Abstract
The nirS nitrite reductase genes were studied in two strains (strains 27 and 28) isolated from two denitrifying reactors and characterized as Thauera according to their 16S rRNA gene sequences. Strain 28 contains a single nirS sequence, which is related to the nirS of Thauera mechernichensis, and strain 27 contains two nirS sequences; one is similar to the nirS sequence from Thauera mechernichensis (gene 2), but the second one (gene 8) is from a separate clade with nirS from Pseudomonas stutzeri, Azoarcus species, Alcaligenes faecalis, and other Thauera species. Both genes were expressed, but gene 8 was constitutively expressed while gene 2 was positively regulated by nitrate.
Denitrification, the respiratory reduction of nitrate to gaseous products, is an important component of the nitrogen cycle. Complete denitrification requires the sequential action of four enzymes: nitrate reductase, nitrite reductase, nitric oxide (NO) reductase, and nitrous oxide (N2O) reductase (8, 9, 13). Respiratory nitrite reduction to NO is catalyzed by a copper nitrite reductase (NirK) or a cytochrome cd1 nitrite reductase (NirS). Since denitrification is widespread among microorganisms belonging to phylogenetically distinct groups of Bacteria and Archaea, functional genes, e.g., nirK and nirS, have been used as markers in ecological studies of marine sediments and soil (cf. references 1 and 2).
We used nirS and nirK as gene markers to study the denitrifier ecology in reactors being developed for high-rate nitrate removal from wastewater. Previous work demonstrated that an upflow anaerobic sludge blanket denitrifying reactor could be developed with the denitrifiers retained in granules for a high-rate process (5, 6). In this work, two upflow anaerobic sludge blanket laboratory-scale reactors (reactors 1 and 2) were running in parallel but seeded with different inocula. During 2 years of reactor operation, 20 strains were isolated at different times from terminal most-probable-number denitrifier tubes and from direct tryptic soy agar (TSA; Difco) plates (5, 6). Ten of these strains, isolated from both reactors at different reactor operation times, presented the same amplified 16S rRNA gene restriction fragment length polymorphism (RFLP) profile and single-strand conformational polymorphism peak (5, 6). Two of these strains (strains 27 and 28), isolated from reactors 1 and 2, respectively, at 12 weeks of operation, were selected and characterized by 16S rRNA gene sequence analysis as belonging to the genus Thauera (5, 6). A high level of DNA-DNA homology between the strains suggested that both belong to the same species (6). This result suggests that organisms belonging to the Thauera genus persisted in the reactors throughout 1 year of operation.
In the present work, the diversity of nitrite reductase genes was studied in the Thauera isolates. Surprisingly, two different nirS gene sequences were found in 9 of the 10 strains. The aim of this work was to characterize both genes and to study their expression in strain 27, the strain carrying both genes.
Nitrite reductase genes analysis from denitrifying isolates.
DNA was extracted from the 10 denitrifying strains previously identified as members of the genus Thauera according to 16S rRNA gene RFLP profile, single-strand conformational polymorphism peak, and 16S rRNA gene sequence (5, 6) by using a Wizard DNA extraction kit (Promega). The nirS and nirK genes were amplified by PCR using specific primers nirK 1F and nirK 5R for nirK and nirS 1F and nirS 6R for nirS (3); positive controls were used with both sets of primers. All the isolates yielded only nirS gene PCR products. This was in accordance with previous results that showed that strains from the genus Thauera contain only nirS nitrite reductase genes (12). The nirS gene amplicons were digested with HhaI, and the fragments were separated in an agarose gel (3%; Methaphor) and then stained with ethidium bromide. Two RFLP patterns were observed. Strain 27 showed a pattern (profile 2) more complex than that of strain 28 (profile 1) (Fig. 1). All the bands of profile 1 were included in profile 2, suggesting that profile 2 was the sum of the profile 1 bands and another set of bands. The other eight isolates, which also had the same 16S rRNA gene RFLP profile as strain 27, presented the same nirS complex pattern as strain 27 (profile 2) (data not shown).
