Abstract
Bacillus sphaericus JG-A12 is a natural isolate recovered from a uranium mining waste pile near the town of Johanngeorgenstadt in Saxony, Germany. The cells of this strain are enveloped by a highly ordered crystalline proteinaceous surface layer (S-layer) possessing an ability to bind uranium and other heavy metals. Purified and recrystallized S-layer proteins were shown to be phosphorylated by phosphoprotein-specific staining, inductive coupled plasma mass spectrometry analysis, and a colorimetric method. We used extended X-ray absorption fine-structure (EXAFS) spectroscopy to determine the structural parameters of the uranium complexes formed by purified and recrystallized S-layer sheets of B. sphaericus JG-A12. In addition, we investigated the complexation of uranium by the vegetative bacterial cells. The EXAFS analysis demonstrated that in all samples studied, the U(VI) is coordinated to carboxyl groups in a bidentate fashion with an average distance between the U atom and the C atom of 2.88 ± 0.02 Å and to phosphate groups in a monodentate fashion with an average distance between the U atom and the P atom of 3.62 ± 0.02 Å. Transmission electron microscopy showed that the uranium accumulated by the cells of this strain is located in dense deposits at the cell surface.
Uranium is a long-lived radionuclide that is an ecological and human health hazard. The mining and processing of uranium for nuclear power plants and nuclear weapons production have resulted in the generation of significant amounts of radioactive wastes. The mobility of this radionuclide is controlled by its interaction with ions, minerals, and microorganisms present in nature. As a consequence of their small size and diverse metabolic activities, bacteria are able to interact intimately with metal ions present in their environment (15). Highly reactive bacterial cell surfaces bind uranium and other metal ions (7). This reactivity arises from the presence of a wide array of ionizable groups, such as carboxylate and phosphate, present in the lipopolysaccharides (LPS) of gram-negative bacterial cell walls (8) and the peptidoglycan, teichuronic acids, and teichoic acids of gram-positive bacteria (9).
The bacterial cell wall may be overlayed by a number of surface structures, which can also interact with metal ions. These may be composed primarily of carbohydrate polymers (e.g., capsules) or proteinaceous surface layers (e.g., S-layers) and may occur singly or in combination (15). The crystalline bacterial cell S-layers represent the outermost cell envelope component of many bacteria and archaea (50). S-layers are generally composed of identical protein or glycoprotein subunits, and they completely cover the cell surface during all stages of bacterial growth and division. Most S-layers are 5 to 15 nm thick and possess pores of identical size and morphology in the range of 2 to 6 nm (6). As porous lattices completely covering the cell surface, the S-layers can provide prokaryotic cells with selective advantages by functioning as protective coats, as structures involved in cell adhesion and surface recognition, and as molecule or ion traps (53).
Strong interest in sorption of U by bacterial surfaces as a method of U immobilization for the bioremediation of uranium-contaminated waters has resulted in numerous macroscopic, microscopic, and spectroscopic sorption studies of U by gram-positive and gram-negative bacteria (19, 26, 40, 42). Microorganisms can mobilize radionuclides and metals through autotrophic and heterotrophic leaching, chelation by microbial metabolites and siderophores, and methylation, which can result in volatilization. Conversely, immobilization can result from sorption to cell components or exopolymers (33), intracellular sequestration, or precipitation as insoluble organic and inorganic compounds, e.g., oxalates, sulfides, or phosphates (10, 49). In the context of bioremediation, solubilization provides a route for removal from solid matrices such as soils, sediments, dumps, and industrial wastes. Alternatively, immobilization processes may enable metals to be transformed in situ into insoluble and chemically inert forms and are particularly applicable to the removal of radionuclides and metals from mobile aqueous phases.
In this paper, we describe the interaction of B. sphaericus JG-A12 cells and its S-layer with uranium. This bacterium was isolated from a uranium mining waste pile near the town of Johanngeorgenstadt in Saxony, Germany. It has been demonstrated that this strain possesses an S-layer protein that differs significantly in its primary structure from those of the other B. sphaericus S-layers studied to date (45, 47). The interaction of this strain with 19 heavy metals (Al, Ba, Cd, Co, Cr, Cs, Cu, Fe, Ga, Mn, Ni, Rb, Si, Sn, Sr, Ti, U, and Zn) was investigated. The results of these studies demonstrated that this strain selectively and reversibly accumulates U, Cu, Pb, Al, and Cd (52).
The aim of the present investigation is to elucidate the nature of the chemical interactions between uranium and the S-layer and cells of B. sphaericus JG-A12 using spectroscopic and microscopic methods. To our knowledge, this is the first study that investigates the coordination of U to bacterial S-layer sheets at a molecular scale.
MATERIALS AND METHODS
Bacterial growth.
B. sphaericus JG-A12 cells were grown to late exponential phase in 6 liters of nutrient broth medium (8 g/liter; Difco), pH 7.0, in 7.5-liter bioreactors (Ochs, Bovenden/Lenglern, Germany). All runs were performed in batch culture at 30°C at a stirring speed of 500 rpm using a magnetic stirrer and airflow at a rate of 3 liters/min. Bacterial growth was monitored by measuring optical densities at 600 nm using Pharmacia Biotech spectrometer Ultrospec 1000 (Amersham Biosciences, Freiburg, Germany). The bacterial cells were harvested by centrifugation at 10,000 × g for 20 min.
Preparation of S-layer protein.
