Abstract
Pasteurella multocida is a highly infectious, facultative intracellular bacterium which causes fowl cholera in birds. This study reports, for the first time, the observed interaction between P. multocida and free-living amoebae. Amoebal trophozoites were coinfected with fowl-cholera-causing P. multocida strain X-73 that expressed the green fluorescent protein (GFP). Using confocal fluorescence microscopy, GFP expressing X-73 was located within the trophozoite. Transmission electron microscopy of coinfection preparations revealed clusters of intact X-73 cells in membrane-bound vacuoles within the trophozoite cytoplasm. A coinfection assay employing gentamicin to kill extracellular bacteria was used to assess the survival and replication of P. multocida within amoebae. In the presence of amoebae, the number of recoverable intracellular X-73 cells increased over a 24-h period; in contrast, X-73 cultured alone in assay medium showed a consistent decline in growth. Cytotoxicity assays and microscopy showed that X-73 was able to lyse and exit the amoebal cells approximately 18 h after coinfection. The observed interaction between P. multocida and amoebae can be considered as an infective process as the bacterium was able to invade, survive, replicate, and lyse the amoebal host. This raises the possibility that similar interactions occur in vivo between P. multocida and host cells. Free-living amoebae are ubiquitous within water and soil environments, and P. multocida has been observed to survive within these same ecosystems. Thus, our findings suggest that the interaction between P. multocida and amoebae may occur within the natural environment.
In recent years, there has been intense interest in the complex relationship between bacteria and free-living amoebae. Subsequently, an increasing number of bacterial pathogens have been found to interact with amoebal species such as Acanthamoeba polyphaga. Amoebae are known to be environmental hosts of several pathogens, such as Legionella spp. (41), Mycobacterium avium (14, 30), Chlamydia pneumoniae (17), Burkholderia pseudomallei (26), and Listeria monocytogenes (34). This relationship has important advantages for the bacteria, such as protection against bactericidal agents and increased survival in adverse environmental conditions (5). In addition, this bacterial-amoebal interaction has significance beyond the dynamics of the ecosystem, as it is thought to lead to endosymbiotic relationships which closely resemble the pathogenesis of chronic bacterial infections in mammalian cells (21, 24).
Pasteurella multocida is a facultative gram-negative pathogen that is the causative agent of several economically significant diseases. These include atrophic rhinitis in swine, hemorrhagic septicemia and pneumonic pasteurellosis in cattle and buffalo, and fowl cholera in all avian species (15, 12, 22). Fowl cholera affects both wild and domesticated birds and levies a heavy environmental toll as well as an economic burden on the poultry industry (22, 39). Among wild birds, the disease has greatest impact on North American wildfowl, killing thousands of birds annually (6). Identification of certain virulence factors (7, 13) and genome-wide analyses (8, 9, 25) have led to the further understanding of pathogenic mechanisms that P. multocida utilizes to cause disease. However, control measures to minimize the impact of fowl cholera have been greatly impeded due to the lack of safe and effective vaccines and because the natural reservoir and transmission route of P. multocida are largely unknown. Possible environmental reservoirs may include water, contaminated soils, and birds that harbor P. multocida (4, 37, 44), and one potential route of transmission may be the aerosolization and subsequent inhalation of P. multocida-laden water droplets (6, 48).
Free-living amoebae, such as Acanthamoeba polyphaga and Hartmanella vermiformis, can be isolated in abundance from both aquatic and soil environments. It has been established that P. multocida can survive and persist within these same environmental niches (4, 6, 10, 37, 43, 48). The presence of P. multocida within these aquatic environments and the need to identify the reservoir and route of transmission of this important avian pathogen warrant an investigation into the bacterium's ability to utilize amoebae as hosts. The current report is the first to demonstrate that fowl-cholera-causing P. multocida strains have the ability to enter and survive within the free-living amoebal species A. polyphaga and H. vermiformis.
MATERIALS AND METHODS
Bacterial strains, plasmid, and growth conditions.
Bacterial strains and the plasmid used in this study are listed in Table 1. All P. multocida strains were grown on heart infusion (HI; Difco, Franklin Lakes, New Jersey) medium at 37°C, and all Escherichia coli strains were cultured on Luria-Bertani medium (Difco) at 37°C. Media were supplemented with the appropriate antibiotics when necessary.
TABLE 1.
Bacterial strains and plasmid used in the study
| Bacterial species and strain or plasmid no. | Relevant strain characteristics | Reference |
|---|---|---|
| P. multocida X-73 | Serogroup A: serotype 1, chicken isolate, reference strain | 40 |
| P. multocida PBA815 | Serogroup A: serotype 3/4, chicken isolate | 50 |
| P. multocida P-1059 | Serogroup A: serotype 3, turkey isolate | 40 |
| P. multocida ACP19 | Contains pVT1303 plasmid | This study |
| Escherichia coli | ||
| VT1300 | Contains pVT1303 plasmid | 33 |
| pVT1303 | Contains gfp gene under control of the A. actinomycetemcomitans ltx promoter | 33 |
Amoebal strains and growth conditions.
