Abstract
A multicopper oxidase gene from Staphylococcus aureus was cloned and overexpressed. Purified recombinant multicopper oxidase oxidized the substrate 3,3′-dimethoxybenzidine in the presence of copper. Disruption of mco showed copper sensitivity and H2O2 resistance, suggesting roles for mco in copper homeostasis and oxidative stress response. Northern blot analysis showed copper-induced mco transcription.
Multicopper oxidases (MCO) are enzymes that are critically involved in copper homeostasis. These enzymes show enhanced oxidase activity for a wide range of substrates (1, 2, 4, 9, 22, 24) and play a role in iron transport in eukaryotes and prokaryotes (9, 23). mco genes have been cloned and identified from animals, plants, insects, fungi, and bacteria (3, 4, 8, 12, 16). However, there is no report of mco in Staphylococcus aureus.
We have been investigating the regulation of metal transport and homeostasis in S. aureus (25, 26). To identify new loci involved in metal homeostasis in S. aureus, we selected streptonigrin-resistant clones from a transposon Tn917-induced mutant library of S. aureus strain ATCC 12600 and characterized mutants by cloning and sequencing. In this study, we identified one of the mutations in the gene encoding multicopper oxidase.
Staphylococcus aureus strains were grown in tryptic soy broth (TSB) or defined medium. Escherichia coli strains were grown in Luria-Bertani broth with the appropriate antibiotic. DNA manipulation, sequencing, cloning, transformation, and overexpression were performed as described earlier (7, 21, 26). DNA sequences were determined with an ABI Prism 310 automated sequencer (Perkin Elmer, Foster City, CA).
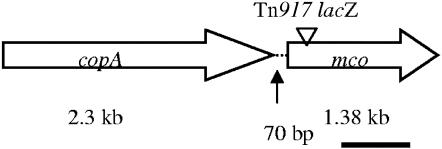
The Tn917-flanking sequence of the mutant was cloned in E. coli as described earlier (21). Nucleotide analysis of the flanking region of the Tn917 insertion of one of the streptonigrin-resistant clones showed sequence homology to putative multicopper oxidases. The complete nucleotide sequence of the parental gene copy of S. aureus showed an open reading frame (ORF) comprising 1,389 bp encoding a hypothetical polypeptide of 462 amino acids with a predicted molecular mass of 52 kDa and pI of 9.68. Nucleotide comparison showed a 99% sequence identity with a gene in the S. aureus EMRSA-16 database. Amino acid comparisons of the translated ORF showed 84% identity to putative MCO from Staphylococcus epidermidis and 26 to 41% identities to laccase, CueO, and ascorbate oxidase, which are members of the MCO family. Therefore, we designated this ORF mco. Seventy nucleotides upstream of mco, an ORF designated copA, a copper ATPase, was also identified (Fig. 1).
FIG. 1.
Schematic representation of the multicopper oxidase and copper ATPase genes. The arrow indicates predicted gene and orientation. The transposon insertion site within the gene is indicated by an inverted triangle. The solid bar indicates the DNA fragment which was used as a probe for the Northern analysis.
The multicopper oxidase gene from S. aureus was cloned and overexpressed in E. coli BL21(DE3). MCO activity was determined by using 3,3′-dimethoxybenzidine as described earlier (5, 15, 18). This enzymatic activity was copper dependent, and the presence of 0.5 mM CuSO4 is optimum for enzymatic activity. The purified MCO showed a specific activity of 9.7 U/mg, compared to 1.6 U/mg in the crude extract (Table 1). The purified MCO also exhibit low levels of ferroxidase (1.58 U/mg) and phenoloxidase (2.3 U/mg) activities compared to those reported for other organisms (11, 13).
TABLE 1.
Overexpression and purification of MCO from E. coli BL21(DE3)(pLysS)a
| Step | Protein (mg) | Total activity (U)b | Sp actc (U/mg) |
|---|---|---|---|
| Crude extract | 21 | 19.9 ± 1.3 | 1.6 ± 0.2 |
| Ni column elute | 0.6 | 42.0 ± 2.0 | 9.7 ± 0.3 |
Enzyme activity was measured using 3,3′-dimethoxybenzidine as a substrate. The reaction was initiated by the addition of 0.472 μmol of 3,3′-dimethoxybenzidine (o-dianisidine) and terminated after 5 and 120 min.
One unit of activity catalyzed the oxidation of 1 μmol of substrate per min.
Specific activity is defined as the μmol of substrate oxidized per min per mg of total protein.