Intact cells were washed once in a buffer containing 50 mM Tris-HCl, 1 mM MgCl2 · 6H2O and 3 mM NaN3, pH 7.5 (referred to as standard buffer). For removing bacterial flagella, the suspension was homogenized in a rotating-blade IKA T8 blender (IKA Labortechnik, Stauffen, Germany) at maximum speed for 10 min on ice. Flagellum-free cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C, resuspended 1:1 in the standard buffer, and mixed with a few crystals of DNase II and RNase. Cells were disintegrated in a Microfluidizer 110L (Microfluidics Cooperation, Newton, Mass.) on ice at a pressure of 9.6 × 107 Pa, with three passes. After removing the unbroken cells by centrifugation at 6,000 × g for 10 min at 4°C, cell wall fragments were washed two times in the standard buffer. Plasma membranes were solubilized in 1% Triton X-100 in the standard buffer solution for 10 min at room temperature, and the remaining cell wall fragments were washed twice. Peptidoglycan was hydrolyzed by incubating the samples in a standard buffer solution containing 0.2 mg/ml lysozyme for 6 h at 30°C. The S-layer fraction was washed several times, resuspended in standard buffer, and stored at 4°C.
Recrystallization of the S-layer.
For solubilization of the S-layer, protein suspensions, each at a concentration of 20 mg/ml, were mixed with a volume of 6 M guanidine hydrochloride in 50 mM Tris (pH 7.2) five times as large as that of the protein suspension and stirred at room temperature for 6 h. Nonprotein components were precipitated by centrifugation at 12,400 × g for 60 min at 4°C. The reassembling of the S-layer proteins was performed by dialyzing the supernatant two times against 2 liters of 10 mM CaCl2 for 24 h at 4°C using dialysis tubes with a molecular weight cutoff of 1,000 to 50,000. Reassembled S-layer sheets were harvested by centrifugation at 12,400 × g for 60 min at 4°C, resuspended in 10 mM CaCl2, and stored at 4°C until use.
Cross-linking of the S-layer.
To stabilize the square lattice structure of the isolated native and recrystallized S-layer sheets, they were incubated with 1-ethyl-3-(N,N′-dimethyl-aminopropyl)-carbodiimide (24) (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) at a concentration of 30 mg/ml in standard buffer for 48 h at 20°C. Cross-linked S-layer sheets were harvested by centrifugation at 12,400 × g for 30 min at 4°C, washed three times with deionized water, and stored at 4°C.
Determination of phosphorus content using ICP-MS.
The surface layer protein was washed 10 times with 30 ml sterile 0.9% NaCl and once with 30 ml sterile ultrapure water. For determination of the phosphorus content of the native surface layer protein, 100 mg of it was resuspended in 1.7 ml sterile ultrapure water in order to achieve a concentration of 60 mg (dry weight) per ml. Samples were treated with 3 to 4 ml 55% (vol/vol) HNO3 and 100 μl 30% (vol/vol) H2O2 and ashed in a microwave (Büchi Labortechnik GmbH, Konstanz, Germany). Sample volumes were adjusted to 20 to 30 ml with ultrapure water, and the phosphorus content was determined by using inductive coupled plasma mass spectrometry (ICP-MS).
Colorimetric determination of phosphorus.
For the determination of phosphorus bound to seryl and threonyl residues in phosphoproteins, a method described previously by Ekman and Jaeger (17) was used. Before the procedure, all glassware was washed with 6 M HCl. The phosphate reagent was prepared as follows: 1 volume of 10% (wt/vol) (NH4)6Mo7O24 · 4H2O in 4 M HCl was mixed with 3 volumes of 0.2% (wt/vol) malachite green. After being stirred for 30 min at room temperature, the solution was filtered. The filter paper and funnel were prewashed in 6 M HCl and drained well before filtering. The first 25% of the filtrate was discarded. For the colorimetric quantification of phosphorus, 100 μl of 2 M NaOH was added to 100 μl of protein suspension, mixed, and incubated for 15 min at 100°C in a water bath. The alkaline hydrolysis was stopped by adding 100 μl of 4.7 M HCl, and samples were cooled to room temperature. After the addition of 100 μl phosphorus reagent, the samples were incubated for at least 20 min at room temperature. The optical density of the solution was measured at 660 nm. The calibration curve was prepared under identical conditions using 0 to 2 nmol K2HPO4 in 100 μl ultrapure water.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out in a Mini Protean II cell (Bio-Rad Laboratories, München, Germany) as described previously by Laemmli (30). The protein analysis was performed using a gel system with a 4% stacking gel and a 10% separation gel. As molecular weight standards, α-2-macroglobulin (181 kDa), bovine serum albumin (68 kDa), egg albumin (43 kDa), carbonic anhydrase (29 kDa), and lysozyme (14.3 kDa) (Sigma-Aldrich Chemical, Deisenhofen, Germany) were used. Gels were stained with Sypro Ruby (Bio-Rad Laboratories, München, Germany) or by use of the GelCode phosphoprotein staining kit (Pierce, Rockford, Ill.).
Phosphoprotein-specific staining of SDS-PAGE gels.
For phosphoprotein-specific staining, the gel was washed in 50 ml of deionized water by gentle agitation for 10 min. Afterwards, the water was replaced by 25 ml of reagent 1 (sulfosalicylic acid solution), and the gel was incubated for 15 min. Reagent 1 was then removed, and 25 ml of reagent 2 (sulfosalicylic acid plus CaCl2 solution) was added. Thirty minutes later, reagent 2 was poured off, and the gel was quickly rinsed with deionized water to eliminate any surface sorbed CaCl2. In a next step, the gel was transferred in 25 ml of reagent 3 (0.5 N NaOH), and the tray was covered with a lid and placed in an oven at 65°C for 20 min. Reagent 3 was replaced by 25 ml of reagent 4 (ammonium molybdate solution) for 10 min with agitation. This step was repeated once. Reagent 4 was then removed from the tray, and the gel was incubated for 20 min in 25 ml of reagent 5 (ammonium molybdate plus HNO3 solution) for 20 min with gentle agitation. Reagent 5 was poured off, 25 ml of reagent 6 (methyl green solution) was added, and the gel was agitated for 20 min. After removing reagent 6, the gel was destained two times for 15 min by adding 25 ml of reagent 1. For complete destaining, the gel was transferred to 25 ml of reagent 7 (7% acetic acid) and incubated overnight with gentle agitation. Solution 7 was replaced once after it turned green.