Acanthamoeba polyphaga strain JAC/S2 (ATCC 50372) and Hartmanella vermiformis (ATCC 50802) were obtained from the American Type Culture Collection (Manassas, VA). Amoebal strains were maintained axenically as confluent monolayers of trophozites in 75-cm2 tissue culture flasks with the appropriate medium. A. polyphaga cells were maintained in modified ATCC 712 PYG (36) culture medium, with lactose substituted for glucose at 35°C, and H. vermiformis cells in ATCC 1034 culture medium supplemented with 10% heat-inactivated fetal bovine serum at 25°C (18). Trophozites were suspended before use by tapping the flask sharply by hand, and cell counts were determined as described previously (36). Amoebae were subcultured into fresh medium every 6 days and plated on HI medium to check for possible bacterial contamination.
Coinfection assay conditions.
The coinfection assay was modified based on the method described by Moffat and Tompkins (36). Briefly, axenically maintained amoebal cells were pelleted and resuspended in modified 1× Acanthamoeba castellanii assay medium (36). In a 24-well plate, 1 ml of the resuspended amoeba was seeded and allowed to form a monolayer for 1 h. The wells were then inoculated with P. multocida (105 CFU/ml) at a multiplicity of infection of 100:1. Immediately after the addition of the bacteria, an aliquot of culture was removed to confirm the CFU in the initial inoculate. P. multocida-infected monolayers were incubated at either 35°C with A. polyphaga or 25°C with H. vermiformis for 24 to 30 h. P. multocida interaction with the amoebae was allowed to continue for 2 h, after which time the A. castellanii medium was aspirated. The wells were washed once with the same medium and replaced with A. castellanii medium containing 100 μg/ml of gentamicin to kill the extracellular bacteria. This treatment had been previously determined to reduce the number of extracellular bacteria by greater than 95% (data not shown). After 2 h, the wells were fully aspirated and the cells were pelleted and resuspended in fresh 1× A. castellanii buffer to remove any remaining gentamicin. The amoebal cells were harvested at this time point, plated onto HI medium, and placed in an incubator at 35°C for 24 h, after which P. multocida cells were enumerated. For most assays, viable count time points were taken at 2-h intervals. All experiments were carried out in triplicate on at least four separate occasions.
Cytochalasin D assays.
Coinfection assays were performed as described above except for the addition of 1 μg of cytochalasin D (Sigma, St. Louis, MO) per ml of 1× A. castellanii buffer to the A. polyphaga 1 h prior to the infection with P. multocida. Cytochalasin D was present for the subsequent duration of the coinfection assays.
Construction of green fluorescent protein (GFP)-expressing P. multocida.
E. coli strain VT1300 was cultured on Luria-Bertani medium and supplemented with 50 μg/ml of spectinomycin. The pVT1303 plasmid was isolated using a method described by previously (32). Employing the ability of P. multocida to uptake DNA naturally, pVT1303 was transformed into wild-type strain X-73. Approximately 1 μg of pVT1303 plasmid was resuspended in 140 μl of A. castellanii buffer and plated onto a HI plate supplemented with 50 μg/ml spectinomycin. The plate was placed in a 37°C incubator for 15 min to allow the DNA to absorb into the agar. A bacterial suspension was prepared by harvesting X-73 cells from an 8-h plate incubated at 37°C, and cells were resuspended in a final concentration of 105 CFU/ml of 1× A. castellanii buffer. Onto the dry agar, 500 μl of the 105-CFU/ml X-73 bacterial suspension was spread plated over the pVT1303 DNA. The plate was incubated again at 37°C for 48 h, enabling the growth of transformants. One P. multocida transformant, strain ACP19, was selected and further cultured onto HI plates with spectinomycin.
Cytotoxicity assay.
The CytoTox 96 nonradioactive cytotoxicity assay (Promega Corp., Madison, WI) was used to measure the level of P. multocida cytotoxicity on amoebal cells. In brief, the standard infection assay was preformed with wild-type P. multocida strain X-73 and A. polyphaga. After 18 h of coinfection, cytotoxicity was measured in cellular lysates and culture supernatants via the CytoTox 96 assay. The cytotoxicity assay and the percentage of cytotoxicity were conducted and calculated, respectively, according to the manufacturer's instructions.
Laser scanning confocal microscopy.
