Abstract
2-Amino-3-methylimidazo[4,5-f]quinoline (IQ) is a mutagenic/carcinogenic compound formed from meat and fish during cooking. Following ingestion, IQ is metabolized mainly by liver xenobiotic-metabolizing enzymes, but intestinal bacteria may also contribute to its biotransformation. The aim of this study was to investigate the metabolism of IQ by the human intestinal microbiota. Following incubation of IQ (200 μM) under anoxic conditions with 100-fold dilutions of stools freshly collected from three healthy volunteers, we quantified residual IQ by high-pressure liquid chromatography (HPLC) analysis and characterized the production of IQ metabolites by in situ 1H nuclear magnetic resonance (1H-NMR) spectroscopic analysis of crude incubation media. In addition, we looked for IQ-degrading bacteria by screening collection strains and by isolating new strains from the cecal contents of human-microbiota-associated rats gavaged with IQ on a regular basis. HPLC and 1H-NMR analyses showed that the three human microbiota degraded IQ with different efficiencies (range, 50 to 91% after 72 h of incubation) and converted it into a unique derivative, namely, 7-hydroxy-IQ. We found 10 bacterial strains that were able to perform this reaction: Bacteroides thetaiotaomicron (n = 2), Clostridium clostridiiforme (n = 3), Clostridium perfringens (n = 1), and Escherichia coli (n = 4). On the whole, our results indicate that bacteria belonging to the predominant communities of the human intestine are able to produce 7-hydroxy-IQ from IQ. They also suggest interindividual differences in the ability to perform this reaction. Whether it is a metabolic activation is still a matter of debate, since 7-hydroxy-IQ has been shown to be a direct-acting mutagen in the Ames assay but not carcinogenic in laboratory rodents.
Evidence is accumulating that heterocyclic aromatic amines (HAs), which are food-borne carcinogens formed from amino acids in meats during cooking, may be involved in the etiology of colon cancer (10). The genotoxic/carcinogenic effect of HAs is closely related to a highly complex metabolism involving xenobiotic-metabolizing enzymes that generate very reactive metabolites as well as detoxified derivatives (17, 18). The endogenous metabolism of HAs by the digestive system has been extensively studied (15, 30, 32). On the other hand, the involvement of the intestinal microbiota in the digestive fate of HAs remains underinvestigated (19), although around 10% of the ingested HAs may reach the colon in their native form and thus come into contact with the resident microbiota (21, 34). Direct binding of HAs to the cell walls of intestinal bacteria has been reported and is currently considered a detoxification mechanism, since it prevents absorption of HAs through the intestinal mucosa (19, 23, 41). On the other hand, results of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ)-induced genotoxic assays in germfree and conventional rodents show that the presence of intestinal microbiota is essential to the induction of DNA damage in the colon and liver cells (13, 16). These findings suggest that intestinal microbiota take part in the bioconversion of HAs into harmful metabolites. Indications exist that hydrolysis of HA-glucuronides by bacterial β-glucuronidase (EC 3.2.1.31) may release mutagenic intermediates (26). Although there is, as yet, no definitive evidence of the involvement of this reaction in vivo, it may account for the higher DNA damage observed by Kassie et al. (16) in the colonocytes and hepatocytes of conventional rats versus human-microbiota-associated (HMA) companions; indeed, it is well known that β-glucuronidase activity is naturally higher in the rat than in the human intestinal microbiota (5). Information on the bacterial metabolism of native HAs is still scarce. The most detailed studies to date were conducted by Wilkins and collaborators, who reported a series of investigations on this topic in the 1980s. They incubated mixed human feces under anoxic conditions with [14C]IQ (Fig. 1), extracted the culture medium with blue cotton, and analyzed the metabolite profile of the extract by thin-layer chromatography; the major metabolite was 7-OH-IQ (Fig. 1), the identity of which was ascertained by 1H nuclear magnetic resonance (1H-NMR) spectroscopy and mass spectrometry (MS) (1). This compound was subsequently detected in human feces following consumption of fried meat (35).
FIG. 1.
Structures of IQ and its 7-hydroxy derivative.
In the present study, we investigated the metabolism of IQ by human intestinal microbiota by using 1H-NMR spectroscopic analysis of unprocessed incubation media to detect potential metabolites in an exhaustive fashion. Indeed, 1H-NMR spectroscopy of crude biological samples allows measurement of a wide range of molecules simultaneously and without a priori hypotheses concerning their chemical structure; it has proved, on many occasions, to be a powerful tool to analyze the microbial degradation of xenobiotics (7, 8). Due to the detection limit of this analytical method, around 1 μM, we used a relatively high concentration of IQ, i.e., 100 to 200 μM, to ensure identification and quantification of IQ metabolites. Interindividual differences occur with regard to the species composition (31) and the metabolic activities (11, 20) of the human intestinal microbiota. Therefore, we examined the bioconversion potentials of fecal samples collected from different subjects. Eventually, we looked for individual bacteria able to metabolize IQ by screening representatives of the gut microbiota belonging to culture collections and by isolating new strains. On the assumption that IQ-degrading microbial populations may be favored by a continuous exposure to the substrate, the isolation procedure was performed on feces of HMA rats consuming IQ on a regular basis.
MATERIALS AND METHODS
Chemicals.
IQ was obtained from Research Chemicals Inc. (Toronto, Canada). For incubation purposes, it was dissolved in (methyl-sulfoxide)-d6 and subsequently diluted using phosphate buffer (0.2 M, pH 7.0) with added yeast extract (0.2 g/liter) (PYE buffer); this stock solution (10 mM) was filter sterilized (Millex-LG 0.22-μm filter; Millipore, St-Quentin-en-Yvelines, France) before use. For gavage of HMA rats, IQ was suspended in corn oil (Sigma, St-Quentin-Fallavier, France) and sterilized by autoclaving (120°C, 20 min); this process does not affect the concentration of IQ (data not shown).