FIG. 1.
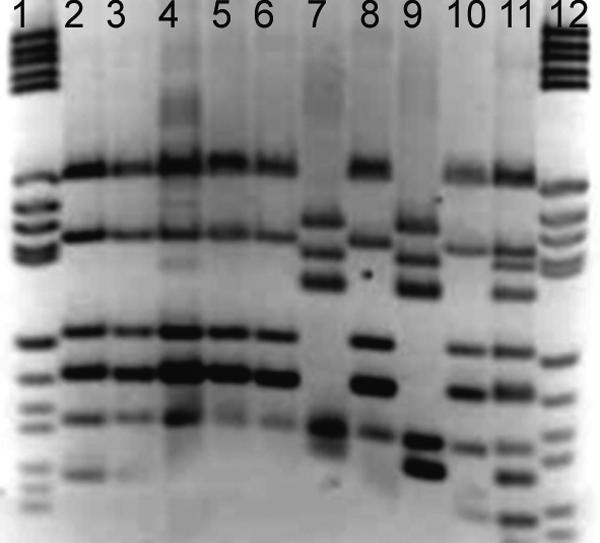
Inverted image from an agarose gel stained with ethidium bromide showing the RFLP patterns obtained from digesting the nirS PCR product from strain 28 (profile 1; lane 10), from strain 27 (profile 2; lane 11), and from the clones obtained from cloning strain 27 nirS. Clones 1, 2, 3, 4, 5, and 7 had profiles similar to that of strain 28 (profile 1; lanes 2 through 6 and 8, respectively), and clones 6 and 8 had profiles that differed (profile 3; lanes 7 and 9, respectively). Molecular weight marker V (Roche) results are shown (lanes 1 and 12).
Evidence of two nirS genes in strain 27.
After purification (QIAquick PCR purification kit; QIAGEN), the nirS PCR products from strain 27 and 28 were sequenced using the nirS 1 forward primer at the Michigan State University Genomics Technology and Support Facility. While a single sequence was obtained for strain 28, a mixture of sequences was obtained for strain 27, suggesting that two genes were amplified by PCR. To ensure that strain 27 was a monoculture, the strain was purified two additional times by streaking on an agar plate (tryptic soy agar; Difco). All colonies were homogenous and identical. DNA was then extracted from a liquid culture (tryptic soy broth; Difco) started from a single colony on the second plate. The same DNA was used to amplify by PCR both the 16S rRNA genes and the nirS genes. A single gene was detected for the 16S rRNA gene, but a mixture of genes was detected for the nirS gene by RFLP analysis. Almost the entire 16S rRNA gene sequence was determined (1,493 nucleotides) from both DNA strands by using the 16S rRNA gene Bacteria primers 9F (5′-GAGTTTGATCMTGGCTCAG-3′), 500F (5′-CTAACTACGTGCCAGCAGC-3′), 1200F (5′-GGAGGAAGGYGGGGAYGA-3′), 1492R (5′-GNTACCTTGTTACGACTT-3′), 1100R (5′-TCGTTGCGGGACTTAAC-3′), 700R (5′-TACGCATTTCACCKCTACA-3′), and 340R (5′-TGCTGCCTCCCGTAGGAGT-3′). This nearly full-length sequence was 99% identical to that of the 16S rRNA gene of Thauera mechernichensis, and the same result was obtained for the 16S rRNA gene partial sequence of strain 28; as 16S rRNA genes from Thauera species are very closely related (98 to 99%), additional taxonomic studies are needed to define strain 27 and strain 28 species.
The nirS PCR product from strain 27 was cloned using a TOPO-TA cloning kit (Invitrogen). Twenty clones were selected, and the inserts were amplified by PCR (using primers from the plasmid vector) and analyzed for RFLP as described above. Two different RFLP patterns were retrieved from the clones; one profile was similar to profile 1 retrieved from strain 28, and a new profile (profile 3) was detected. The profile from strain 27 (profile 2) was highly similar to the sum of the two different clone profiles (Fig. 1).