To confirm that ACP19 expressed the GFP protein and exhibited fluorescence, X-73 and ACP19 cells were wet mounted onto microscope slides and examined using a three-dimensional (3D) laser scanning confocal microscope TCS SP2 (Leica Microsystems, Bannockburn, IL) at magnifications of up to 4,000×. The filters used to visualize GFP were dichroic filters, the photomultiplier settings ranged from 2.0 to 2.9, and the excitation energy setting was RSP 500. Once fluorescence was confirmed, a standard coinfection assay was then performed using ACP19 and monolayers of A. polyphaga. After 18 h postinfection, amoebal cells were removed, washed in phosphate-buffered saline, treated with 25% formaldehyde solution for 30 min, and washed twice more in phosphate-buffered saline to allow the samples to be visualized. The samples were wet mounted and viewed with a 3D laser scanning confocal microscope TCS SP2 at magnifications of up to 4,000× as described above.
Transmission electron microscopy.
A standard coinfection assay was set up with A. polyphaga and X-73 and allowed to incubate for 18 h. Uninfected monolayers of A. polyphaga were incubated for 18 h and used as a control group. In both cases, the amoebae were harvested and fixed for electron microscopy. Samples were individually fixed in a primary fixation solution of 1 M HEPES and 2.5% glutaraldehyde for 2 h. The samples were then washed twice for 15 min in fresh 1 M HEPES to remove the excess glutaraldehyde. The samples were moved into the secondary fixation solution containing 1% OsO4 (Sigma) and allowed to incubate overnight. A series of dehydration washes was then preformed utilizing an increasing concentration of ethanol. Once sufficiently dehydrated, three washes with propylene oxide were conducted. Finally, samples were embedded in resin using an Eponate 12 embedding medium kit (Pella, Inc. Redding, CA). Sectioning was preformed with an RMC MT 7000 Ultramicrotome (Boeckeler Instruments, Inc., Tucson, AZ), and ultrathin sections were placed on copper grids (400 square mesh) and poststained with uranyl acetate and Reynold's lead citrate. Sections were examined with a Hitachi (Pleasanton, CA) H-600 transmission electron microscope operating at 75 kV.
RESULTS
Expression of GFP in P. multocida.
To date, the potential for P. multocida to express green fluorescent protein had not been investigated. P. multocida strain X-73 was transformed with the Actinobacillus actinomycetemcomitans shuttle vector pVT1303, which contained a modified gfp gene (33). The resultant construct ACP19 was examined for green fluorescence using a 3D laser scanning confocal microscope. ACP19 clearly exhibited green fluorescence, thus confirming the expression of the gfp gene within P. multocida (Fig. 1A). This result was the first to demonstrate the expression of GFP in a Pasteurella species. It was also a crucial step in the initial study of the amoebal-P. multocida interaction and it demonstrated the use of A. actinomycetemcomitans shuttle vectors in P. multocida. The A. actinomycetemcomitans ltx promoter was used to drive the expression of the gfp gene in pVT1303, and closer examination of the ltx promoter region revealed a conserved σ70 promoter that most likely enabled the expression of gfp within the P. multocida background. The GFP-expressing strain ACP19 can be utilized as a reporter for future P. multocida intracellular studies.
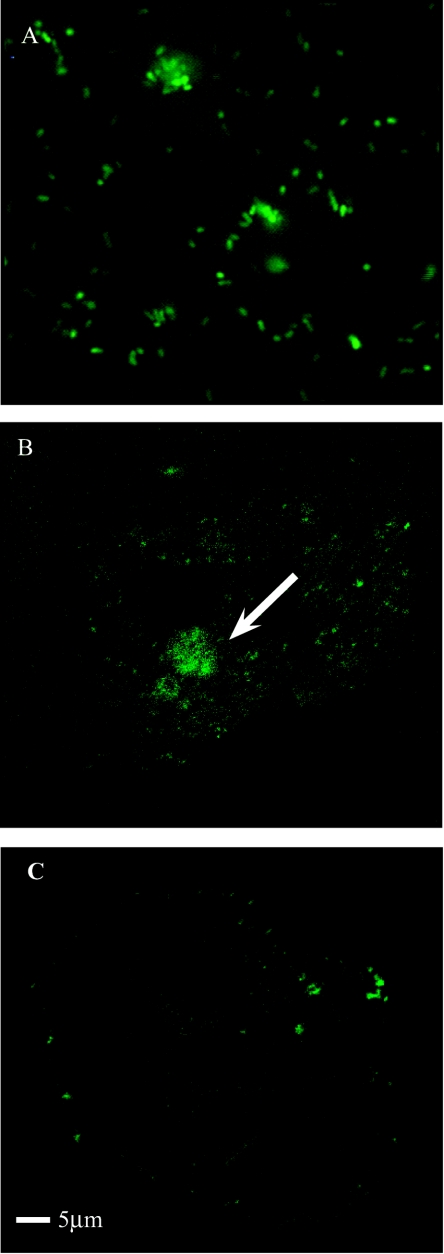
FIG. 1.
Expression of GFP within P. multocida ACP19 cells. (A) ACP19 cells exhibiting green fluorescence; (B) A. polyphaga cells coinfected with ACP19, arrow indicating internalized GFP-fluorescingACP19 cells within A. polyphaga 18 h postinfection; (C) uninfected A. polyphaga cells. Cells were examined using a 3D laser scanning confocal microscope TCS SP2 at a magnification of up to 4,000×.