The constituents of the culture media, namely, tryptone, brain heart infusion (BHI), Bi Tek agar, and yeast extract, were obtained from Difco (Le Pont-de-Claix, France). Horse blood came from BioMérieux (Marcy-l'Etoile, France), and glycerol was purchased from VWR (Fontenay-sous-Bois, France). Acetonitrile for high-pressure liquid chromatography (HPLC) analyses came from Merck (Nogent-sur-Marne, France). Hemin and all other salts and amino acids came from Sigma. Tetradeuterated sodium trimethylsilylpropionate and deuterium oxide used for NMR analyses were purchased from Eurisotop (Saint-Aubin, France).
Collection and preparation of human fecal samples.
Fresh stools from three healthy volunteers between 26 and 44 years of age were used. Donors were on a Western-type diet, and none had history of digestive pathology or had received antibiotics for the last 3 months. Stools collected in sterile plastic boxes were kept under anoxic conditions by using Anaerocult A (Merck) and stored at 4°C for a maximum of 6 h before processing. They were transferred into an anaerobic glove box, where they were diluted 100-fold in sterile PYE buffer and thoroughly homogenized with an Ultra-Turrax blender. Fecal suspensions were then incubated with IQ as described in “Incubation conditions for bacterial resting-cell suspensions” below.
Strains from culture collections.
Nine strains were chosen from the collection of Unité d'Ecologie et de Physiologie du Système Digestif (INRA, Jouy-en-Josas, France). All of them originated from human feces or intestinal contents and had been purchased from the American Type Culture Collection or isolated locally (Table 1). They were strictly anaerobic gram-negative bacilli (Bacteroides), gram-positive bacilli (Bifidobacterium, Clostridium, Eggertella, and Eubacterium), and gram-positive cocci (Ruminococcus). The cells were stored deep-frozen at −80°C as stock suspensions in appropriate media supplemented with sterile glycerol (20%, vol/vol). They were grown in modified BHI broth (BHI, 37 g/liter; yeast extract, 5 g/liter; cysteine, 0.5 g/liter; hemin, 5 mg/liter; pH 7.0), except for Eggertella lenta, which was grown in modified TY broth (tryptone, 30 g/liter; yeast extract, 20 g/liter; cysteine, 0.5 g/liter; hemin, 5 mg/liter; pH 7.0) with added arginine (1 g/liter). Cultures were incubated at 37°C in an anaerobic glove box for 10 h. After incubation, 50-ml volumes were collected and centrifuged (8,000 × g, 10 min, 4°C). Supernatants were discarded; cells were washed twice with PYE buffer and resuspended in the same buffer. Each resting-cell suspension was then incubated with IQ as described in “Incubation conditions for bacterial resting-cell suspensions” below.
TABLE 1.
Abilities of individual bacterial strains originating from the human digestive tract to convert IQ to 7-OH-IQa
| Bacterial species and strain | Origin | Source or reference | % of initial IQ degradedb |
|---|---|---|---|
| Bacteroides thetaiotaomicron | |||
| ATCC 29148 | Human feces | 29 | 74 |
| 9-3JE | HMA rat cecumc | This studyd | 52 |
| Bacteroides vulgatus ATCC 8482 | Human feces | 29 | 0 |
| Bifidobacterium bifidum B536 | Infant feces | 14 | 0 |
| Bifidobacterium longum ATCC 15707 | Adult intestine | 29 | 0 |
| Clostridium clostridiiforme | |||
| 5-1JD1 | HMA rat cecum | This study | 76 |
| 9-2JB | HMA rat cecum | This study | 60 |
| 10-2JD2 | HMA rat cecum | This study | 50 |
| Clostridium nexile ATCC 27757 | Human feces | 29 | 0 |
| Clostridium perfringens G22 | Human feces | F. Marcille, unpublished data | 100 |
| Eggertella lenta γ12 | HMA rat cecum | 2 | 0 |
| Escherichia coli | |||
| 2-1JC | HMA rat cecum | This study | 61 |
| 8-1JA | HMA rat cecum | This study | 80 |
| 8-1JC | HMA rat cecum | This study | 61 |
| 10-2JB | HMA rat cecum | This study | 47 |
| Eubacterium rectale ATCC 33656 | Human feces | 29 | 0 |
| Ruminococcus gnavus FRE1 | Human feces | 12 | 0 |
For incubation, resting-cell suspensions in PYE buffer were supplemented with 100 μM IQ (anoxic conditions, 37°C, 150 rpm).
At the end of incubation (72 h), the IQ concentration was determined by HPLC analysis.
HMA, human microbiota associated.
Among the 135 strains isolated from the mixed cecal contents of HMA rats and assayed for IQ degradation, only the 8 biodegradative strains are indicated in this table.
Human-microbiota-associated rats.
Two 3-month old, germfree, male Fischer 344 rats were provided by the breeding facility of Unité d'Ecologie et de Physiologie du Système Digestif (INRA, Jouy-en-Josas, France). Throughout the study, they were housed in a flexible-film isolator (La Calhène, Vélizy, France) and kept in a room that was maintained at constant temperature (21°C ± 1°C) and humidity (50% ± 5%) with a 12-h light/dark cycle. They were given free access to autoclaved tap water and a pelleted semisynthetic diet (22) sterilized by gamma irradiation at 45 kGy (SAFE-U.A.R., Augy, France). Inoculation with the fecal microbiota of one of the human subjects was performed as described by Roland et al. (25). After 3 weeks of acclimatization to the diet and to the bacterial status treatments, rats were gavaged every 2 days for 2 weeks with a sterile suspension of IQ in corn oil (90 mg/kg) (16). Rats were subsequently killed by CO2 asphyxiation. Cecal contents were collected, pooled, and immediately transferred into an anaerobic glove box, where they were diluted 100 times in sterile PYE buffer and thoroughly homogenized with an Ultra-Turrax blender. A part of the cecal suspension was incubated with IQ as described in “Incubation conditions for bacterial resting-cell suspensions” below; the other part was used for isolation of IQ-degrading bacterial strains as described in “Isolation and identification of IQ-degrading bacterial strains” below.