Preparation of biological samples for X-ray absorption spectroscopy (XAS) analyses.
Bacterial cells grown to the late exponential phase were harvested by centrifugation at 10,000 × g for 20 min at 4°C and washed three times with 0.01 M NaClO4 to remove the contaminants and the interfering ingredients of the growth medium. The washed cells and S-layers were resuspended and shaken for 48 h in 35 ml of 9 × 10−4 M UO2(NO3)2 · 6H2O, pH 4.5. We used 0.01 M NaClO4 as a background electrolyte.
After contact with the uranium solution, the cells were harvested and washed with 0.01 M NaClO4. The pelleted samples were dried in a vacuum incubator at 30°C for 24 h and powdered.
Reference samples.
Two solutions, one of U(VI) and another one of U(IV), each at a concentration of 4 × 10−2 M in 1 M HClO4, served as reference samples for the uranium oxidation states. The stock solution of U(VI) was obtained by dissolving Na2U2O7 · 6H2O in 7 M HClO4. Part of this solution was reduced electrochemically to U(IV) at a mercury pool cathode. The uranium oxidation state in these solutions was confirmed by UV/visible spectroscopy.
The U-oxygen axial atom (Oax), U-equatorial oxygen atom (Oeq), U-C, and U-P bond length values found in this work were compared to those of solid U(VI) reference compounds to characterize the coordination of the uranium species formed on the cells and S-layer of B. sphaericus JG-A12. Good agreement was found with the structures of calcium uranyl phosphate (meta-autunite, Ca[UO2PO4]2 · 6H2O), a uranyl phosphate mineral phase from Autun/France and a solution of uranyl triacetate [UO2(CH3COO)34−] at pH 4.48. Therefore, extended X-ray absorption fine-structure (EXAFS) measurements of these compounds were performed. The compositional and structural identity of meta-autunite to the compounds reported in the literature (22, 32, 35) was checked by energy-dispersive X-ray fluorescence spectroscopy and powder X-ray diffraction (XRD) analysis.
XAS measurement.
Uranium LIII edge XAS spectra were collected at room temperature in fluorescence and in transmission mode at the Rossendorf Beamline at the European Synchrotron Radiation Facility, Grenoble, France (36), using the Si(111) double-crystal monochromator. The energy was calibrated by measuring the Y K-edge transmission spectrum of an yttrium foil and defining the first inflection point as 17,038 eV. The biological/uranyl samples were measured as dry samples. Eight spectra for each sample were recorded. The region from about 45 eV below to 60 eV above the absorption edge of each scan was isolated for the X-ray near-edge structure (XANES) analysis. The pre-edge background was then subtracted, and the absorption coefficient was normalized to equal intensity at 17,230 eV so that all the spectra could be plotted on the same scale in the figure.
The EXAFS oscillations were isolated from the raw, averaged data by removal of the pre-edge background, approximated by a first-order polynomial, followed by μ0 removal via spline-fitting techniques and normalization using a Victoreen function. Dead-time correction was applied. The ionization energy for the U LIII electron, E0, was arbitrarily defined as 17,185 eV for all averaged spectra. The EXAFS spectra were analyzed according to standard procedures using the program EXAFSPAK (20). The theoretical scattering phase and amplitude functions used in data analysis were calculated using FEFF8 (3). All fits included the four-legged multiple-scattering path of the uranyl group U-Oax-U-Oax. The coordination number (N) of this multiple-scattering path was linked to N of the single-scattering (SS) path U-Oax. The radial distance (R) and Debye-Waller (σ2) factor of the multiple-scattering path were linked at twice the R and σ2 of the SS path U-Oax, respectively (25). During the fitting procedure, N of the U-Oax SS path was held constant at 2. The amplitude reduction factor was held constant at 1.0 for the FEFF8 calculation and EXAFS fits. The shift in threshold energy, ΔE0, was varied as a global parameter in the fits.
Sample preparation for electron microscopy.
Transmission electron microscopy (TEM) is a useful technique that can help to localize and to identify uranium deposited within or around microbial cells.
After incubation with uranium, the bacterial cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 2 h at 4°C and then washed three times with the same cacodylate buffer. The cell pellets were fixed for 60 min at 4°C in 1% OsO4 in cacodylate buffer before being dehydrated with ethanol and embedded in Spurr resin. The samples were thin sectioned (0.25 μm) using a diamond knife on a Reichert Ultracut S ultramicrotome, and the sections were supported on copper grids and coated with carbon. Samples were examined with a high-resolution Philips CM 200 transmission electron microscope at an acceleration voltage of 200 kV under standard operating conditions with the liquid nitrogen anticontaminator in place.
Energy-dispersive X-ray (EDX) analysis can provide elemental information via the analysis of X-ray emissions caused by a high-energy electron beam. EDX analysis was also performed at 200 kV using a spot size of 70 Å and a live counting time of 200 s. For selected-area electron diffraction, we used the Philips CM 200 transmission electron microscope in the diffraction mode with a camera length of 1,000 mm and an exposure time between 15 and 20 seconds.
RESULTS
Determination of phosphorus content.
The phosphorus content of all samples studied in this work was determined in order to quantify the bioavailable phosphorus that could serve as a possible binding site for uranium. The phosphorus in cells, cell walls, and S-layer sheets of B. sphaericus JG-A12 was measured by ICP-MS (Table 1). To confirm the results obtained by the ICP-MS measurements, the P content of the S-layer protein from B. sphaericus JG-A12 was also analyzed by use of a colorimetric method described previously by Ekman and Jäger (17) (Table 1). Results of the B. sphaericus JG-A12 S-layer analyses were compared to those of analyses performed with the S-layer from the reference strain, B. sphaericus NCTC 9602.