To determine if P. multocida entered into and survived within amoebae, ACP19 was employed in a standard coinfection assay with A. polyphaga. By using the 3D laser scanning confocal microscope, the intracellular location of ACP19 was visualized within the amoebal cytoplasm (Fig. 1B). When compared to the uninfected A. polyphaga (Fig. 1C), the infected amoebal cells showed a concentration of fluorescence within the cytoplasm which was likely due to a number of ACP19 cells. The GFP has been shown not to be affected by the fixing of the specimens that was required for this type of microscopy (27).
Localization of P. multocida within A. polyphaga.
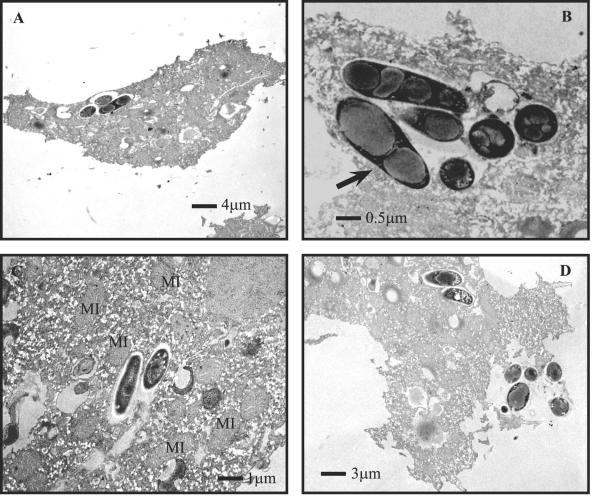
From the results gained using ACP19 in coinfection assays, it appeared that the bacteria were located in the amoebal cytoplasm. In order to verify whether the P. multocida X-73 cells coinfected with A. polyphaga or H. vermiformis trophozoites were indeed intracellular, a standard coinfection assay was terminated at 18 h postinfection and amoebal cells were examined via transmission electron microscopy (Fig. 2). P. multocida X-73 cells were visualized in membrane-bound vacuoles within the cytoplasm of the amoebal cell (Fig. 2A through C). Within the vacuoles, bacteria appeared to be dividing (Fig. 2B), lysing, and exiting their amoebal host (Fig. 2D). In addition, vacuoles that contained X-73 were consistently associated with mitochondria (Fig. 2B and 2C).
FIG. 2.
Transmission electron microscopy of A. polyphaga trophozites infected with P. multocida X-73. (A through C) Micrographs show intravacuolar P. multocida at 18 h postinfection. MI indicates mitochondria, and arrow indicates a P. multocida cell that appears to be in the process of cell division. (D) Intracellular X-73 cells that have lysed and exited A. polyphaga cells.
Survival of P. multocida within A. polyphaga.
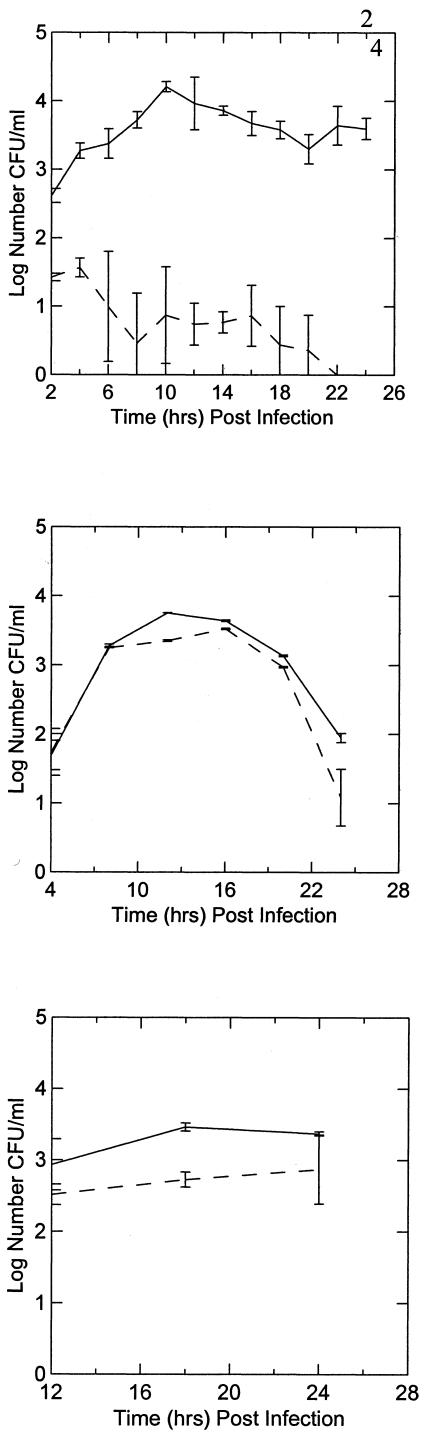
A quantitative coinfection assay was carried out with P. multocida X-73 to determine the effect amoebae have on the survival and growth of the bacteria (Fig. 3, top panel). In the presence of A. polyphaga, there was a steady and significant increase in the number of recovered viable P. multocida X-73 cells from 1.8 × 102 to 1.7 × 104 (Fig. 3, top panel, solid line). X-73 coinfected with A. polyphaga showed a limited lag phase, as our initial time point represents 2 h postinfection. In contrast, viable counts of P. multocida X-73 cells cultured alone progressively decreased to nondetectable levels after 20 h (Fig. 3, top panel, dashed line). The A. castellanii buffer used in the assays was a nutrient-limiting medium (36) that does not support the growth of P. multocida.