Although multicopper oxidase is an important enzyme in many organisms, analysis of the S. aureus database indicated the presence of MCO homologues only in an S. aureus EMRSA-16 strain. We used Southern blot analysis to search for homologous sequences in various S. aureus strains whose genomes have not been sequenced. The mco gene hybridized with a 2.5-kb HindIII DNA fragment of three strains (ATCC 12600, H, and Wood) out of seven laboratory strains. However, the most commonly used S. aureus strains, RN450 and COL, did not show any sequence homology with mco, suggesting that it plays a dispensable role in staphylococci.
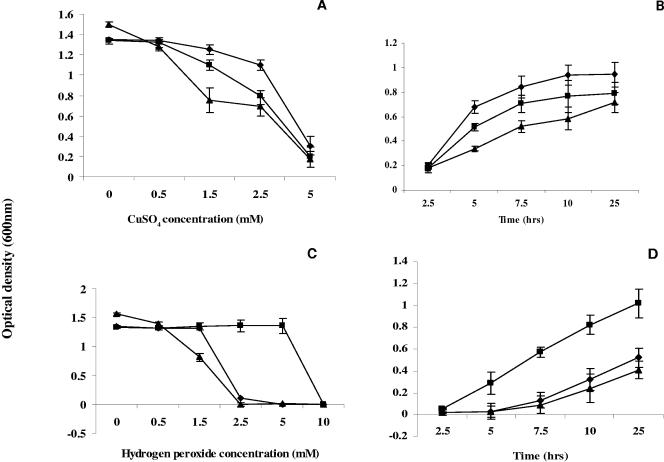
The insertion of Tn917 in the multicopper oxidase gene resulted in streptonigrin tolerance and copper sensitivity. As shown in Fig. 2A and B, the mutant strain grows slowly in medium containing more than 1.5 mM CuSO4. We also checked the mutant's sensitivities to iron, nickel, cobalt, and other metallic ions. So far, we found the mco mutant sensitive only to copper and cobalt (data not shown).
FIG. 2.
Effects of copper and hydrogen peroxide on growth. Overnight cultures were diluted 1:500 in TSB with different concentrations of CuSO4 (A) or H2O2 (C) and incubated at 37°C with shaking. Cell growth was monitored by measuring optical density at 600 nm for 18 h. Growth curves with 2.5 mM CuSO4 (B) and 1.5 mM H2O2 (D) are shown. Overnight cultures were diluted 1:500 in TSB with 1.5 mM H2O2 or 2.5 mM CuSO4. Cell growth was monitored by measuring absorbance at 600 nm at various intervals of incubation at 37°C with shaking. Symbols: ⧫, wild type; ▪, mco mutant; ▴, mco complemented strain. Each point represents the mean value ± standard deviation (represented by bar) of three experiments.
A role of mco in the oxidative stress response has been proposed previously (10, 16). To test whether a mutation in mco has any impact on the oxidative tolerance of S. aureus, the mco mutant and the parent cells were grown in TSB containing various concentrations of H2O2 and methyl viologen (paraquat). The mco mutant cells were able to grow in the medium containing 5 mM H2O2, whereas the parent cells were unable to grow (Fig. 2C). However, the tolerance levels of the mco mutant and the parent strains to paraquat, another oxidative agent, were similar. Additional experiments using catalase assay activity gels (7) showed that the higher hydrogen peroxide tolerance of the mco mutant was not due to the induced expression of the kat gene, which encodes the catalase (data not shown).
In the presence of heavy metals, MCO catalyzes the formation of H2O2. It has been shown that cupric ions and ceruloplasmin, a multicopper oxidase family protein in human serum, have the capacity to oxidize the substrate with the production of superoxide anions and H2O2 (17). Moreover, the production of H2O2 in the oxidation of heavy metals by laccase, an MCO in Stropharia rugosoannulata, has been reported previously (20). However, the precise role of multicopper oxidase in the hydrogen peroxide tolerance phenotype of the mco mutant is unknown.
Complementation studies were performed to provide genetic evidence that copper sensitivity and H2O2 tolerance are due to the transposon insertion within the mco. A 1.8-kb BamHI-HindIII fragment containing mco was cloned in the shuttle vector pLI50 (14) and transformed into an mco mutant strain. As shown in Fig. 2C and D, the complemented strain became H2O2 sensitive as predicted; however, it unexpectedly remained copper sensitive (Fig. 2A and B). To explain this result, we hypothesize that the mutation within the mco gene had an additional unanticipated negative effect upon copA expression, resulting in higher intracellular copper levels, which would cause copper sensitivity. In order to test this hypothesis, we measured the intracellular copper contents of the mutant, wild-type, and complemented strains by using an atomic absorption spectrophotometer as described previously (19). The intracellular copper contents in the mutant and complemented strains were 5 pmol and 10 pmol/108 cells, respectively, while the copper content of the wild type was less than 1 pmol/108 cells. These results suggest that the failure to complement the copper-sensitive phenotype may be due to an inability of the cell to efflux copper out of the cell.