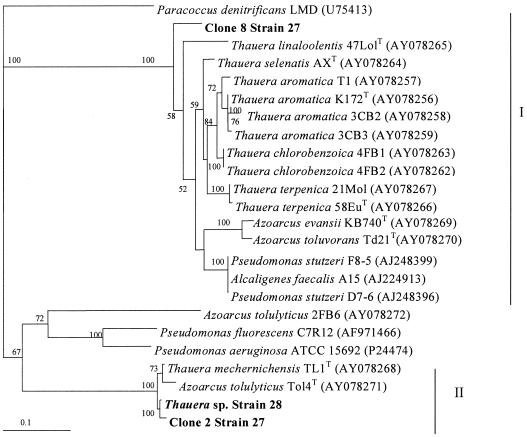
Two clones, each representing an RFLP pattern, were selected, and the sequence of the inserts was determined as described above. Comparison of the gene sequences with the NCBI database using a Blastn search showed that sequences from clones 1 and 2 (profile 1) (510 and 641 nucleotides, respectively) had a high level (98%) of similarity to the nirS gene from T. mechernichensis. The sequences from clones 6 and 8 (510 and 892 nucleotides, respectively) (profile 3) had homologies to the nirS gene sequences from strain D7-6 (suggested to be Pseudomonas stutzeri by 16S rRNA gene sequencing) (87% homology), from Alcaligenes faecalis strain A15 (87%), and also from other Thauera nirS genes (Thauera selenatis strain AXT [86%] and Thauera chlorobenzoica strain 4FB1 [86%]). As was reported by Song and Ward in 2003 (12), Thauera NirS sequences were phylogenetically positioned in two different clades. All the species of Thauera with the exception of Thauera mechernichensis were positioned in the same branch (clade 1) with Pseudomonas stutzeri, Alcaligenes faecalis, Azoarcus evansii, and Azoarcus toluvorans. T. mechernichensis NirS sequences were positioned, regardless of their taxonomies, in another branch (clade 2) with Azoarcus tolulyticus and were distantly related to the NirS sequences from Pseudomonas fluorescens and Pseudomonas aeruginosa.
Phylogenetic analysis was performed using 180 amino acid positions from strain 28 NirS and the two NirS sequences from strain 27 (clone 2 and clone 8) after alignment to the most related sequences using Clustal W. Analysis was done with the Phylip 3.5 software package (7) using a Dayhoff PAM matrix (4) and neighbor-joining methods (10). Seqboot was used to obtain the confidence level in 100 data sets (Fig. 2). The strain 27 clone 2 NirS sequence was closely related to the strain 28 NirS sequence, and both were positioned in the same clade as the T. mechernichensis NirS sequence was. The clone 8 NirS sequence was positioned in a separate clade with NirS sequences from other Thauera species as well as from Pseudomonas, Azoarcus, and Alcaligenes strains (Fig. 2). Similar results were obtained from nirS PCR product cloning and RFLP analysis from strain 39, isolated 8 months later from the other reactor, showing that the same two nirS genes were present in strains 27 and 39.
FIG. 2.
Phylogenetic tree of NirS sequences on the basis of 180 amino acids showing the NirS positions of strain 28 and those of two clones from strain 27 (clone 2 and clone 8) and the most related NirS sequences. The two clades referred to in the text (clade 1 [I] and clade 2 [II]) are indicated to the right of the figure. The scale bar represents 10 amino acid differences in 100. Bootstrap values higher than 50% are presented next to the nodes.
Expression of two nirS genes from strain 27.