FIG. 3.
The top panel shows the viable counts of P. multocida X-73 alone (dashed line) or coinfecting A. polyphaga (solid line) recovered from coinfection assays. The middle panel shows the effects of cytochalasin D added to the A. polyphaga cells 1 h before the addition of X-73. The solid line depicts the A. polyphaga not treated with cytochalasin D, and the dashed line indicates the pretreated amoeba. Viable counts were taken at 4, 8, 12, 16, and 20 h postinfection. The bottom panel shows P. multocida strains P-1059 (dashed line) and PBA815 (solid line) recovered from coinfection assays with A. polyphaga. Viable counts were taken at 12, 18, and 24 h postinfection. As observed with X-73, there was a decline in the number of recoverable P. multocida cells when cultured without A. polyphaga (data not shown). All assays were conducted in triplicate on at least four separate occasions. The graphs were generated using the Systat10 program. Error bars represent the 75% confidence intervals in the averages of log CFUs recovered per ml.
The cell number increase in the first 10 h of the coinfection was assumed to be the initial internalization and subsequent growth of P. multocida within the amoebal cells (Fig. 3, top panel). After this initial period, there was approximately 1 log less recoverable P. multocida X-73 until 22 h postinfection. This decline may be attributed to the lysing of the amoebal cells by P. multocida, as cytotoxicity studies (see below) and electron micrographs provided evidence of P. multocida lysing and escaping the amoebae (Fig. 2D). Once P. multocida have lysed their amoebal host cells and exit into the surrounding medium, they become susceptible to the gentamicin treatment and the viable cell count decreases. This constant decrease was observed until 20 h postinfection, after which an increase in recoverable P. multocida was observed. Even though P. multocida was shown not to survive within A. castellanii buffer, the coinfection assays were conducted in the presence of gentamicin to ensure that the viable bacteria enumerated were derived from within the amoebal cells. This is further supported by both fluorescence and transmission electron microscopy that show the presence of P. multocida localized within the amoebal cells (Fig. 1 and 2).
In order to show that interactions and infections of free-living amoeba are not limited to A. polyphaga, we also conducted the same coinfection assays with the environmental amoeba H. vermiformis and found the interaction kinetics to be very similar to those observed with A. polyphaga (data not shown).
Cytotoxic effect of P. multocida.
Transmission electron microscope observations (Fig. 2D) and the kinetics of the P. multocida-amoebal interaction (Fig. 3, top panel) strongly suggested that the decrease seen in viable cell count (starting after 10 h postinfection) was a direct result of P. multocida lysing the amoebal host cells. To further confirm and quantify the degree of cytotoxicity, a nonradioactive cytotoxicity assay was conducted. Assay results indicated that, compared to amoebae in the absence of P. multocida, amoebae coinfected with the bacterium showed 33% (± 1.6%) lysis 18 h postinfection.
Effects of a cytoskeletal inhibitor on P. multocida infection.
To initiate studies of the specific interaction between P. multocida and the amoebal cells, the ability of P. multocida to infect the amoebae in the presence of the cytoskeletal inhibitor cytochalasin D was assessed. Cytochalasin D inhibits actin polymerization and therefore inhibits endocytosis. A. polyphaga cells were preincubated with cytochalasin D prior to their addition to a 24-h coinfection assay. Although it appears at first that cytochalasin D did have some inhibitory effect (Fig. 3, middle panel) on the intracellular growth of P. multocida, it is probable that this was not directly due to the inhibition of endocytosis but rather the effect cytochalasin D has on the morphology of the amoebal cells themselves. Moffat and Tompkins (36) reported that the inhibitor itself caused the amoebae to round up and lift up from the bottom of the assay wells, which resulted in the loss of amoeba during the assay procedure, resulting in fewer amoebae and thus fewer P. multocida X-73 cells able to be recovered. Microscopic observation of amoebal cells preincubated with cytochalasin D showed a rounded morphology (data not shown), indicating that the slight decrease in X-73 recovery (Fig. 3, middle panel) may not be due directly to the inhibition of endocytosis but rather the loss of amoebal cells during the assay.