All procedures were carried out in accordance with the European guidelines for the care and use of laboratory animals.
Isolation and identification of IQ-degrading bacterial strains.
The 100-fold suspension derived from the mixed cecal contents of HMA rats gavaged with IQ was diluted using serial 10-fold dilutions (10−2 to 10−10) in modified TY broth supplemented with IQ (100 μM). Dilutions were incubated at 37°C under anoxic conditions for 3 days and assayed at 24-h intervals for residual IQ. At the same time intervals, samples from all dilutions were spread onto modified TY agar plates supplemented with horse blood (5%, vol/vol) and with IQ (100 μM) to maintain a continuous exposure of the bacteria to the substrate. Following incubation at 37°C under anoxic conditions, five colonies per plate that differed, whenever possible, in size, shape, and color were picked, subcultured in modified TY broth supplemented with IQ (100 μM), and then stored as stock cultures at −80°C after addition of glycerol (40%, vol/vol). One hundred thirty-five viable strains were obtained during this process; they were named after the dilution (10−2, 10−3, 10−4, etc.) and the day of incubation (1, 2, or 3) from which they originated. Subcultures intended for strain storage were sampled at the initial and final times of incubation (3 days) for analysis of residual IQ in order to detect IQ-degrading strains. Further investigation of IQ metabolism by the biodegradative strains was performed on resting-cell suspensions prepared in PYE buffer and incubated with IQ (for details, see “Strains from culture collections” above and “Incubation conditions for bacterial resting-cell suspensions” below).
Identification of the biodegradative strains was performed phenotypically by microscopic examination, Gram staining, and determination of biochemical characteristics (API Systems, BioMérieux) and genetically by sequence comparison of the amplification products of the 16S rRNA genes. Total DNA was extracted from 48-h cultures in modified TY broth by using the Wizard genomic DNA purification kit (Promega, Charbonnières, France). Primers W001 (3, 6) and 23S1 (GenBank accession no. J01695) were used to amplify the 16S rRNA-encoding genes, including the intergenic region located between the 16S rRNA and 23S rRNA genes. The PCR product (about 2.3 kb) was purified using the QIAGEN QIAquick PCR purification kit (QIAGEN, Courtaboeuf, France). The sequence reaction was performed by using the primers SP3, SP4, and SP5 (6, 36) and the ABI Prism Big Dye Terminator version 2.0 kit (Applera, Courtabœuf, France). Sequences were analyzed using a 96-capillary 3700 sequencer (Perkin-Elmer, Courtaboeuf, France). Each clone sequence was identified by comparison with the Ribosomal Database Project II (http://rdp.cme.msu.edu).
Incubation conditions for bacterial resting-cell suspensions.
Bacterial resting-cell suspensions in PYE buffer were supplemented with IQ and transferred into glass vials that were tightly closed with butyl rubber stoppers and sealed with aluminum caps to maintain an anoxic environment. The vials were subsequently incubated in a shaking water bath (150 rpm) at 37°C for 72 h. The IQ concentration was 200 μM for resting-cell suspensions derived from human feces and the cecal contents of HMA rats and 100 μM for resting-cell suspensions of individual strains. Incubation media were sampled at intervals by sterile puncture through the butyl rubber stoppers. Samples were centrifuged (8,000 × g, 10 min, 4°C), and the supernatants were kept at −20°C until HPLC and 1H-NMR analyses.
Three types of controls were used, namely, (i) sterile PYE buffer supplemented with IQ to check the stability of the substrate over time, (ii) resting-cell suspensions devoid of IQ to check the absence of coeluting peaks, and (iii) autoclaved resting-cell suspensions (120°C, 20 min) supplemented with IQ to ascertain that the disappearance of the substrate could be attributed to the metabolic activity of viable cells and not to a passive adsorption on bacterial cell walls (23, 41).
HPLC analysis.
IQ and its hydroxy derivative, 7-OH-IQ, were analyzed by HPLC using the method described by Rafter and Gustafsson (24) with slight modifications. Briefly, culture and resting cell supernatants were injected with an 2690 autosampler (Waters, Milford, MA) onto a reversed-phase column packed with LiChrospher 100 RP-18e (5 μm, 25 cm) (Merck) and equipped with a LiChrospher 100 RP-18e precolumn (5 μm) (Merck). Samples were eluted at 1.0 ml/min for 20 min with a 0 to 30% acetonitrile linear gradient in 0.02 M phosphate buffer. IQ and 7-OH-IQ were monitored at 252 nm using a 996 photodiode array detector (Waters). Under these conditions, IQ eluted at 10.6 min, while 7-OH-IQ eluted at 9.2 min. Data were collected and peaks integrated using the Millenium32 chromatography manager software (Waters). Identification of IQ was based on the identities of the retention time and absorption spectrum with those of an authentic standard (Research Chemicals Inc.), and quantification was achieved using a standard curve from 0 to 500 μM. 7-OH-IQ is not commercially available; nevertheless, the identity of the peak could be ascertained by comparing its absorption spectrum with those reported in the literature (4). Furthermore, relative amounts of 7-OH-IQ produced over time and between samples could be compared by integrating the peak areas.
1H-NMR analysis.