Preliminary experiments with strain 27 grown on tryptic soy broth under denitrifying conditions showed that of the 20 cDNA clones examined following reverse transcription of the RNA, 12 had profile 1 (clone 2) and 8 had profile 3 (clone 8) (data not shown). Since transcription to RNA occurred from both genes, we quantified expression of both nirS genes under different growth conditions. For that, the amount of each mRNA was quantified by TaqMan real-time PCR. Specific primers and TaqMan probes were designed for each gene using the Primer Express (PE Applied Biosystems) program (Table 1). The optimal primer/probe concentration was determined using controls (150 nM final concentration for strain 27 clone 2 and 300 nM for strain 27 clone 8 primers and probes). The gene products were then quantified using the TaqMan real-time PCR kit in two steps as described above and an ABI Prism 7700 sequence detection system (PE Applied Biosystems). The fluorescence increase was monitored using a standard PCR cycle (95°C for 10 min and 40 cycles of the following two steps: 95°C for 15 s and 60°C for 1 min), with the PCR cycle threshold (CT) determined in each case. A standard curve of CT versus the number of gene copies was prepared for each gene by using different concentrations of nirS genes from clone 2 and from clone 8. Negative controls with no template DNA were run for each reaction. DNA concentration was determined by calculating the absorbance at 260 nm. The number of copies/ng DNA of nirS PCR product was calculated considering that one nirS copy has 890 bp, the average mass of 1 bp is 600 g/mol, and 1 mol of nirS genes has the Avogadro number of copies.
TABLE 1.
Primers and fluorescent probes designed in this work used in the real-time TaqMan PCR quantification
| Primer or probea | Sequence | Gene locationb |
|---|---|---|
| St27clone2F | 5′-CGTGGCCGCCATCATC-3′ | 873-887 |
| St27clone2R | 5′-GCCGGTTTCCTTCACATTGA-3′ | 917-936 |
| St27clone2P (TaqMan probe) | 5′-CCTCGCACTTCAACCCGGAGTTCTT-3′ | 890-914 |
| St27clone8F | 5′-ATCCGCAGTTCGGTCCG-3′ | 1163-1179 |
| St27clone8R | 5′-AGATCAGCGAAACCACGTCG-3′ | 1206-1225 |
| St27clone8P (TaqMan probe) | 5′-TGGGCAACGGGTCACCTGGGT-3′ | 1183-1203 |
TaqMan probes were doubly labeled with 5′ 6-carboxyfluorescein and 3′ 6-carboxytetramethylrhodamine.
Gene location according to Pseudomonas stutzeri ZoBell nirS sequence (GenBank accession number X56813).
After optimization of the real-time PCR protocol, expression of the two nirS genes was studied under three different culture conditions: (i) aerobic conditions with no nitrate, (ii) aerobic conditions with 10 mM nitrate, and (iii) anaerobic conditions (Ar) with 10 mM nitrate. All treatments were performed in duplicate. Cultures were grown in rich liquid medium (tryptic soy broth; Difco) and incubated with agitation (200 rpm) at 37°C, and samples were taken. The RNA was extracted immediately using an RNeasy Mini extraction kit (QIAGEN, Chatsworth, Calif.). The integrity of the RNA was verified in a 0.8% agarose gel stained with ethidium bromide. Genomic DNA traces were eliminated by RNase-free DNase treatment followed by a phenol-chloroform purification (11). The cDNA concentration was measured by detecting absorbance at 260 nm. The real-time PCR was performed in triplicate for each dilution of each cDNA sample using the two primer/probe sets described above. The number of copies of each transcript was determined from the standard curve.
The T. mechernichensis-like gene (clone 2) showed a two-order-of-magnitude increase in expression with nitrate in both aerobic and anaerobic conditions (Table 2). This result was consistent with previous work that shows that nirS in Pseudomonas species is positively regulated by N oxides (9). Oxygen had less effect on the expression of this gene. Surprisingly, the other strain 27 nirS gene (clone 8) was highly expressed independently of nitrate or oxygen, suggesting a constitutive expression (Table 2). These results show the presence of a constitutively expressed nirS in addition to the nitrate-regulated nirS gene in strain 27. To our knowledge, this is the first report of this property in any denitrifying strain. The presence of this dual nirS strain throughout a year in two high-nitrate-removal-rate granulated wastewater treatment reactors started with different inocula suggests that the nitrate induction of one gene and the constitutive expression of the other at a higher level are important to the ecological success of this organism and to the efficiency of denitrification in this type of environment. This constructed denitrifying ecosystem has unique features, namely, high nitrate loading and removal rates (0.9 g N-NO3−/liter/day) and a high number of aggregated cells. The fact that a second gene is from a very different clade and is not found in the other strain (strain 28) suggests that it may have been horizontally transferred to Thauera, resulting in an increased competitive ability under these conditions.