TABLE 1.
Comparison of phosphorus contents of cells, cell wall, recrystallized S-layers from B. sphaericus strain JG-A12, and recrystallized S-layers from B. sphaericus NCTC 9602 determined by ICP-MS and colorimetric measurements
| Sample | Phosphorus content (nmol/mg [dry wt])
|
|
|---|---|---|
| ICP-MS | Colorimetric methoda | |
| B. sphaericus JG-A12 cells | 3,040 ± 100 | − |
| B. sphaericus JG-A12 cell wall | 460 ± 40 | − |
| B. sphaericus JG-A12 S-layer | 4.81 ± 0.05 | 1.93 ± 0.02 |
| B. sphaericus NCTC 9602 S-layer | 0.78 ± 0.03 | 0.35 ± 0.02 |
−, not determined.
The cells had the highest phosphorus content of all examined samples. Many structures in the bacterial cells contain this element. Gram-positive bacteria like Bacillus may possess teichoic acids as part of their cell wall (4, 23). In addition, the phospholipids of the plasma membrane and the phosphodiester or phosphonate compounds of energy metabolism and signal transduction cascades make a contribution to the total quantity of phosphorus. The sample with the second-highest P content was the cell wall fraction, followed by the S-layer samples. As evident from the results presented in Table 1, the S-layer of the U waste pile isolate B. sphaericus JG-A12 contains six times more phosphorus than the S-layer of the reference strain, B. sphaericus NCTC 9602. The quantification of phosphorus by ICP-MS resulted in higher amounts of phosphorus than those obtained by the colorimetric method. The differences in P content could be due to the different detection limits of the two methods.
Phosphoprotein-specific staining following SDS-PAGE.
Because a contamination of the S-layer fraction with phosphorus-containing cell wall components cannot be completely excluded from the results of the ICP-MS measurements and the colorimetric determination of phosphorus, phospho-specific staining following SDS gel electrophoresis was undertaken to verify that S-layer proteins of B. sphaericus JG-A12 and NCTC 9602 contain phosphorus.
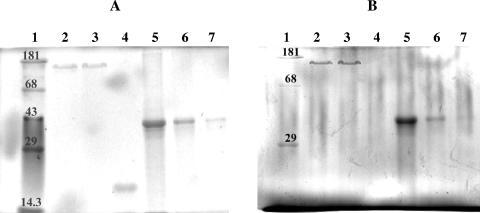
All of the different protein samples were analyzed using conventional staining of SDS-PAGE gels. As highly phosphorylated proteins cannot be stained by Coomassie brilliant blue or silver staining, the reference gel was stained with the fluorescent dye Sypro Ruby. As evident from the results shown in Fig. 1A, neither purified S-layer protein sample showed any signs of protein contamination (Fig. 1A, lanes 2 and 3). An SDS-PAGE gel prepared in parallel was stained with the GelCode Phosphoprotein Staining kit (Pierce, Rockford, Ill.) (Fig. 1B) for the detection of phosphoproteins. The positive staining of the S-layer proteins from B. sphaericus JG-A12 and from B. sphaericus NCTC 9602 confirmed phosphorylation (Fig. 1B, lanes 2 and 3).
FIG. 1.
SDS-PAGE protein analyses. (A) Total protein staining with Sypro Ruby. Ten micrograms of S-layer protein of B. sphaericus JG-A12 (lane 2), 10 μg of S-layer protein of B. sphaericus NCTC 9602 (lane 3), 2 μg nonphosphorylated protein soybean trypsin inhibitor (lane 4), and the highly phosphorylated protein phosvitine in different concentrations of 10 μg (lane 5), 2 μg (lane 6), and 0.5 μg (lane 7) are shown. In lane 1, molecular mass standards (in kilodaltons) were applied. (B) Phosphoprotein-specific staining. Lane 1, molecular mass standard (molecular weights in kilodaltons); lane 2, 10 μg B. sphaericus JG-A12 S-layer protein; lane 3, 10 μg B. sphaericus NCTC 9602 S-layer protein; lane 4, 10 μg soybean trypsin inhibitor (negative control); lanes 5 to 7, phosvitine (10, 2, and 0.5 μg) (positive control).
TEM.
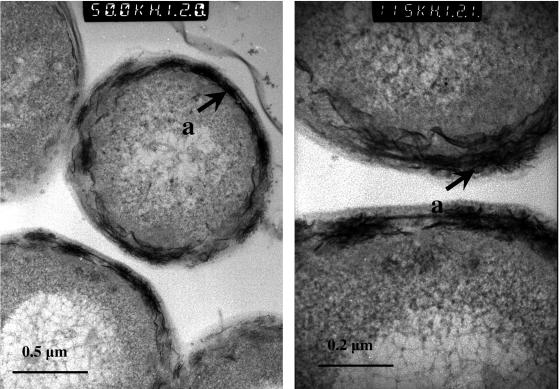
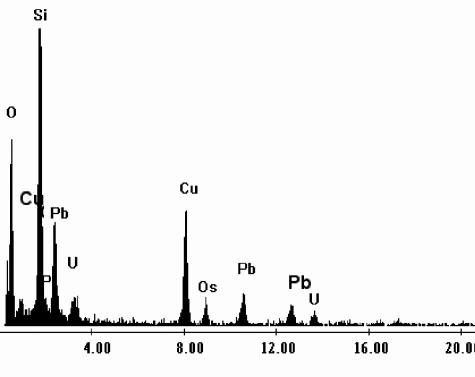
TEM observation of B. sphaericus JG-A12 cells exposed to 0.9 mM U solution (Fig. 2) revealed the presence of electron-dense accumulations on the cell surface (cytoplasmic membranes, peptidoglucan, and S-layer). The EDX spectrum derived from these deposits (Fig. 3) indicated that they are composed of oxygen (O), phosphorus (P), and uranium (U). The high copper (Cu) peak is from the electron microscopy grid used to support the specimen. The lead (Pb) peak originated from the lead citrate solution which was used to improve the visualization of the uranium-treated thin cell sections. The presence of the Si peak can be attributed to impurities from the culture medium and/or from the glass material of the flasks in which the cells were grown.