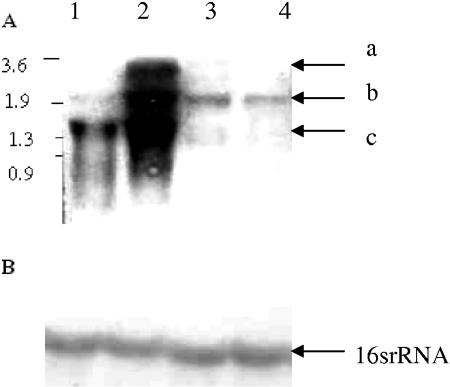
Northern blot analysis was performed to examine the effect of copper on mco transcription. As shown in Fig. 3 (lane 2), three transcripts (1.3, 2.0, and 3.5 kb) were induced by elevated concentrations of copper ions (200 μM) in the parent strain, but no induction occurred in the mutant strain (Fig. 3, lane 4). The three different transcripts might represent the products from different genes or of specific degradation. The 3.5-kb transcript likely corresponds to a transcript containing both mco and copA. The nucleotide sequence analysis indicated that mco and copA genes in S. aureus are separated by 70 bp and have no significant sequence identity. Therefore, hybridization of all three transcripts with mco genes is puzzling and possibly indicative of a complex transcriptional regulation that needs further investigation.
FIG. 3.
Northern blot analysis. (A) Ten micrograms of total RNA was electrophoresed on formaldehyde agarose gels (1.2%) and transferred to a nitrocellulose membrane. The blot was probed with radiolabeled mco. Lane 1, RNA extract from the wild type grown in defined medium (DM); lane 2, RNA extract from the wild type grown in DM with 200 μM CuSO4; lane 3, RNA extract from mutant grown in DM; and lane 4, RNA extract from the mutant grown in DM with 200 μM CuSO4. Arrows indicate the multiple transcripts of approximately 1.3 (a), 2.0 (b), and 3.5 (c) kb. Molecular size markers (in kilobases) are indicated on the left. (B) Same blot, probed with radiolabeled 16S rRNA gene.
In conclusion, we have identified an mco gene which encodes an enzyme that shows oxidase activity in S. aureus. The expression of this gene can be induced by copper. A transposon-induced mutant showed copper and cobalt sensitivity and H2O2 tolerance. Since mco was not found in all strains of S. aureus, this suggests that it plays a dispensable role in staphylococci. It has been suggested that the mco role in copper homeostasis may be compensated for by other gene systems (6). Further studies are required to determine the role of mco in heavy metal homeostasis and oxidative response.
Nucleotide sequence accession number.
The complete nucleotide sequence of the parental gene copy of S. aureus has been deposited to the GenBank database under accession no. AY259130 gi 33391259.
Acknowledgments
We are grateful to James Webb, Illinois State University, for his help in copper measurement and Chia Lee, University of Kansas, for providing plasmid pLI50. We thank Anthony Otsuka for critical reading of the manuscript.
This work has been supported by a Grant-in-Aid from the American Heart Association—Greater Midwest Affiliate to R.K.J.