TABLE 2.
mRNA real-time PCR quantification of the two nirS genes under three different culture conditionsa
| Culture conditions | Mean mRNA nirS clone 2 copy number/ ng DNA (SD)b | mRNA nirS clone 8 copy number/ ng DNA (SD)c |
|---|---|---|
| Aerobic, no nitrate | 1.14 × 104 (3.74 × 103) | 2.08 × 107 (9.30 × 105) |
| Aerobic, nitrate | 1.17 × 106 (4.40 × 105) | 9.16 × 107 (1.82 × 107) |
| Anaerobic, nitrate | 4.62 × 105 (3.67 × 105) | 1.66 × 107 (1.17 × 107) |
Data are means and standard deviations of two culture replicates which are comprised of triplicate real-time PCR measurements for each sample.
Determined using the primers St27clone2F and St27clone2R and the TaqMan probe St27clone2P designed for targeting clone 2 sequence (see Table 1).
Determined using the primers St27clone8F and St27clone8R and the TaqMan probe St27clone8P designed for targeting clone 8 sequence (see Table 1).
The use of functional genes as markers in ecological studies usually assumes only one gene type per population, but the possible occurrence of two or more would lead to an overstatement of diversity when using terminal RFLP or clone library data.
Nucleotide sequence accession numbers.
EMBL GenBank accession numbers for partial nirS from strain 27 clones are: for clone 1, AY838757; for clone 2, AY838762; for clone 6, AY838758; and for clone 8, AY838759. That for partial nirS from strain 28 is AY829012, that for the 16S rRNA gene complete sequence from strain 27 is AY838760, and that for the 16S rRNA gene partial sequence from strain 28 is AY838761.
Acknowledgments
This work was partially supported by an International Fellowship for Latin America from the American Society for Microbiology that provided C.E. the opportunity to work at the Center for Microbial Ecology and by DOE grant no. DE-FG02-98ER62535 (BI-OMP).
REFERENCES
- 1.Avrahami, S., R. Conrad, and G. Braker. 2003. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braker, G., H. Ayala del Río, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dayhoff, M. O. 1972. Atlas of protein sequence and structure. National Biomedical Research Foundation, Washington, D.C.
- 5.Etchebehere, C., A. Cabezas, P. Dabert, and L. Muxí. 2003. Evolution of the bacterial community during granules formation in denitrifying reactors followed by molecular, culture-independent techniques. Water Sci. Technol. 48(6):75-79. [PubMed] [Google Scholar]
- 6.Etchebehere, C., M. I. Errazquin, A. Cabezas, M. J. Pianzzola, M. Mallo, G. Ottonello, L. Borzacconi, and L. Muxí. 2002. Sludge bed development in denitrifying reactors using different inocula—performance and microbiological aspects. Water Sci. Technol. 45(10):365-370. [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1997. PHYLIP phylogeny inference package (version 3.2). Department of Genetics, University of Washington, Seattle.
- 8.Knowles, R. 1982. Denitrification. Microbiol. Rev. 46:43-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 10.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. J. Mol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Song, B., and B. B. Ward. 2003. Nitrite reductase genes in halobenzoate degrading denitrifying bacteria. FEMS Microbiol. Ecol. 43:349-357. [DOI] [PubMed] [Google Scholar]
- 13.Zumft, W. G. 1992. The denitrifying prokaryotes, p. 554-582. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer Verlag, New York, N.Y.