All 1H-NMR spectra were recorded on a Bruker Avance 500 spectrometer at 500.13 MHz at 298 K, using 5-mm-diameter tubes. Water resonance was suppressed by the classical double-pulsed field gradient echo sequence, WATERGATE, and 256 scans were collected (relaxation delay, 4 s; acquisition time, 4.67 s; spectral window of 7,003 Hz; 65,536 data points). A 1-Hz line broadening was applied before Fourier transformation, and a baseline correction was performed on spectra before integration with Bruker software. Tetradeuterated sodium trimethylsilylpropionate constituted an internal reference for chemical shift (0 ppm) and quantification. The method for quantification of IQ and metabolites was as previously described (7). Several 1H-NMR spectra were recorded to assign signals coming from bacterial cells and/or PYE buffer (mainly yeast extract). A calibration curve of IQ chemical shifts versus concentration was prepared before interpretation of kinetics. Moreover, comparison of water resonance suppression by WATERGATE sequence and by presaturation was carried out to check that quantitative conditions were reached. Whatever the method used, areas of signals were underestimated, due to long relaxation properties, but the shift was reproducible and equal for IQ and 7-OH-IQ.
2D 1H-NMR.
Two-dimensional (2D) phase-sensitive (time proportional phase incrementation) total correlation spectroscopy (TOCSY) experiments with water resonance suppression by a WATERGATE sequence (put at the end of the sequence) were used to assign all members of a coupled spin network. Spectral widths were adjusted in both dimensions to encompass all 1H signals of interest. The “mixing period” (corresponding to several cycles of MLEV-17 spinlock sequence) was 20 to 90 ms. The responses of eight scans for each of 512 t1 increments were acquired. Zero-filling in t1 and sine window function in both dimensions were applied prior to 2D Fourier transformation.
MS analysis.
MS analysis was performed using an HP 5989 B mass spectrometer equipped with an atmospheric pressure chemical ionization interface. Supernatants were diluted in methanol or acetonitrile. Sample injections were carried out by a Rheodyne valve type at 100 μl/min. The solvent and analytes were ionized through the combined action of applying a high electric field (1.5 kV) and a pneumatic nebulization (80 lb/in2 nitrogen flux). The charged droplets thus formed were heated (375°C) and shrank due to evaporation. These highly charged microdroplets were directed toward the quadrupole analyzer through a charged capillary (100 V). The data were acquired in full-scan mode in a range of 20 to 300 amu. Both positive and negative modes were used, but only positive mode allowed observation of IQ and 7-OH-IQ peaks.
RESULTS
Metabolism of IQ by human feces and by the cecal contents of HMA rats (HPLC analysis).
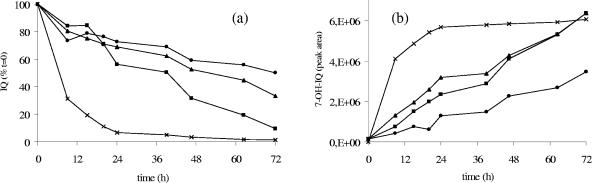
Figure 2 depicts the kinetics of IQ degradation and 7-OH-IQ formation measured by HPLC analysis in the resting-cell incubations prepared with the feces of the three human volunteers and with the mixed cecal contents of the two HMA rats. All three human feces degraded IQ, although with different efficiencies. Indeed, the amount of IQ degraded over 72 h ranged from 50 to 90% of the initial quantity. Furthermore, the t50s (the times required for degradation of half of the initial substrate amount) were 39, 51, and 72 h for the high-, intermediate-, and low-degrading microbiota, respectively. HMA rats were inoculated with the fecal microbiota of the subject with high degrading capacity. In this regard, it must be emphasized that the 100-fold dilution of the mixed cecal contents of those animals degraded IQ with a higher efficiency than the original microbiota; the substrate was almost completely degraded within 72 h, and the t50 was only 6 h.
FIG. 2.
Kinetics of IQ degradation (a) and 7-OH-IQ formation (b) in resting-cell suspensions derived from human feces (▪, •, and ▴) and from the mixed cecal contents of HMA rats (×). The initial IQ concentration was 200 μM, and incubation was in a shaking water bath (anoxic conditions, 37°C, 72 h, 150 rpm). IQ and 7-OH-IQ concentrations were determined by HPLC analysis.
7-OH-IQ formation accompanied IQ degradation in each resting-cell incubation. Interindividual differences between the kinetics of 7-OH-IQ formation largely reflected those between the kinetics of IQ degradation. The low-degrading human microbiota produced the lowest quantity of 7-OH-IQ. Nevertheless, peak areas of the hydroxy derivative measured in the incubation media of the medium- and high-degrading human microbiota and of the HMA rats' mixed cecal contents all tended to be similar at the end of incubation. This phenomenon may be due to the loss of linearity between 7-OH-IQ amount and peak area at high concentrations.
Metabolism of IQ by individual bacterial strains (HPLC analysis).
Among the nine collection strains that were assayed in the present experiment, only two strains were able to degrade IQ under resting-cell conditions as shown by HPLC analysis (Table 1). These strains were Bacteroides thetaiotaomicron ATCC 29148 and Clostridium perfringens G22, which consumed 74 and 100% of the initial amount of IQ (100 μM), respectively.
The serial 10-fold dilutions of the mixed cecal contents of HMA rats all degraded IQ to some extent (range, 50 to 100%) (data not shown). Among the 135 strains isolated from these dilutions, only eight strains were able to degrade IQ as measured by HPLC analysis of culture supernatants (data not shown). This ability was confirmed under resting-cell conditions similar to those applied to the collection strains; the amounts of IQ degraded by each strain after 72 h of incubation are presented in Table 1. The biodegradative strains were isolated mainly from the highest cecal dilutions, namely, 10−8 to 10−10, and hence belong to the predominant microbial communities. They were members of the species Bacteroides thetaiotaomicron (n = 1), Clostridium clostridiiforme (n = 3), and Escherichia coli (n = 4).
In situ 1H-NMR and MS analyses of human feces, HMA rats' cecal contents, and individual bacterial strains incubated with IQ.