Other P. multocida strains infect A. polyphaga.
In order to determine if other fowl-cholera-causing isolates of P. multocida could also enter into and survive within an amoeba host, we employed two other serogroup A P. multocida strains in standard coinfection assays. The interaction of strains P-1059, serotype 3, and PBA815, serotypes 3 and 4, were very similar to those seen with X-73 (Fig. 3, bottom panel).
DISCUSSION
Since the initial studies with Legionella pneumophila, bacterial-amoebal interactions have been observed with a number of pathogens including Chlamydia spp. (17), Mycobacterium spp. (14, 30, 46), Francisella tularensis (1), Burkholderia spp. (26, 35), and Salmonella spp. (47). These observations have enabled further understanding of bacterial environmental reservoirs, routes of transmission, and virulence mechanisms employed during host cell infection. Interactions between members of the Pasteurellaceae family and free-living amoebae have not been previously reported. This study has observed the interaction between fowl-cholera-causing P. multocida strains and free-living amoebae.
The interaction between P. multocida and amoebae shares many important characteristics observed when other pathogens infect and grow within amoebal cells. Under assay conditions, P. multocida X-73 cultured alone eventually resulted in no recoverable viable cells whereas a steady increase in X-73 viable counts was observed during coinfection with amoebae (Fig. 3, top panel). These recovered cells were presumed to be internalized within the amoebae, as the assay used gentamicin to eliminate noninternalized bacteria at any given time point. Furthermore, the bacterium was unable to survive within the assay medium alone and thus sustained survival of P. multocida could occur only in the presence of amoebae. These data were further supported by electron microscopy studies (Fig. 2). Internalized P. multocida cells were found residing in spacious membrane-bound vacuoles within the amoebal cytoplasm, which is a hallmark characteristic of bacteria that are able to infect and survive within amoebae (for a review, see reference 23). Such vacuoles differ from the tight-fitting vacuoles associated with bacteria ingested as a food source (35). Although the nature of the P. multocida-containing vacuoles has not yet been identified, two other observations support the possible replication of P. multocida within these vacuoles. Intracellular P. multocida examined via microscopy appeared to show the initiation of replication (Fig. 2B), and more importantly, mitochondria were found to be associated with the P. multocida-containing vacuoles (Fig. 2B and 2C). In both amoebae and macrophages, vacuoles that contain replicating L. pneumophila are associated with mitochondria and then endoplasmic reticulum (2, 21). In addition, F. tularensis (1), Simkania negevensis (28), and rickettsial species (19) were also observed in intraamoebal vacuoles associated with mitochondria. In light of the above observations, we propose that P. multocida, like other pathogens, be classified as an amoeba-resistant bacterium (23). According to Greub and Raoult (23), an amoeba-resistant bacterium, by definition, is able to enter, multiply within and exit its amoebal host.
The cytoskeletal inhibitor cytochalasin D had no dramatic effect on the entry or replication of P. multocida within the amoebal cell. In contrast, two reports demonstrated that the invasion of P. multocida into bovine endothelial cells and MDCK cells could be reduced up to 60% by pretreatment with cytochalasin D (20, 38). Interestingly, these conflicting results were also observed with L. pneumophila, in that cytochalasin D had little effect on invasion into amoebal cells (29, 36) but it inhibited the invasion of host cells (16, 29). Consequently, this suggests that, like L. pneumophila, P. multocida does not enter the amoebae via normal endocytosis and may instead use an alternative pathway.
Most bacterial pathogens able to infect amoebae have also been shown to invade and infect host cells (1, 14, 17, 23, 26). The invasive ability of P. multocida has long been debated, and only a few studies have investigated the intracellular potential of this pathogen. Virulent atrophic rhinitis and fowl-cholera-causing strains were reported to invade and survive within various host cell lines such as turkey kidney epithelial cells and avian mononuclear phagocytic cells (20, 31, 38, 49). P. multocida rabbit strains were shown to enter nasal epithelial and endothelial cells and reside within membrane-bound vacuoles in the cytoplasms of these cells (3). The observed relationship between amoebae and P. multocida further supports the above reports and strongly indicates that this pathogen is capable of intracellular invasion and survival within host cells. Furthermore, like L. pneumophila (21, 45), genes utilized for invasion by P. multocida may be similar for both the host cell and amoebae.
The importance of free-living amoebae in soil and water ecosystems has been well established. However, increasing attention has now been focused on the potential role of amoeba as reservoirs and vehicles for the transmission of bacterial pathogens. P. multocida has been found in a number of aquatic and soil environments (4, 10, 37, 43, 48). Although the bacterium has been recovered from the soil and water around enzootic fowl cholera sites (6, 37, 43), it has been proposed that these ecosystems do not play a major role in harboring the organism (42). However, since P. multocida can be found in the same environments as the amoebae, interactions between the two are likely. Further work is needed to establish if amoebae within the environment harbor virulent P. multocida and if this allows bacterial transmission to the host in a manner similar to that in which L. pneumophila is transmitted to humans via infected amoebae (11). Nevertheless, these data provide evidence that P. multocida cells enter, survive, replicate within, and escape from free-living amoebae. These data enhance our understanding of the environmental reservoirs and modes of transmission of this important animal pathogen.