FIG. 2.
Transmission electron micrographs of a thin section of B. sphaericus JG-A12 cells treated with uranium. The metal accumulated is located on the cell surface (a).
FIG. 3.
Energy-dispersive X-ray spectrum of the uranium accumulated at the cell surface of B. sphaericus JG-A12.
XANES.
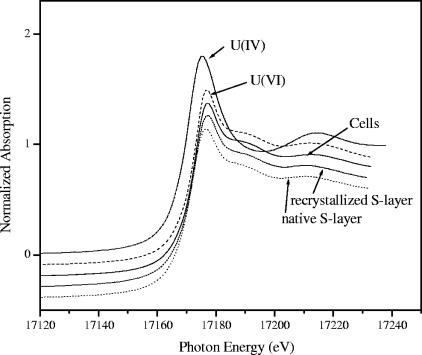
XANES spectra (Fig. 4) of the uranium complexes formed by cells and S-layer of B. sphaericus JG-A12 contained a XANES peak at 17,188 eV which has previously been attributed to uranium in the 6+ oxidation state (25). In addition, the intensity maximum for the absorption edge occurred at the position characteristic for U(VI). Both observations indicate that uranium is present in the samples as U(VI).
FIG. 4.
Normalized uranium LIII-edge XANES spectra of 0.04 M U(IV) in 1 M HClO4, 0.04 M U(VI) in 1 M HClO4, cells, and native and recrystallized S-layer of B. sphaericus JG-A12. The spectra were normalized to equal intensity at 17,230 eV.
EXAFS.
Information on the local environment of uranium atoms in the uranium bacterial samples was obtained by analysis of the EXAFS data.
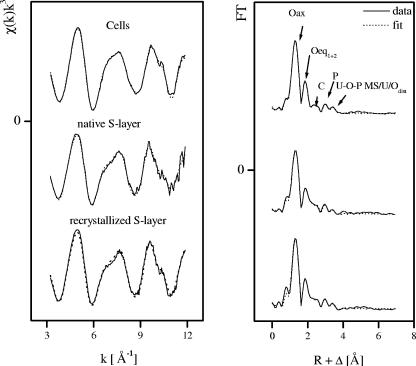
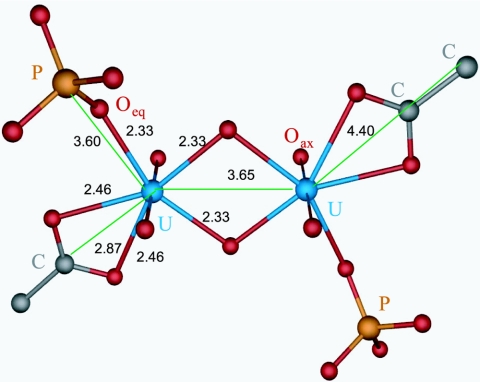
The k3-weighted χ spectra determined from EXAFS analyses of the uranium species formed at pH 4.5 by the cells, and by the native and recrystallized S-layer of B. sphaericus JG-A12, are presented in Fig. 5 along with the best fits obtained from the fitting procedure. Solely on the basis of the raw data, the spectra showed a strong similarity. There was close agreement among the three samples in the phases as well in the amplitude of all the features in k space. Similarly, the R-space plots of the Fourier-transformed EXAFS shown were also in close agreement. The Fourier transforms (FTs) represent a pseudoradial distribution function of the uranium near-neighbor environment. The peaks appeared at lower R values relative to the true near-neighbor distances as a result of the EXAFS phase shift, which is different for each neighboring atom (φ = 0.2 to 0.5 Å). The theoretical phase and amplitude functions used in data analysis were calculated with FEFF8 using the model described in the legend of Fig. 6, which contains fragments of two molecules: meta-autunite and uranyl triacetate. This model includes two oxygen axial (Oax) atoms, two shells of equatorial oxygen atoms (Oeq1 and Oeq2), a shell of carbon (C) atoms, a U-Oax-U-Oax multiple-scattering path from the two tightly bound Oax atoms, a shell of phosphorus (P) atoms, a multiple-scattering path from the Oeq atoms to the P atoms, and a shell of uranium atoms at approximately 3.65 Å.
FIG. 5.
Uranium LIII-edge k3-weighted EXAFS spectra (left) and corresponding FT (right) of the uranium complexes formed by the S-layer and vegetative cells of B. sphaericus JG-A12.
FIG. 6.
Model used for the fit of cell and S-layer uranium spectra (fragments of two molecules, meta-autunite and uranyl triacetate). In this model, the uranium has two oxygen axial atoms (Oax) at distance of 1.77 Å and two types of equatorial oxygen atoms (Oeq1 at a distance of 2.33 Å and Oeq2 at a distance of 2.46 Å). The uranium is coordinated to carboxyl groups in a bidentate fashion, with an average distance between the U atom and the C atom of 2.87 Å, and to phosphate groups in a monodentate fashion, with an average distance between the U atom and the P atom of 3.60 Å. In addition, there is a U-U bond distance of 3.65 Å.
The uranyl unit consists of a uranium center with a formal charge of +6 coordinated to two doubly bonded oxygen atoms to form a linear dioxo cation, UO22+. This unit is highly stable and binds to other ligands via the formation of U-O bonds in a plane perpendicular to the axis of the uranyl ion. The “equatorial” oxygen coordination number varies from 4 to 6 depending on the chemical environment, and these equatorial bonds are the sole means of complexation available for uranyl units under normal conditions.
