Abstract
Six strains of multidrug-resistant Stenotrophomonas maltophilia were isolated from cultured yellowtail. The strains were divided into two clusters based on the 16S rRNA genes, and all of them contained L1 metallo-β-lactamase and L2 β-lactamase genes. Differences in the intercluster divergence between the lactamase genes suggest that horizontal transfer of the genes occurred.
Stenotrophomonas maltophilia is a multidrug-resistant bacterium that has been isolated from numerous terrestrial environments (3, 7, 12, 18) and recently has emerged as a nosocomial pathogen in clinical environments (8, 13, 19, 30). Although the phylogenetic relationship between environmental and clinical strains has been examined, no clear distinction has been established yet.
We isolated six strains of S. maltophilia, which were identified with API 20NE (BioMerieux, La Balm, France), from yellowtail (Seriola quinqueradiata) cultured in a fish farm located at the southern tip of Japan (14). All six strains grew on Muller-Hinton agar (Becton Dickinson Microbiology Systems, Cockeysville, MD) supplemented with either ampicillin (32 μg ml−1) or panipenem (32 μg ml−1). The strains were also resistant to cefotaxime and ceftazidime, as determined by susceptibility tests with Sensi-Disks (Showa, Tokyo, Japan). Although the strains grew in a seawater-based medium, the growth rates in this medium were 0.6-fold lower than those with no NaCl. Hence, we concluded that the strains are not indigenous to marine environments. The resistance to ampicillin seemed to be a factor that could induce the presence of S. maltophilia in the fish farm, since medication with ampicillin was used in the previous year according to the fish farmer. Samuelsen et al. have reported persistence of drug-resistant bacteria even in the absence direct selection (28). NaCl may have been a factor in the survival. Resistance to panipenem, cefotaxime, and ceftazidime probably did not play a role in the distribution of the strains, since none of these antibiotics can be used in aquaculture. Rather, the resistance to these drugs suggests that the strains were derived from clinical environments.
16S rRNA gene sequences of the six strains were determined for the phylogenetic analysis by a method described previously (14). The phylogenetic analysis divided the strains into two distinct clusters, cluster A (strains BL-9, BL-10, BL-15, and BL-16) and cluster B (strains BL-12 and BL-13), and both of the clusters were included in group I described by Hauben et al., which is comprised mainly of clinical and environmental strains (15).
PCR amplification with forward primer 5′-CACACCTGGCAGATCGGCAC-3′ and reverse primer 5′-GCCGCATCCGCGTAGGC-3′ at an annealing temperature of 65°C was used to amplify L1 metallo-β-lactamase (L1), which can degrade carbapenems (23, 24, 33). L2 serine β-lactamase (L2), which cannot degrade carbapenems (23, 32), was PCR amplified with forward primer 5′-CGATTCCTGCAGTTCAGT-3′ and reverse primer 5′-CGGTTACCTCATCCGATC-3′ at an annealing temperature of 55°C. No differences were found in the amplification profiles of the six strains, and none of the other lactamase genes were amplified, although primers were designed for imiS (34), cphA (20), IMP (22), blaB (5), cfiA (17, 31), ccrA (26), VIM (25), TEM (29), SHV (27), CTX (4), Toho (16), CME (6), and CEF (9).
The sequences of the L1 and L2 genes were determined by using a Big Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.). The sequences of the genes of reference strains S. maltophilia K279a (1), LMG10888 (= IAM1411), LMG10877 (= ATCC 17676), and LMG10879 (= IAM1566) were also determined, but the sequences of the genes of LMG10883 and LMG10871 were not determined because of a lack of PCR amplification.
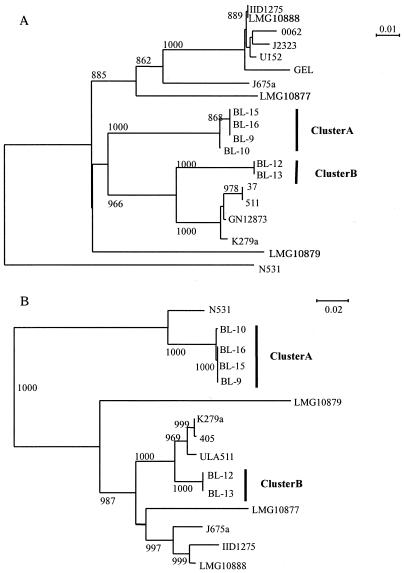
Figure 1A shows the clustering of the bacteria inferred from the 459-bp nucleotide sequence (nucleotide positions 168 to 626) of the L1 gene. While the fish farm S. maltophilia strains were divided into the same clusters, clusters A and B, the clustering pattern differed slightly from that based on the 16S rRNA genes (data not shown). The LMG10888 strain, which was closely related to cluster B based on the 16S rRNA gene analysis, was excluded from the group containing clusters A and B. The levels of similarity of nucleotide sequences between clusters A and B were 89.4 to 89.6%, which were lower than the level of similarity found for 16S rRNA genes (99.6%). The levels of divergence of the L1 genes from the purified strain IID1275 gene were 13% for cluster A and 15% for cluster B, which were within the range reported previously (8 to 20%) (1).