Acknowledgments
This work was partially supported by a SEED grant from the U.S. Department of Agriculture and Cooperative State Research, Education and Extension Service.
We thank Gregory Mayer, University of Wisconsin—Parkside, for his advice on statistical analysis and Ben Adler, Monash University, Australia, for his helpful suggestions for the manuscript. Heather Owen of the University of Wisconsin—Milwaukee Electron Microscope Laboratory is gratefully acknowledged for aiding in preparing all microscopy work and for the use of all the microscopes employed in this work.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Haddawi, M. H., S. Jasni, M. Zamri-Saad, A. R. Mutalib, R. Son, and A. R. Sheikh-Omar. 2000. Ultrastructural observation of nasal and pulmonary intracellular Pasteurella multocida A:3 in rabbits. Vet. Res. Commun. 24:153-167. [DOI] [PubMed] [Google Scholar]
- 4.Backstrand, J. M., and R. G. Botzler. 1986. Survival of Pasteurella multocida in soil and water in an area where avian cholera is enzootic. J. Wildl. Dis. 22:257-259. [DOI] [PubMed] [Google Scholar]
- 5.Barker, J., and M. R. W. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 6.Boltzer, R. G. 1991. Epizootiology of avian cholera in wildfowl. J. Wildl. Dis. 3:367-395. [DOI] [PubMed] [Google Scholar]
- 7.Bosch, M., E. Garrido, M. Llagostera, A. M. P. de Rozas, I. Badiola, and J. Barbe. 2002. Pasteurella multocida exbB, exbD and tonB genes are physically linked but independently transcribed. FEMS Microbiol. Lett. 210:201-208. [DOI] [PubMed] [Google Scholar]
- 8.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2004. Genomic-scale analysis of Pasteurella multocida gene expression during growth within liver tissue of chickens with fowl cholera. Microbes Infect. 6:290-298. [DOI] [PubMed] [Google Scholar]
- 10.Bredy, J. P., and R. G. Botzler. 1989. The effects of six environmental variables on Pasteurella multocida populations in water. J. Wildl. Dis. 25:232-239. [DOI] [PubMed] [Google Scholar]
- 11.Brieland, J. K., J. C. Fantone, D. G. Remick, M. LeGendre, M. McClain, and N. C. Engleberg. 1997. The role of Legionella pneumophila-infected Hartmannella vermiformis as an infectious particle in a murine model of Legionnaire's disease. Infect. Immun. 65:5330-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter, G. R., and M. C. L. De Alwis. 1989. Haemorrhagic septicaemia, p. 131-160. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press Limited, London, England.
- 13.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Alwis, M. C. L. 1992. Pasteurellosis in production animals: a review, p. 11-22. In B. E. Patten, T. L. Spencer, R. B. Johnson, D. Hoffmann, and L. Lehane (ed.), Pasteurellosis in production animals. ACIAR, Canberra, Australia.
- 16.Elliott, J. A., and W. C. Winn, Jr. 1986. Treatment of alveolar macrophages with cytochalasin D inhibits uptake and subsequent growth of Legionella pneumophila. Infect. Immun. 51:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essig, A., M. Heinemann, U. Simnacher, and R. Marre. 1997. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl. Environ. Microbiol. 63:1396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields, B. S., J. M. Barbaree, G. N. Sanden, and W. E. Morrill. 1990. Virulence of a Legionella anisa strain associated with Pontiac fever: an evaluation using protozoan, cell culture, and guinea pig model. Infect. Immun. 58:3139-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K. H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero, M., L. De Martino, U. Pagnini, M. Pisciotta, and E. Galdiero. 2001. Interactions between bovine endothelial cells and Pasteurella multocida: association and invasion. Res. Microbiol. 152:57-65. [DOI] [PubMed] [Google Scholar]
- 21.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glisson, J. R. 1992. Current poultry health problems. Poult. Dig. November:10-16. [Google Scholar]
- 23.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harb, O. S., and J. A. Kwaik. 2000. Interaction of Legionella pneumophila with protozoa provides lessons. ASM News 66:609-616. [Google Scholar]
- 25.Harper, M., J. D. Boyce, I. W. Wilkie, and B. Adler. 2003. Signature-tagged mutagenesis of Pasteurella multocida identifies mutants displaying differential virulence characteristics in mice and chickens. Infect. Immun. 71:5440-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inglis, T. J., P. Rigby, T. A. Robertson, N. S. Dutton, M. Henderson, and B. J. Chang. 2000. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect. Immun. 68:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, M. R., V. M. Factor, and S. S. Thorgeirsson. 1998. Regulation of Cyclin G1 during murine hepatic regeneration following Dipin-induced DNA damage. Hepatology 28:537-546. [DOI] [PubMed] [Google Scholar]