The FT of the EXAFS spectra of the samples studied in this section had five significant peaks (Fig. 5). The first two peaks arose from the scattering contribution of the Oax at 1.3 Å and the Oeq at 1.8 Å in the FT. The amplitude of the entire EXAFS spectrum was influenced mainly by these two strong contributions. They were considered major components. The weaker peaks seen above 2 Å in the FT were considered minor components. It should be mentioned that the major components also included the four-legged multiple-scattering path U-Oax-U-Oax along the uranyl chain, since it has previously been observed to contribute significantly to the uranyl EXAFS signal (2). The coordination number (N) of this multiple-scattering path was linked to N of the SS path U-Oax. The radial distance (R) and Debye-Waller (σ2) factor of the multiple-scattering path were linked at twice the R and σ2 of the SS path U-Oax, respectively. During the fitting procedure, N of the U-Oax SS path was held constant at 2.
The third and the fourth FT peaks, which appeared at 2.3 and 3 Å, were the result of the back-scattering from carbon and phosphorus, respectively.
The weak FT peak at 3.4 Å could have arisen from the following: (i) a twofold degenerated three-legged multiple-scattering path, U-Oeq1-P (model A), (ii) a multiple-scattering path, U-Oeq1-P and U-distal oxygen atom (Odist) SS (model B), or (iii) an SS path, U-U (model C).
In order to determine whether this peak arose from SS path U-U, we measured the same samples at a low temperature, and we compared the resulting spectra to those obtained at room temperature. The intensity of the corresponding FT peak did not change (data not shown), indicating that it originated from the contribution of single scattering or/and multiple scattering of a light atom. At low temperature, the disorder due to thermal vibrations is reduced, and therefore, the contribution of heavy atoms (e.g., U) to the EXAFS amplitude is enhanced (56). We therefore excluded model C from further discussion.
During the fitting procedure according to models A and B, the coordination number (N) of the U-Oeq1-P multiple-scattering path was linked at twice the coordination number of the U-P single-scattering path.
In order to isolate the minor components from the whole EXAFS spectrum, the difference technique was used (54). This method can increase the sensitivity in the determination of the EXAFS structural parameters of more distant coordination shells of the uranium complexes. Briefly, the difference technique consists of the following steps. First, the raw k3-weighted EXAFS spectrum is fitted using a structural model that contains only the SS and multiple-scattering paths of the major components, i.e., Oax and Oeq1,2. The difference between this fit and the experimental EXAFS spectrum, i.e., the residual spectrum, contains the contributions of the minor components and experimental noise. Second, the residual spectrum is Fourier filtered in the range of 1.8 to 4.0 Å to separate the minor scattering contributions from the experimental noise.
In addition to the difference technique, we also fitted the raw EXAFS data in a seven-shell model. It included both the major and minor components according to cases A and B. The results for the scattering paths U-Oax, U-Oeq1, U-Oeq2, U-C, and U-P; the U-Oeq1-P multiple-scattering path; and U-Odist (Tables 2 and 3) are very similar to the results obtained by the difference technique. Small differences in N and σ2 between the fits of raw and filtered residual EXAFS spectra could be due to nonseparable U-Oeq1,2 contributions, which superimpose onto the U-P contribution.
TABLE 2.
Structural parameters of the uranium complexes formed by the S-layer and cells of B. sphaericus JG-A12 (model A)
| Sample | Shell | Na | R (Å)b | σ2 (Å2)c | ΔE (eV) | Errord |
|---|---|---|---|---|---|---|
| Cells | U-Oax | 2e | 1.76 | 0.0023 | −10.9 | 0.028 |
| U-Oeq1 | 3.1 (6) | 2.30 | 0.0085 | |||
| U-Oeq2 | 2.7 (2) | 2.45 | 0.0085 | |||
| U-C | 1.3 (2) | 2.89 | 0.0038f | |||
| U-P | 1.7 (1) | 3.62 | 0.0016 | |||
| U-Oeq1-P (MS) | 3.4 | 3.74 | 0.0016 | |||
| Native S-layer | U-Oax | 2e | 1.78 | 0.0028 | −9.5 | 0.084 |
| U-Oeq1 | 1.8 (3) | 2.32 | 0.0057 | |||
| U-Oeq2 | 3.1 (6) | 2.49 | 0.0057 | |||
| U-C | 1.8 (4) | 2.88 | 0.0038f | |||
| U-P | 2.2 (4) | 3.63 | 0.0052 | |||
| U-Oeq1-P (MS) | 4.4 | 3.75 | 0.0052 | |||
| Recrystallized S-layer | U-Oax | 2e | 1.78 | 0.0021 | −8.6 | 0.071 |
| U-Oeq1 | 2.4 (3) | 2.34 | 0.0058 | |||
| U-Oeq2 | 3.2 (4) | 2.50 | 0.0058 | |||
| U-C | 2.1 (4) | 2.87 | 0.0038f | |||
| U-P | 2.1 (3) | 3.64 | 0.0040 | |||
| U-Oeq1-P (MS) | 4.2 | 3.76 | 0.0040 |
Errors in coordination numbers are ±25%, and standard deviations, as estimated by EXAFSPAK, are given in parentheses.
Errors in distance are ±0.02 Å.
Debye-Waller factor.
Error is given as the normalized fit error, Σ [χdata(k)k3 − χfit(k)k3]2/(P − F) (P is the number of data points, and F is the number of variables).
Value fixed for calculation.
Value taken from Table 6.
TABLE 3.