FIG. 1.
Phylogenetic relationships of S. maltophilia strains based on the genes encoding L1 metallo-β-lactamase (A) and L2 β-lactamase (B). The branching pattern was generated by the neighbor-joining method. The numbers at the nodes indicate bootstrap values greater than 800. The DNA sequences for L1 were obtained from strains N531 (accession no. AJ272109), K279a (AJ251814), J657a (AJ251815), IID1275 (X75074), GN12873 (AF010282), U152 (AJ289083), J2323 (AJ289084), 0062 (AJ289082), 37 (AJ289085), 511 (AJ289086), and GEL (AJ289081). The DNA sequences for L2 were obtained from strains N531 (accession no. AJ272110), K279a (AJ251816), J657a (AJ251817), IID1275 (Y08562), ULA511 (AF299368), and 405 (AJ506737).
Figure 1B shows the clustering of the bacteria inferred from the 870-bp sequence (nucleotide positions 34 to 933) of the L2 gene. The branching pattern differed from the patterns based on the16S rRNA gene and L1 gene analyses, although the isolated strains were divided into the same two clusters, clusters A and B. Cluster B and the reference strains formed a large group which did not contain cluster A. The level of similarity of the nucleotide sequences between clusters A and B was 74.2%, which was considerably lower than the levels of similarity determined for the L1 and 16S rRNA genes. The levels of divergence from the IID1275 L2 gene were 27% for cluster A and 9% for cluster B in the present study.
Genetic analyses of the L1 metallo-β-lactamase and the L2 β-lactamase indicated that the levels of change in the L1 and L2 genes in the fish farm strains from the IID1275 genes are not consistent between clusters A and B. Also, the difference in the L2 gene between clusters A and B was far larger than the difference in the L1 gene. Hence, it is very likely that the L1 or L2 gene was externally acquired in either cluster A or B if the evolutionary rates are similar for the L1 and L2 genes. Avison et al. presented data on the localization of the L1 and L2 genes in cells (1, 2). Minkwitz and Berg have suggested that there is a reservoir of S. maltophilia strains in the natural environment (21). This hypothesis does not contradict the suggestion that multiple or independent acquisitions of S. maltophilia occurred from a variety of sites both within and outside hospitals (10, 11). If drug resistance obtained via horizontal transfer plays a key role in the distribution in the clinical environment, it is very likely that different lineages of S. maltophilia spread as nosocomial pathogens.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AB194303 to AB194327.
Acknowledgments
We sincerely thank Matthew B. Avison for providing the S. maltophilia K279a strain and H. Ishizuka, M. Ozawa, and Y. Kakimoto for technical assistance.