- 28.Kahane, S., B. Dvoskin, M. Mathias, and M. G. Friedman. 2001. Infection of Acanthamoeba polyphaga with Simkania negevensis and S. negevensis survival within amoebal cysts. ppl. Environ. Microbiol. 67:4789-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King, C. H., B. S. Fields, E. B. Shotts, Jr., and E. H. White. 1991. Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect. Immun. 59:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishna-Prasad, B. N., and S. K. Gupta. 1978. Preliminary report on engulfment and retention of mycobacteria by trophozoites of axenically grown Acanthamoeba castellanii Douglas. Curr. Sci. 47:245-247. [Google Scholar]
- 31.Lee, M. D., R. E. Wooley, and J. R. Glisson. 1994. Invasion of epithelial cell monolayers by turkey strain Pasteurella multocida. Avian Dis. 38:72-77. [PubMed] [Google Scholar]
- 32.Le Gouill, C., J. L. Parent, M. Rola-Pleszczynski, and J. Stankova. 1994. Analysis of recombinant plasmids by a modified alkaline lysis method. Anal. Biochem. 219:164. [DOI] [PubMed] [Google Scholar]
- 33.Lippmann, J. E., E. H. Froeliger, and P. M. Fives-Taylor. 1999. Use of the Actinobacillus actinomycetemcomitans leukotoxin promoter to drive expression of the green fluorescent protein in an oral pathogen. Oral Microbiol. Immunol. 15:321-325. [DOI] [PubMed] [Google Scholar]
- 34.Ly, T. M., and H. E. Muller. 1990. Ingested Listeria monocytogenes survive and multiply in protozoa. J. Med. Microbiol. 33:51-54. [DOI] [PubMed] [Google Scholar]
- 35.Marolda, C. L., B. Hauroder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 36.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, J. I., and C. J. Brand. 1984. Persistence of Pasteurella multocida in Nebraska wetlands under epizootic conditions. J. Wildl. Dis. 20:90-94. [DOI] [PubMed] [Google Scholar]
- 38.Rabier, M. J., N. K. Tyler, N. J. Walker, L. M. Hansen, D. C. Hirsh, and F. Tablin. 1997. Pasteurella multocida enters polarized epithelial cells by interacting with host F-actin. Vet. Microbiol. 54:343-355. [DOI] [PubMed] [Google Scholar]
- 39.Rhoades, K. R., and R. B. Rimler. 1989. Fowl cholera, p. 95-113. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press Limited, London, England.
- 40.Rimler, R. B. 1990. Comparisons of Pasteurella multocida lipopolysaccharides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to determine relationship between group B and E hemorrhagic septicemia strains and serologically related group A strains. J. Clin. Microbiol. 28:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuel, M. D., D. J. Shadduck, and D. R. Goldberg. 2004. Are wetlands the reservoir for avian cholera? J. Wildl. Dis. 40:377-382. [DOI] [PubMed] [Google Scholar]
- 43.Samuel, M. D., D. J. Shadduck, D. R. Goldberg, M. A. Wilson, D. O. Joly, and M. A. Lehr. 2003. Characterization of Pasteurella multocida isolates from wetland ecosystems during 1996 to 1999. J. Wildl. Dis. 69:798-807. [DOI] [PubMed] [Google Scholar]
- 44.Samuel, M. D., D. R. Goldberg, D. J. Shadduck, J. I. Price, and E. G. Cooch. 1997. Pasteurella multocida serotype 1 isolated from a Lesser Snow Goose: evidence of a carrier state. J. Wildl. Dis. 33:332-335. [DOI] [PubMed] [Google Scholar]
- 45.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tezcan-Merdol, D., M. Ljungstrom, J. Winiecka-Krusnell, E. Linder, L. Engstrand, and M. Rhen. 2004. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 70:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson, C. M. A., N. Chanter, and C. M. Wathes. 1992. Survival of toxigenic Pasteurella multocida in aerosols and aqueous liquids. Appl. Environ. Microbiol. 58:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truscott, W. M., A. T. Cheung, and D. C. Hirsh. 1990. Reduced microbicidal activity of peripheral mononuclear phagocytic cells infected with Pasteurella multocida. Vet. Microbiol. 21:283-290. [DOI] [PubMed] [Google Scholar]
- 50.Wilkie, I. W., S. E. Grimes, D. O'Boyle, and A. J. Frost. 2000. The virulence and protective efficacy for chickens of Pasteurella multocida administered by different routes. Vet. Microbiol. 72:57-68. [DOI] [PubMed] [Google Scholar]