Structural parameters of the uranium complexes formed by the S layer and cells of B. sphaericus JG-A12 (model B)
| Sample | Shell | Na | R (Å)b | σ2 (Å2)c | ΔE (eV) | Errord |
|---|---|---|---|---|---|---|
| Cells | U-Oax | 2e | 1.76 | 0.0023 | −11.6 | 0.021 |
| U-Oeq1 | 4.3 (4) | 2.31 | 0.0120 | |||
| U-Oeq2 | 2.7 (3) | 2.45 | 0.0120 | |||
| U-C | 1.5 (1) | 2.88 | 0.0038f | |||
| U-P | 2.1 (2) | 3.62 | 0.0029 | |||
| U-Oeq1-P (MS) | 4.2 | 3.74 | 0.0029 | |||
| U-Odist | 1.7 (4) | 4.02 | 0.0097e | |||
| Native S layer | U-Oax | 2e | 1.78 | 0.0028 | −9.6 | 0.075 |
| U-Oeq1 | 1.9 (3) | 2.32 | 0.0060 | |||
| U-Oeq2 | 3.2 (7) | 2.48 | 0.0060 | |||
| U-C | 1.8 (4) | 2.88 | 0.0038f | |||
| U-P | 2.4 (6) | 3.62 | 0.0059 | |||
| U-Oeq1-P (MS) | 4.8 | 3.74 | 0.0059 | |||
| U-Odist | 1.6 (7) | 4.09 | 0.0097e | |||
| Recrystallized S layer | U-Oax | 2e | 1.78 | 0.0020 | −8.5 | 0.066 |
| U-Oeq1 | 2.0 (2) | 2.34 | 0.0030 | |||
| U-Oeq2 | 2.7 (3) | 2.50 | 0.0030 | |||
| U-C | 1.7 (3) | 2.87 | 0.0038f | |||
| U-P | 2.3 (3) | 3.65 | 0.0047 | |||
| U-Oeq1-P (MS) | 4.6 | 3.78 | 0.0047 | |||
| U-Odist | 2.2 (8) | 4.05 | 0.0097e |
Errors in coordination numbers are ±25%, and standard deviations, as estimated by EXAFSPAK, are given in parentheses.
Errors in distance are ±0.02 Å.
Debye-Waller factor.
Error is given as the normalized fit error, Σ [χdata(k)k3 − χfit(k)k3]2/(P − F) (P is the number of data points, and F is the number of variables).
Value fixed for calculation.
Value taken from Table 6.
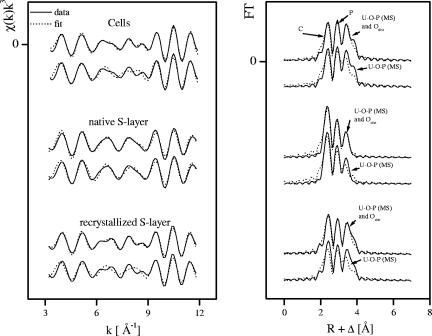
Tables 4 and 5 summarize the structural parameters of the minor components determined according to models A and B, respectively. Figure 7 shows the best fit to the Fourier-filtered residual EXAFS spectra.
TABLE 4.
EXAFS fit results for C, P, and U-O-P multiple-scattering shells from fits to residual spectra (model A)
| Sample | Shell | Na | R (Å)b | σ2 (Å2)c | ΔE (eV)d |
|---|---|---|---|---|---|
| Cells | U-C | 1.2 (1) | 2.89 | 0.0038e | −10.9 |
| U-P | 2.2 (1) | 3.64 | 0.0016 | ||
| U-Oeq1-P (MS) | 4.4 | 3.76 | 0.0016 | ||
| Native S-layer | U-C | 1.5 (1) | 2.88 | 0.0038e | −9.5 |
| U-P | 2.0 (1) | 3.62 | 0.0045 | ||
| U-Oeq1-P (MS) | 4.0 | 3.74 | 0.0045 | ||
| Recrystallized S-layer | U-C | 1.1 (1) | 2.89 | 0.0038e | −8.6 |
| U-P | 1.5 (1) | 3.65 | 0.0022 | ||
| U-Oeq1-P (MS) | 3.0 | 3.77 | 0.0022 |
TABLE 5.
EXAFS fit results for C, P, and U-O-P multiple-scattering shells and U-Odist SS shells from fits to residual spectra (model B)
| Sample | Shell | Na | R (Å)b | σ2 (Å2)c | ΔE (eV)d |
|---|---|---|---|---|---|
| Cells | U-C | 1.2 (1) | 2.89 | 0.0038e | −11.6 |
| U-P | 2.2 (3) | 3.63 | 0.0040 | ||
| U-Oeq1-P (MS) | 4.4 | 3.77 | 0.0040 | ||
| U-Odist | 2.8 (3) | 4.01 | 0.0097f | ||
| Native S-layer | U-C | 1.5 (1) | 2.89 | 0.0038e | −9.6 |
| U-P | 2.1 (1) | 3.62 | 0.0050 | ||
| U-Oeq1-P (MS) | 4.2 | 3.74 | 0.0050 | ||
| U-Odist | 1.4 (4) | 4.09 | 0.0097f | ||
| Recrystallized S-layer | U-C | 1.2 (1) | 2.90 | 0.0038e | −8.5 |
| U-P | 1.3 (1) | 3.64 | 0.0018 | ||
| U-Oeq1-P (MS) | 2.6 | 3.77 | 0.0018 | ||
| U-Odist | 2.2 (2) | 4.06 | 0.0097f |
FIG. 7.
Residual EXAFS of the uranium complexes formed by vegetative cells and native and recrystallized S-layer proteins of B. sphaericus JG-A12 (produced by subtraction of U-Oax, U-Oeq1,2, and U-Oax multiple-scattering shells) and its Fourier transform corresponding to the region between 1.8 and 4 Å. The peak at 3.4 Å was modeled as (A) U-O-P (multiple scattering) and U-Odist (SS) and (B) U-O-P (multiple scattering).