Before interpretation of the kinetics monitored by in situ 1H-NMR, assignment of signals coming from the bacterial cells or from the PYE buffer used for resting-cell incubations (especially those from yeast extract) was performed. This was done by comparing control samples, namely, resting-cell suspensions devoid of IQ, sterile PYE buffer supplemented with IQ, and autoclaved resting-cell suspensions, with supernatants collected from experimental samples. Because IQ chemical shifts change with concentration, calibration curves of IQ chemical shifts versus concentration had to be prepared.
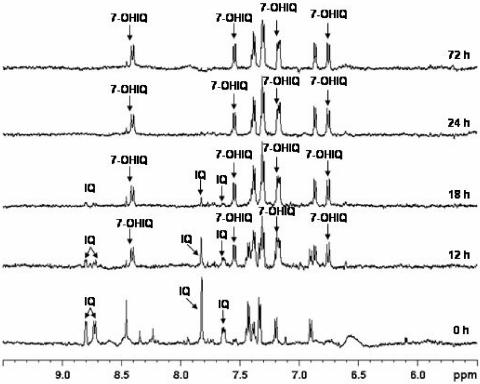
Sometimes signals from “background” compounds overlapped signals of interest; in such cases, 2D 1H-1H TOCSY experiments were performed to increase the resolution and thus unambiguously assign signals belonging to IQ and IQ transformation products. This approach allowed us to observe solely one transformation product, whether the sample was human feces, cecal contents of HMA rats, or individual strains, and regardless of the initial concentration of IQ (100 μM versus 200 μM). This compound was identified as 7-OH-IQ from analysis of 2D TOCSY spectral patterns and from analysis of mass spectra of supernatants recorded for several samples without any purification. Such mass spectra showed two main peaks at m/z 199 and m/z 215, corresponding to the [M + H]+ adducts of IQ and 7-OH-IQ, respectively. The complete assignment of 1H NMR spectra is shown in Fig. 3.
FIG. 3.
Examples of 1H-NMR spectra. Data are kinetics of IQ bioconversion into 7-OH-IQ by a resting-cell suspension derived from the mixed cecal contents of HMA rats. The initial IQ concentration was 200 μM, and incubation was in a shaking water bath (anoxic conditions, 37°C, 72 h, 150 rpm). Signals not marked by an arrow correspond to compounds of the incubation medium (PYE buffer) or to compounds formed by bacterial cells independently of IQ metabolism.
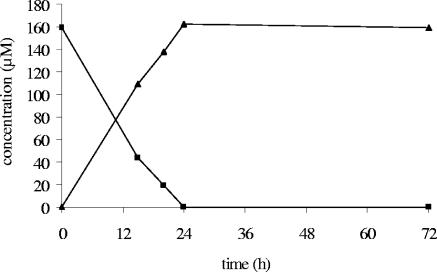
On this basis, and insofar as signals were well resolved, quantification of IQ and 7-OH-IQ could be achieved. This was the case for resting-cell incubations of individual bacterial strains and of the mixed cecal contents derived from HMA rats. It was found that hydroxylation at position 7 was the sole transformation occurring under these conditions; 7-OH-IQ accumulated over time and accounted for all the IQ degraded (Fig. 4). It was not possible to precisely quantify IQ and 7-OH-IQ in resting-cell incubations of human feces since their signals were broadened, probably because of an interaction with unknown biomacromolecules.
FIG. 4.
Kinetics of IQ (▪) bioconversion into 7-OH-IQ (▴) determined by in situ 1H-NMR analysis of resting-cell suspensions derived from the mixed cecal contents of HMA rats. Incubation was in a shaking water bath (anoxic conditions, 37°C, 72 h, 150 rpm).
DISCUSSION
By analyzing crude incubation media with 1H-NMR spectroscopy, we have shown that human fecal microbiota and individual bacteria isolated from them can transform IQ into a hydroxy derivative, namely, 7-OH-IQ. These findings agree with the original discovery by Bashir et al. (1), who found 7-OH-IQ as the major detectable metabolite following incubation of IQ with human fecal microbiota. However, since those authors analyzed the metabolite profile on a selective extract of the incubation medium, they could not rule out the possibility that other derivatives could have been released yet not recovered in the extract (1). In this connection, Carman et al. (4) hypothesized that a reductive deamination of IQ could occur, since removal of amino groups is a common reaction among the intestinal microbiota (28). Today, in situ 1H-NMR spectroscopy is one of the most powerful analytical methods for providing structural and quantitative information on the metabolic state of a biological medium. Therefore, we can assert here that 7-OH-IQ is unambiguously the unique metabolite produced by bacterial conversion of IQ, at least under our experimental conditions. According to Carman et al., the formation of 7-OH-IQ from IQ is a reversible reaction; indeed, incubation of IQ with the most active bacterial strain that they isolated, Eubacterium moniliforme VPI 13480, always led to an equilibrium in which IQ and its 7-hydroxy derivative coexisted in equal amounts, regardless of the initial concentration of IQ (10 or 250 μM) (4). We did not confirm this phenomenon, since we found that 7-OH-IQ accumulated over time, whether the parent compound was incubated with a whole intestinal microbiota or with individual bacteria and regardless of its initial concentration (100 or 200 μM); in some instances, namely, upon incubation with the mixed cecal contents of HMA rats and with the strain Clostridium perfringens G22, IQ even totally disappeared from the incubation medium.
