the relative importance of major groups of fungi and other soil microbes has been challenging for soil ecologists. Because the primers used to generate clone libraries for this study have been shown to have broad specificity across the fungi, metazoa, and protozoa, we would expect the composition of our libraries to be roughly proportional to the number of genomes of each group present in our sample. This is confirmed by the overall consistency found between ITS and SSU clone libraries obtained using independent priming sites despite differences between these regions in selective constraints. We did find the Chytridiomycota and Zygomycota to be underrepresented in the ITS libraries (0% and 1.5%, respectively) relative to SSU libraries (4% each). While this may reflect differences in primer specificity, it should be noted that 7% of our fungal ITS sequences could not be identified to phylum and may belong to these groups which are underrepresented in public databases relative to other fungal phyla (40).
While sample diversity was high relative to our sampling effort, we were able to detect compositional differences at higher taxonomic scales. Many of the taxa found in the litter layer, such as Mycena species, are known decomposers, while mycorrhizal Basidiomycota sequences, including Russula species, were most common in the O, A, and B layers. The B horizon samples contained a higher proportion of unidentifiable sequences and were considerably less diverse than the other horizons, which may reflect a reduction in niche breadth with soil depth.
Implications for fungal diversity.
We found a remarkably large number of species from just a few grams of soil, but this is only a fraction of the estimated number of fungal species worldwide. Based on extrapolations from the ratio of fungi to vascular plants in the British Isles to the a worldwide vascular plant species richness estimate of 270,000 species, global fungal richness has been estimated to be 1.5 million species (17). Based on our ITS-based estimates of soil fungal richness (491 and 616 for P and MH, respectively) and the vascular plant richness of these plots (26 and 48, respectively; R. Peet, personal communication), this calculation yields global soil fungal species richness estimates ranging from 3.5 to 5.1 million. These values are almost certainly underestimates because the richness estimators continue to increase with increasing sampling and because of the conservative 97% ITS similarity clustering used to designate OTUs.
Given the results of the present study, along with data suggesting that endophytic (3) and insect-associated (41) fungi also represent vast amounts of formerly uncharted fungal diversity, the true magnitude of fungal diversity could be orders of magnitude higher than previously suggested.
Supplementary Material
Acknowledgments
We thank Jason Stajich for assistance with bioinformatics, Dave Carmine for help with the DFMO database, and R. Peet, D. Urban, and N. Christensen for providing plant census data from their “Long-term succession on the North Carolina Piedmont” project funded by NSF grants DEB97-07551 and DEB-7804043.
This work was supported in part by National Science Foundation grant MCB-00-84207 to R.V., J.M., and J. E. Johnson.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, I. C., C. D. Campbell, and J. I. Prosser. 2003. Diversity of fungi in organic soils under a moorland-Scots pine (Pinus sylvestris L.) gradient. Environ. Microbiol. 5:1121-1132. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, A. E., Z. Maynard, G. S. Gilbert, P. D. Coley, and T. A. Kursar. 2000. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 3:267-274. [Google Scholar]
- 4.Bridge, P. D., and B. M. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154. [Google Scholar]
- 5.Bruns, T. D., T. J. White, and J. W. Taylor. 1991. Fungal molecular systematics. Annu. Rev. Ecol. Syst. 22:525-564. [Google Scholar]
- 6.Buchan, A., S. Y. Newell, J. I. L. Moreta, and M. A. Moran. 2002. Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern US salt marsh. Microb. Ecol. 43:329-340. [DOI] [PubMed] [Google Scholar]
- 7.Chazdon, R. L., R. K. Colwell, J. S. Denslow, and M. R. Guariguata. 1998. Statistical methods for estimating species richness of woody regeneration in primary and secondary rain forests of northeastern Costa Rica, p. 285-309. In F. Dallmeier and J. A. Comiskey (ed.), Forest biodiversity research, monitoring, and modeling: conceptual background and Old World case studies. Parthenon, Paris, France.
- 8.Christensen, N. L., and T. MacAller. 1985. Soil mineral nitrogen transformations during succession in the Piedmont of North Carolina. Soil Biol. Biochem. 17:675-681. [Google Scholar]
- 9.Colwell, R. K. 2001. EstimateS 6.0b1: statistical estimation of species richness and shared species from samples. Guide and application published at http://purl.oclc.org/estimates.