REFERENCES
- 1.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 beta-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avison, M. B., C. S. Higgins, P. J. Ford, C. J. von Heldreich, T. R. Walsh, and P. M. Bennett. 2002. Differential regulation of L1 and L2 beta-lactamase expression in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 49:387-389. [DOI] [PubMed] [Google Scholar]
- 3.Aznar, R., E. Alcaide, and E. Garay. 1992. Numerical taxonomy of pseudomonads isolated from water, sediment and eels. Syst. Appl. Microbiol. 14:235-246. [Google Scholar]
- 4.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellais, S., D. Aubert, T. Naas, and P. Nordmann. 2000. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 44:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellais, S., L. Poirel, T. Naas, D. Girlich, and P. Nordmann. 2000. Genetic-biochemical analysis and distribution of the Ambler class A beta-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob. Agents Chemother. 44:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg, G., P. Marten, and G. Ballin. 1996. Stenotrophomonas maltophilia in the rhizosphere of oilseed rape—occurrence, characterization and interaction with phytopathogenic fungi. Microbiol. Res. 151:19-27. [Google Scholar]
- 8.Bingen, E. H., E. Denamur, N. Y. Lambert-Zechovsky, A. Bourdois, P. Mariani-Kurkdjian, J. P. Cezard, J. Navarro, and J. Elion. 1991. DNA restriction fragment length polymorphism differentiates crossed from independent infections in nosocomial Xanthomonas maltophilia bacteremia. J. Clin. Microbiol. 29:1348-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denton, M., V. Keer, and P. M. Hawkey. 1999. Correlation between genotype and beta-lactamases of clinical and environmental strains of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 43:555-558. [DOI] [PubMed] [Google Scholar]
- 11.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elad, Y., I. Chet, and R. Baker. 1987. Increased growth response of plants induced by rhizobacteria antagonistic to soilborne pathogenic fungi. Plant Soil 98:325-339. [Google Scholar]
- 13.Friedman, N. D., T. M. Korman, C. K. Fairley, J. C. Franklin, and D. W. Spelman. 2002. Bacteraemia due to Stenotrophomonas maltophilia: an analysis of 45 episodes. J. Infect. 45:47-53. [DOI] [PubMed] [Google Scholar]
- 14.Furushita, M., T. Shiba, T. Maeda, M. Yahata, A. Kaneoka, Y. Takahashi, K. Torii, T. Hasegawa, and M. Ohta. 2003. Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl. Environ. Microbiol. 69:5336-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauben, L., L. Vauterin, E. R. Moore, B. Hoste, and J. Swings. 1999. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 49:1749-1760. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A beta-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khushi, T., D. J. Payne, A. Fosberry, and C. Reading. 1996. Production of metal dependent beta-lactamases by clinical strains of Bacteroides fragilis isolated before 1987. J. Antimicrob. Chemother. 37:345-350. [DOI] [PubMed] [Google Scholar]
- 18.Lambert, B., L. Frederik, L. Van Rooyen, F. Gossele, Y. Papon, and J. Swings. 1987. Rhizobacteria of maize and their antifungal activities. Appl. Environ. Microbiol. 53:1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manfredi, R., A. Nanetti, M. Ferri, and F. Chiodo. 1998. Xanthomonas maltophilia: an emerging pathogen in patients with HIV disease. Int. J. STD AIDS 9:201-207. [DOI] [PubMed] [Google Scholar]
- 20.Massidda, O., G. M. Rossolini, and G. Satta. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J. Bacteriol. 173:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minkwitz, A., and G. Berg. 2001. Comparison of antifungal activities and 16S ribosomal DNA sequences of clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 39:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, R., R. S. Miles, and S. G. Amyes. 1994. Biochemical properties of inducible beta-lactamases produced from Xanthomonas maltophilia. Antimicrob. Agents Chemother. 38:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne, D. J. 1993. Metallo-beta-lactamases—a new therapeutic challenge. J. Med. Microbiol. 39:93-99. [DOI] [PubMed] [Google Scholar]
- 25.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen, B. A., Y. Gluzman, and F. P. Tally. 1990. Cloning and sequencing of the class B beta-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob. Agents Chemother. 34:1590-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, L. B., L. L. Carias, A. M. Hujer, M. Bonafede, R. Hutton, C. Hoyen, and R. A. Bonomo. 2000. High-level expression of chromosomally encoded SHV-1 beta-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuelsen, O. B., V. Torsvik, and A. Ervik. 1992. Long-range changes in oxytetracycline concentration and bacterial resistance toward oxytetracycline in a fish farm sediment after medication. Sci. Total Environ. 114:25-36. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Pescador, R., M. S. Stempien, and M. S. Urdea. 1988. Rapid chemiluminescent nucleic acid assays for detection of TEM-1 beta-lactamase-mediated penicillin resistance in Neisseria gonorrhoeae and other bacteria. J. Clin. Microbiol. 26:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senol, E. 2004. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J. Hosp. Infect. 57:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J. S., and M. H. Malamy. 1990. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J. Bacteriol. 172:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh, T. R., A. P. MacGowan, and P. M. Bennett. 1997. Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1460-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh, T. R., L. Hall, S. J. Assinder, W. W. Nichols, S. J. Cartwright, A. P. MacGowan, and P. M. Bennett. 1994. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218:199-201. [DOI] [PubMed] [Google Scholar]
- 34.Walsh, T. R., W. A. Neville, M. H. Haran, D. Tolson, D. J. Payne, J. H. Bateson, A. P. MacGowan, and P. M. Bennett. 1998. Nucleotide and amino acid sequences of the metallo-beta-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob. Agents Chemother. 42:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]