Strong interindividual variations occurred between the three human microbiota with regard to their IQ-degrading capabilities. Indications of such variations have been provided by Hirayama et al., who observed that preincubation of IQ with human feces increased its direct mutagenicity in the Ames assay, although to different extents depending on the donor (13). They concluded that the ability of human microbiota to generate direct-acting mutagens from IQ varied with individuals. Rumney et al. also noticed variations in the extent of IQ degradation following incubation in phosphate buffer with feces from three different human donors (27). However, the differences were slight compared to those reported in the present experiment, perhaps because of a much lower (eightfold) concentration of IQ. Interindividual differences in microbial secondary metabolic activities are not uncommon. In another context, we have shown that the rate of conversion of sinigrin, a sulfur compound commonly consumed via cruciferous vegetables, into its anticarcinogenic allyl isothiocyanate derivative depended on the origin of the fecal microbiota that was used to colonize a human large intestinal model (21). Another striking example is the bimodal distribution of the bacterially borne digestive ability to convert cholesterol into coprostanol among human populations; the U.S. population is thus divided into a vast majority of high converters and a minority of low to inefficient converters (38). Otherwise we found that the IQ-degrading ability of the human fecal microbiota was well preserved in the HMA rat model, as observed by Rumney et al. (27). However, unlike those authors, we observed that IQ was metabolized faster by the cecal contents of HMA rats than by the feces of the human donor. It is very likely that the repeated dosing of the HMA rats with IQ was responsible for this enhancement; indeed, such a metabolic adaptation has been reported previously with other xenobiotics, such as flavonoids or cyclamate (28). This alteration may result from quantitative shifts among bacterial groups, so that IQ-degrading populations become more abundant. Alternatively, there may be adaptive changes in the IQ-degrading groups which increase their metabolic capabilities. The fact that the yield of isolation of IQ-degrading bacteria from the mixed cecal contents of the HMA rats was rather low (8 out of 135) supports the latter hypothesis.
The 10 IQ-degrading bacterial strains that we discovered in culture collections (n = 2) and in the mixed cecal contents of the HMA rats (n = 8) belong to four different species, namely, Bacteroides thetaiotaomicron, Clostridium clostridiiforme, Clostridium perfringens, and Escherichia coli. All of them converted IQ solely into 7-OH-IQ, regardless of the extent of substrate consumption (range, 47 to 100%). Bacteroides thetaiotaomicron is well known for its highly diverse and adaptive metabolic capabilities (9, 40). As for the genus Clostridium and the species Escherichia coli, they have been shown to be able to metabolize other polycyclic compounds such as flavonoids (2, 39). In their investigations on the microbial metabolism of IQ, Carman et al. have focused their research of bioactive bacteria on individual representatives of the genus Eubacterium belonging to various environments, e.g., blood, mouth, and feces (4). They found that 14 strains out of 60 were able to perform the conversion of IQ into 7-OH-IQ. We have tested the only strain that is still available in culture collections, i.e., Eubacterium saburreum ATCC 33271 (VPI 11763), which was isolated from human dental plaque. However, we failed to reproduce its IQ-converting capability. This may be due to the difference in incubation conditions, i.e., resting cells in phosphate yeast buffer in the present experiment versus growing cells in peptone yeast broth in the pioneering work by Carman et al. (4). On the whole, the present study shows that the conversion of IQ to 7-OH-IQ is performed by bacteria belonging to the predominant communities of the human colon (Bacteroides and Clostridium) as well as by minor populations (Escherichia coli).
So far, the metabolic pathway leading from IQ to 7-OH-IQ remains unknown. Liver cytochrome P450 in humans and rats is able to realize several hydroxylations of the IQ molecule, but never at position 7 (33). Furthermore, Rumney et al. have clearly shown that this metabolite cannot be produced by feces of germfree rats, while it is always produced by feces of conventional or HMA rats (27). Consequently, the metabolic reaction leading to the addition of a hydroxyl group at position 7 seems to be carried out exclusively by bacteria. Among the bioactive strains that we have identified, Bacteroides thetaiotaomicron ATCC 29148 (type strain of the species) is of particular interest since its genome has been recently sequenced (GenBank accession no. NC004663). Of the 4,779 predicted proteins in its proteome, 1,997 (42%) have homology to proteins with no known function or no appreciable homology to entries in public databases (40). Proteins involved in the conversion of IQ to its 7-hydroxy derivative are likely to belong to this group of proteins having unknown functions. An interesting feature is that the highest number of best hits between protein sequences from Bacteroides thetaiotaomicron and from other members of the human intestinal microbiota is with Clostridium perfringens, the most efficient IQ converter identified in the present study. We can thus reasonably assume that a thorough analysis of proteins common to both species may help to characterize the metabolic pathway leading from IQ to 7-OH-IQ.
7-OH-IQ has been investigated in the past for its potential mutagenic/genotoxic activity. It is a direct-acting mutagen in the Ames assay, and some reports suggest that addition of S9 liver fraction may increase its mutagenicity (4, 37). Nevertheless, when administered by intrarectal infusion to adult rats or by oral gavage and intraperitoneal injections to newborn mice, 7-OH-IQ does not induce cancer, either in the colon or in sites remote from the digestive tract (37). It therefore seems that, in marked contrast with IQ, 7-OH-IQ would not be genotoxic or carcinogenic in laboratory rodents and would hence be considered a benign metabolite. On the other hand, it has been shown that the intestinal microbiota is essential to the induction of DNA damage by IQ (13, 16), and recent investigations (S. Rabot, F. Kassie, F. Ferk, et al., submitted for publication) indicate that Bacteroides thetaiotaomicron ATCC 29148 activates an HA mix representative of fried meat to DNA-damaging metabolites in a gnotobiotic rat model. Such contrasting data highlight the necessity of identifying the metabolites produced by bacteria from the main food-borne HAs in situ in the human colon and of further evaluating their genotoxic/carcinogenic activity.
Acknowledgments
This work was supported by the European Community under the RTD program “Quality of Life and Management of Living Resources,” QLK1-CT99-01197, entitled “Heterocyclic Amines in Cooked Foods—Role in Human Health.” Christèle Humblot acknowledges a Ph.D. grant from the French Ministry of Education and Research.
We thank Bertrand Légeret for MS analysis and Karine Gloux and Catherine Philippe for helpful advice about HPLC analysis. Many thanks also go to Rosa Durao for breeding the germfree rats, to Aurélia Bruneau for technical assistance with the human-microbiota-associated rats, and to Jamila Anba and Chantal Bridonneau for management of the long-term storage of the collection strains. Acknowledgments also go to Gérard Corthier, Maria-José Flores-Sanabria, and Joël Doré for critical reading of the manuscript. Special thanks go to Alan J. Duncan for his wise comments and for English language amendments.