- 10.Daniell, T. J., R. Husband, A. H. Fitter, and J. P. W. Young. 2001. Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiol. Ecol. 36:203-209. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 12.Dettman, J. R., F. M. Harbinski, and J. W. Taylor. 2001. Ascospore morphology is a poor predictor of the phylogenetic relationships of Nurospora and Gelasinospora. Fungal Genet. Biol. 34:49-61. [DOI] [PubMed] [Google Scholar]
- 13.Edel-Hermann, V., C. Dreumont, A. Pérez-Piqueres, and C. Steinberg. 2004. Terminal restriction fragment length polymorphism analysis of ribosomal RNA genes to assess changes in fungal community structure in soil. FEMS Microbiol. Ecol. 47:397-404. [DOI] [PubMed] [Google Scholar]
- 14.Frey, S. D., M. Knorr, J. L. Parrent, and R. T. Simpson. 2004. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For. Ecol. Manag. 196:159-171. [Google Scholar]
- 15.Gardes, M., and T. D. Bruns. 1996. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above- and below-ground views. Can. J. Bot. 74:1572-1583. [Google Scholar]
- 16.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 17.Hawksworth, D. L. 1991. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol. Res. 95:641-655. [Google Scholar]
- 18.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, J., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannesson, H., and J. Stenlid. 2003. Molecular markers reveal genetic isolation and phylogeography of the S and F intersterility groups of the wood-decay fungus Heterobasidion annosum. Mol. Phylogenet. Evol. 29:94-101. [DOI] [PubMed] [Google Scholar]
- 21.Kirk, P. M., P. F. Cannon, J. C. David, and J. A. Stalpers. 2001. Ainsworth & Bisby's dictonary of the fungi, 9th ed. CABI Bioscience, Cambridge, United Kingdom.
- 22.Kuninaga, S., T. Natsuaki, T. Takeuchi, and R. Yokosawa. 1997. Sequence variation of the rDNA ITS regions within and between anastomosis groups in Rhizoctonia solani. Curr. Genet. 32:237-243. [DOI] [PubMed] [Google Scholar]
- 23.LoBuglio, K. F., M. L. Berbee, and J. W. Taylor. 1996. Phylogenetic origins of the asexual mycorrhizal symbiont Cenococcum geophilum Fr and other mycorrhizal fungi among the ascomycetes. Mol. Phylogenet. Evol. 6:287-294. [DOI] [PubMed] [Google Scholar]
- 24.Maddison, D. R., and W. P. Maddison. 2000. MacClade 4: Analysis of phylogeny and character evolution, version 4.0. Sinauer Associates, Sunderland, Mass.
- 25.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 27.O'Donnell, K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 90:919-938. [Google Scholar]
- 28.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 29.Rossman, A. Y., R. E. Tulloss, T. E. O'Dell, and R. G. Thorn. 1998. Protocols for an all taxa biodiversity inventory of fungi in a Costa Rican conservation area. Parkway Publishers, Boone, N.C.
- 30.Schadt, C. W., A. P. Martin, D. A. Lipson, and S. K. Schmidt. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359-1361. [DOI] [PubMed] [Google Scholar]
- 31.Scorzetti, G., J. W. Fell, A. Fonseca, and A. Statzell-Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 32.Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz, C. Dagdigian, G. Fuellen, J. G. Gilbert, I. Korf, H. Lapp, H. Lehväslaiho, C. Matsalla, C. J. Mungall, B. I. Osborne, M. R. Pocock, P. Schattner, M. Senger, L. D. Stein, E. Stupka, M. D. Wilkinson, and E. Birney. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straatsma, G., F. Ayer, and S. Egli. 2001. Species richness, abundance, and phenology of fungal fruit bodies over 21 years in a Swiss forest plot. Mycol. Res. 105:515-523. [Google Scholar]
- 34.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates Inc., Sunderland, Mass.
- 35.Taylor, D. L., and T. D. Bruns. 1999. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: minimal overlap between the mature forest and resistant propagule communities. Mol. Ecol. 8:1837-1850. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, postion-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Elsas, J. D., G. F. Duarte, A. Keijzer-Wolters, and E. Smit. 2000. Analysis of the dynamics of fungal communities in soil via fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J. Microbiol. Methods 43:133-151. [DOI] [PubMed] [Google Scholar]
- 38.Vandenkoornhuyse, P., S. L. Baldauf, C. Leyval, J. Straczek, and J. P. W. Young. 2002. Extensive fungal diversity in plant roots. Science 295:2051. [DOI] [PubMed] [Google Scholar]
- 39.Viaud, M., A. Pasquier, and Y. Brygoo. 2000. Diversity of soil fungi studied by PCR-RFLP of ITS. Mycol. Res. 104:1027-1032. [Google Scholar]
- 40.Vilgalys, R. 2003. Taxonomic misidentification in public DNA databases. New Phytol. 160:4-5. [DOI] [PubMed] [Google Scholar]
- 41.Weir, A., and P. M. Hammond. 1997. Laboulbeniales on beetles: host utilization patterns and species richness of the parasites. Biodivers. Conserv. 6:701-719. [Google Scholar]
- 42.White, T. J., T. D. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-324. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press Inc., New York, N.Y.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.