REFERENCES
- 1.Askwith, C. C., and J. Kaplan. 1998. Site-directed mutagenesis of the yeast multicopper oxidase Fet3p. J. Biol. Chem. 273:22415-22419. [DOI] [PubMed] [Google Scholar]
- 2.Brouwers, G. J., J. P. de Vrind, P. L. Corstjens, P. Cornelis, C. Baysse, and E. W. de Vrind-de Jong. 1999. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 65:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Silva, D., S. Davis-Kaplan, J. Fergestad, and J. Kaplan. 1977. Purification and characterization of Fet3 protein, a yeast homologue of ceruloplasmin. J. Biol. Chem. 272:14208-14213. [DOI] [PubMed] [Google Scholar]
- 4.Eck, R., S. Hundt, A. Hartl, E. Roemer, and W. Kunkel. 1999. A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology 145:2415-2422. [DOI] [PubMed] [Google Scholar]
- 5.Feldman, S. L., J. S. V. Hunter, A. Zgirski, V. M. Chidambaram, and E. Frieden. 1982. Comparison of the catalytic oxidation of cysteine and o-dianisidine by cupric ion and ceruloplasmin. J. Inorg. Biochem. 17:51-60. [DOI] [PubMed] [Google Scholar]
- 6.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horsburgh, M. A., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR control oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hullo, M.-F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huston, W. M., M. P. Jennings, and A. G. McEwan. 2002. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45:1741-1750. [DOI] [PubMed] [Google Scholar]
- 10.Inoue, K., T. Akaike, Y. Miyamoto, T. Okamoto, T. Sawa, M. Otagiri, S. Suzuki, T. Yoshimura, and H. Maeda. 1999. Nitrosothiol formation catalyzed by ceruloplasmin. Implication for cytoprotective mechanism in vivo. J. Biol. Chem. 274:27069-27075. [DOI] [PubMed] [Google Scholar]
- 11.Kim, C., W. W. Lorenz, J. T. Hoopes, and J. F. Dean. 2001. Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J. Bacteriol. 183:4866-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, J. S., M. H. Kim, M. H. Joe, S. S. Song, I. S. Lee, and S. Y. Choi. 2002. The sctR of Salmonella enterica serovar Typhimurium encoding a homologue of MerR protein is involved in the copper-responsive regulation of cuiD. FEMS Microbiol. Lett. 23:99-103. [DOI] [PubMed] [Google Scholar]
- 13.Larrondo, L. F., L. Salas, F. Melo, R. Vicuña, and D. Cullen. 2003. A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl. Environ. Microbiol. 69:6257-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, C. Y., J. J. Schmidt, A. D. Johnson-Winegar, L. Spero, and J. J. Iandolo. 1987. Sequence determination and comparison of the exfoliative toxin A and toxin B genes from Staphylococcus aureus. J. Bacteriol. 169:3904-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann, H. P., K. H. Schosinsky, and M. F. Beeler. 1974. Standardization of serum ceruloplasmin concentrations in international enzyme units with o-dianisidine dihydrochloride as substrate. Clin. Chem. 20:1564-1567. [PubMed] [Google Scholar]
- 16.Lim, S. Y., M. H. Joe, S. S. Song, M. H. Lee, J. W. Foster, Y. K. Park, S. Y. Choi, and I. S. Lee. 2002. cuiD is a crucial gene for survival at high copper environment in Salmonella enterica serovar Typhimurium. Mol. Cells 31:177-184. [PubMed] [Google Scholar]
- 17.Lipsky, P. E. 1984. Immunosuppression by D-penicillamine in vitro: inhibition of human T lymphocyte proliferation by copper- or ceruloplasmin-dependent generation of hydrogen peroxide and protection by monocytes. J. Clin. Investig. 73:53-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 17:30670-30677. [DOI] [PubMed] [Google Scholar]
- 19.Radford, D. S., M. A. Kihlken, G. P. Borrelly, C. R. Harwood, N. E. Le Brun, and J. S. Cavet. 2003. CopZ from Bacillus subtilis interacts in vivo with a copper exporting CPx-type ATPase CopA. FEMS Microbiol. Lett. 220:105-112. [DOI] [PubMed] [Google Scholar]
- 20.Schlosser, D., and C. Höfer. 2002. Laccase-catalyzed oxidation of Mn2+ in the presence of natural Mn3+ chelators as a novel source of extracellular H2O2 production and its impact on manganese peroxidase. Appl. Environ. Microbiol. 68:3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scybert, S., R. Pechous, S. Sitthisak, M. J. Nadakavukaren, B. J. Wilkinson, and R. K. Jayaswal. 2003. NaCl-sensitive mutant of Staphylococcus aureus has a Tn917-lacZ insertion in its ars operon. FEMS Microbiol. Lett. 222:171-176. [DOI] [PubMed] [Google Scholar]
- 22.Solomon, E. I., U. M. Sundaram, and T. E. Machonkin. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 7:2563-2606. [DOI] [PubMed] [Google Scholar]
- 23.Spizzo, T., C. Byersdorfer, S. Duesterhoeft, and D. Eide. 1997. The yeast FET5 gene encodes a FET3-related multicopper oxidase implicated in iron transport. Mol. Gen. Genet. 256:547-556. [DOI] [PubMed] [Google Scholar]
- 24.Stoj, C., and D. J. Kosman. 2003. Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function. FEBS Lett. 554:422-426. [DOI] [PubMed] [Google Scholar]
- 25.Xiong, A., and R. K. Jayaswal. 1998. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J. Bacteriol. 180:4024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterization of a ferric uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146:659-668. [DOI] [PubMed] [Google Scholar]