REFERENCES
- 1.Bashir, M., D. G. Kingston, R. J. Carman, R. L. van Tassell, and T. D. Wilkins. 1987. Anaerobic metabolism of 2-amino-3-methyl-3H-imidazo[4,5-f]quinoline (IQ) by human fecal flora. Mutat. Res. 190:187-190. [DOI] [PubMed] [Google Scholar]
- 2.Brézillon, C. 2001. Ph.D. thesis. Métabolisme endogène et bactérien des catéchines, comopsés polyphénoliques majeurs du vin rouge. Etudes in vitro et in vivo chez le rat à flore humaine. Institut National Agronomique Paris-Grignon, Paris, France.
- 3.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carman, R. J., R. L. Van Tassell, D. G. Kingston, M. Bashir, and T. D. Wilkins. 1988. Conversion of IQ, a dietary pyrolysis carcinogen, to a direct-acting mutagen by normal intestinal bacteria of humans. Mutat. Res. 206:335-342. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick, R. W., S. E. George, and L. D. Claxton. 1992. Role of the gastrointestinal mucosa and microflora in the bioactivation of dietary and environmental mutagens or carcinogens. Drug Metab. Rev. 24:425-492. [DOI] [PubMed] [Google Scholar]
- 6.Cibik, R., E. Lepage, and P. Talliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional french cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 7.Combourieu, B., P. Besse, M. Sancelme, J. P. Godin, A. Monteil, H. Veschambre, and A. M. Delort. 2000. Common degradative pathways of morpholine, thiomorpholine, and piperidine by Mycobacterium aurum MO1: evidence from 1H-nuclear magnetic resonance and ionspray mass spectrometry performed directly on the incubation medium. Appl. Environ. Microbiol. 66:3187-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Combourieu, B., L. Elfoul, A. M. Delort, and S. Rabot. 2001. Identification of new derivatives of sinigrin and glucotropaeolin produced by the human digestive microflora using 1H NMR spectroscopy analysis of in vitro incubations. Drug Metab. Dispos. 29:1440-1445. [PubMed] [Google Scholar]
- 9.Comstock, L. E., and M. J. Coyne. 2003. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. Bioessays 25:926-929. [DOI] [PubMed] [Google Scholar]
- 10.Felton, J. S., M. G. Knize, C. P. Salmon, M. A. Malfatti, and K. S. Kulp. 2002. Human exposure to heterocyclic amine food mutagens/carcinogens: relevance to breast cancer. Environ. Mol. Mutagen. 39:112-118. [DOI] [PubMed] [Google Scholar]
- 11.Gérard, P., F. Béguet, P. Lepercq, L. Rigottier-Gois, V. Rochet, C. Andrieux, and C. Juste. 2004. Gnotobiotic rats harboring human intestinal microbiota as a model for studying cholesterol-to-coprostanol conversion. FEMS Microbiol. Ecol. 47:337-343. [DOI] [PubMed] [Google Scholar]
- 12.Gomez, A., M. Ladire, F. Marcille, M. Nardi, and M. Fons. 2002. Characterization of ISRgn1, a novel insertion sequence of the IS3 family isolated from a bacteriocin-negative mutant of Ruminococcus gnavus E1. Appl. Environ. Microbiol. 68:4136-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirayama, K., P. Baranczewski, J. E. Akerlund, T. Midtvedt, L. Moller, and J. Rafter. 2000. Effects of human intestinal flora on mutagenicity of and DNA adduct formation from food and environmental mutagens. Carcinogenesis 21:2105-2111. [DOI] [PubMed] [Google Scholar]
- 14.Hudault, S., C. Bridonneau, P. Raibaud, C. Chabanet, and M. F. Vial. 1994. Relationship between intestinal colonization of Bifidobacterium bifidum in infants and the presence of exogenous and endogenous growth-promoting factors in their stools. Pediatr. Res. 35:696-700. [DOI] [PubMed] [Google Scholar]
- 15.Kaderlik, K. R., G. J. Mulder, R. J. Turesky, N. P. Lang, C. H. Teitel, M. P. Chiarelli, and F. F. Kadlubar. 1994. Glucuronidation of N-hydroxy heterocyclic amines by human and rat liver microsomes. Carcinogenesis 15:1695-1701. [DOI] [PubMed] [Google Scholar]
- 16.Kassie, F., S. Rabot, M. Kundi, M. Chabicovsky, H. M. Qin, and S. Knasmüller. 2001. Intestinal microflora plays a crucial role in the genotoxicity of the cooked food mutagen 2-amino-3-methylimidazo [4,5-f]quinoline. Carcinogenesis 22:1721-1725. [DOI] [PubMed] [Google Scholar]
- 17.Kassie, F., V. M. Sundermann, R. Edenharder, K. L. Platt, F. Darroudi, E. Lhoste, C. Humblot, E. Muckel, M. Uhl, M. Kundi, and S. Knasmüller. 2003. Development and application of test methods for the detection of dietary constituents which protect against heterocyclic aromatic amines. Mutat. Res. 523-524:183-192. [DOI] [PubMed] [Google Scholar]
- 18.King, R. S., F. F. Kadlubar, and R. J. Turesky. 2000. In vivo metabolism, p. 90-112. In T. Sugimura (ed.), Food borne carcinogens. Wiley, New York, N.Y.
- 19.Knasmüller, S., H. Steinkellner, A. M. Hirschl, S. Rabot, E. C. Nobis, and F. Kassie. 2001. Impact of bacteria in dairy products and of the intestinal microflora on the genotoxic and carcinogenic effects of heterocyclic aromatic amines. Mutat. Res. 480-481:129-138. [DOI] [PubMed] [Google Scholar]
- 20.Krul, C., C. Humblot, C. Philippe, M. Vermeulen, M. van Nuenen, R. Havenaar, and S. Rabot. 2002. Metabolism of sinigrin (2-propenyl glucosinolate) by the human colonic microflora in a dynamic in vitro large-intestinal model. Carcinogenesis 23:1009-1016. [DOI] [PubMed] [Google Scholar]
- 21.Krul, C., A. Luiten-Schuite, R. Baandagger, H. Verhagen, G. Mohn, V. Feron, and R. Havenaar. 2000. Application of a dynamic in vitro gastrointestinal tract model to study the availability of food mutagens, using heterocyclic aromatic amines as model compounds. Food Chem. Toxicol. 38:783-792. [DOI] [PubMed] [Google Scholar]
- 22.Lhoste, E. F., V. Ouriet, S. Bruel, J. P. Flinois, C. Brezillon, J. Magdalou, C. Cheze, and L. Nugon-Baudon. 2003. The human colonic microflora influences the alterations of xenobiotic-metabolizing enzymes by catechins in male F344 rats. Food Chem. Toxicol. 41:695-702. [DOI] [PubMed] [Google Scholar]
- 23.Orrhage, K., E. Sillerstrom, J. A. Gustafsson, C. E. Nord, and J. Rafter. 1994. Binding of mutagenic heterocyclic amines by intestinal and lactic acid bacteria. Mutat. Res. 311:239-248. [DOI] [PubMed] [Google Scholar]
- 24.Rafter, J. J., and J. A. Gustafsson. 1986. Metabolism of the dietary carcinogen TRP-P-1 in rats. Carcinogenesis 7:1291-1295. [DOI] [PubMed] [Google Scholar]
- 25.Roland, N., S. Rabot, and L. Nugon-Baudon. 1996. Modulation of the biological effects of glucosinolates by inulin and oat fibre in gnotobiotic rats inoculated with a human whole faecal flora. Food Chem. Toxicol. 34:671-677. [DOI] [PubMed] [Google Scholar]
- 26.Rumney, C. J., and I. R. Rowland. 1992. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 31:299-331. [DOI] [PubMed] [Google Scholar]
- 27.Rumney, C. J., I. R. Rowland, and I. K. O'Neill. 1993. Conversion of IQ to 7-OHIQ by gut microflora. Nutr. Cancer 19:67-76. [DOI] [PubMed] [Google Scholar]
- 28.Scheline, R. R. 1973. Metabolism of foreign compounds by gastrointestinal microorganisms. Pharmacol. Rev. 25:451-523. [PubMed] [Google Scholar]
- 29.Skerman, V. B. D., V. McGowan, and P. H. A. Sneath. 1980. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30:225-420. [Google Scholar]
- 30.Snyderwine, E. G., R. J. Turesky, K. W. Turteltaub, C. D. Davis, N. Sadrieh, H. A. Schut, M. Nagao, T. Sugimura, U. P. Thorgeirsson, R. H. Adamson, and S. S. Thorgeirsson. 1997. Metabolism of food-derived heterocyclic amines in nonhuman primates. Mutat. Res. 376:203-210. [DOI] [PubMed] [Google Scholar]
- 31.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turesky, R. J., N. P. Lang, M. A. Butler, C. H. Teitel, and F. F. Kadlubar. 1991. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis 12:1839-1845. [DOI] [PubMed] [Google Scholar]
- 33.Turesky, R. J., W. G. Stillwell, P. L. Skipper, and S. R. Tannenbaum. 1993. Metabolism of the food-borne carcinogens 2-amino-3-methylimidazo-[4,5-f]quinoline and 2-amino-3,8-dimethylimidazo[4,5-f]-quinoxaline in the rat as a model for human biomonitoring. Environ. Health Perspect. 99:123-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turteltaub, K. W., J. S. Vogel, C. E. Frantz, and N. Shen. 1992. Fate and distribution of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in mice at a human dietary equivalent dose. Cancer Res. 52:4682-4687. [PubMed] [Google Scholar]
- 35.Van Tassell, R. L., R. J. Carman, D. G. Kingston, and T. D. Wilkins. 1989. Bacterial metabolism in humans of the carcinogen IQ to the direct acting mutagen hydroxy-IQ. Microb. Ecol. Health Dis. 2:123-129. [Google Scholar]
- 36.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisburger, J. H., A. Rivenson, J. Reinhardt, C. Aliaga, J. Braley, L. M. Dolan, G. M. Williams, E. Zang, D. G. Kingston, and M. Bashir. 1994. Genotoxicity and carcinogenicity in rats and mice of 2-amino-3,6-dihydro-3-methyl-7H-imidazolo[4,5-f]quinolin-7-one: an intestinal bacterial metabolite of 2-amino-3-methyl-3H-imidazo[4,5-f]quinoline. J. Natl. Cancer Inst. 86:25-30. [DOI] [PubMed] [Google Scholar]
- 38.Wilkins, T. D., and A. S. Hackman. 1974. Two patterns of neutral steroid conversion in the feces of normal North Americans. Cancer Res. 34:2250-2254. [PubMed] [Google Scholar]
- 39.Winter, J., M. R. Popoff, P. Grimont, and V. D. Bokkenheuser. 1991. Clostridium orbiscindens sp. nov., a human intestinal bacterium capable of cleaving the flavonoid C-ring. Int. J. Syst. Bacteriol. 41:355-357. [DOI] [PubMed] [Google Scholar]
- 40.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X. B., and Y. Ohta. 1993. Microorganisms in the gastrointestinal tract of the rat prevent absorption of the mutagen-carcinogen 3-amino-1,4-dimethyl-5H-pyrido(4,3-b)indole. Can. J. Microbiol. 39:841-845. [DOI] [PubMed] [Google Scholar]