In all samples, fitting showed that U(VI) has two Oax at a distance of 1.76 to 1.78 Å and Oeq at 2.31 to 2.50 Å. The Oeq shell is split into two components, with the first component (U-Oeq1) at distance of 2.30 to 2.34 Å and the second component (U-Oeq2) at a somewhat longer distance (2.45 to 2.50 Å). These two shells were not represented as separate peaks in the FT since their distances span an R range that was not large enough to be discerned as individual peaks in an EXAFS spectrum for which Δk = 8.7 Å−1 in accordance with ΔR ≥ π/(2Δk).
The U-Oeq1 bond distance is within the range of previously reported values for the oxygen atom of the phosphate bound to uranyl (22, 26, 40). The longer equatorial oxygen bond length (2.45 to 2.50 ± 0.02 Å) is similar to previously reported values for the oxygen atom of the carboxyl bound to uranyl (2.45 to 2.51 Å) (14, 26).
As evident in the residual EXAFS (Fig. 7), all spectra contained strong FT components at (i) 2.3 Å, (ii) 3 Å, and (iii) 3.4 Å. These shells were interpreted as arising from the following: (i) C at 2.88 Å or P at 3.02 Å, (ii) P at 3.6 Å, and (iii) multiple-scattering U-Oeq1-P at 3.72 Å or U-Odist at 3.90 to 4.10 Å.
The EXAFS corresponding to the 2.3 Å FT peak was a better fit with one to two carbon atoms at a distance of 2.88 Å than with a phosphorus shell. A survey of the interatomic uranium-carbon distances in solid-phase ring-forming complexes with oxygenated organic ligands found in the Cambridge Structure Database (1) produced the following summary: 2.8 to 2.9 Å for four-membered rings, 3.2 to 3.4 Å for five-membered rings, and 3.2 to 3.6 Å for six-membered rings (no results were available for larger rings). The 2.88-Å U-C distance is most consistent with a four-membered ring resulting from a bidentate complex with a carboxylate group. This distance has also been reported for bidentate coordination of carbonate to uranyl and actinyl complexes (5, 12, 48). In addition, this bond distance is similar to that found in the EXAFS spectrum of the reference sample (uranyl triacetate) measured in this work (Fig. 8 and Table 6).
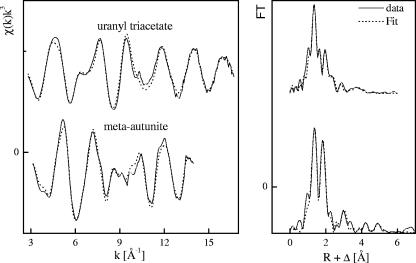
FIG. 8.
Uranium LIII-edge k3-weighted EXAFS spectra (left) and corresponding FT (right) of the reference compounds (meta-autunite and uranyl triacetate).
TABLE 6.
Structural parameters of the uranium reference compounds used in this work
| Sample | Shell | Na | R (Å)b | σ2 (Å2)c | ΔE (eV) |
|---|---|---|---|---|---|
| Uranyl triacetate | U-Oax | 2d | 1.78 | 0.0014 | −8.6 |
| U-Oeq | 6.0 (4) | 2.47 | 0.0085 | ||
| U-C1 | 3.1 (5) | 2.87 | 0.0038 | ||
| U-C2 | 3.1 | 4.39 | 0.0038 | ||
| Meta-autunitee | U-Oax | 2.2 (2) | 1.76 | 0.0045 | −11.0 |
| U-Oeq | 3.9 (2) | 2.29 | 0.0026 | ||
| U-P | 2.3 (3) | 3.60 | 0.008d | ||
| U-U | 2.7 (6) | 5.20 | 0.008d |
Errors in coordination numbers are ±25%, and standard deviations, as estimated by EXAFSPAK, are given in parentheses.
Errors in distance are ±0.02 Å.
Debye-Waller factor.
Value fixed for calculation.
See reference 22.
In this study, bidentate coordination refers to an inner-sphere complex where two oxygen atoms of a single carboxyl functional group are shared with a uranyl ion.
The implication of carboxyl groups in the interaction with uranium is supported by infrared spectroscopy studies (data not shown). FEFF8 calculations predict that the triangular U-C-Oeq2 multiple-scattering path (effective bond distance ≈ 3.29 Å) can also produce an FT peak at ca. 2.3 Å but that the EXAFS oscillations for these paths are about π radians out of phase with the C single-scattering EXAFS and hence do not fit the residual EXAFS (i.e., other shells fit and were subtracted from the spectra). Furthermore, the amplitude of the triangular paths should be much smaller than (about one-fifth) those of U-C SS paths. For these reasons, the triangular U-C-Oeq multiple-scattering path is hereafter omitted from discussions.
Approximately 30 to 50% of the 3-Å FT peak amplitude can be accounted for as linear multiple scattering in the uranyl transdioxo unit. In all samples, an additional P shell at the later FT peak (3 Å) was required to fit the spectra. This U-P distance is consistent with monodentate coordination of phosphate to the uranyl equatorial plane as it was found in meta-autunite (Fig. 8 and Table 6). This topology is common in crystalline uranyl phosphates (11) and was reported for U(VI) sorbed by bacterial cells (22, 26, 40) and by plants (21).
A very weak, but real, frequency that has an FT peak at 3.4 Å was observed in the residual EXAFS for all samples. Attempts to fit this shell with the U-Odist SS path or with U-Oeq1-P multiple scattering resulted in significantly poorer qualities of fit, leading us to prefer the fit in which both paths are included.
DISCUSSION
The speciation and bonding environment of U in bacteria and their surface structures have been examined with a variety of spectroscopic techniques such as Fourier-transform infrared spectroscopy (37), Raman spectroscopy (27), and time-resolved laser-induced fluorescence spectroscopy (27, 37, 44). In addition, synchrotron-based techniques such as X-ray absorption spectroscopy have been used to determine the oxidation state (XANES) and to identify the number of atoms and their distances in the local structural environment (EXAFS) of U within a variety of microbial samples (22, 26, 38-41). In addition, XAS is among the few analytical methods that can provide information on the chemical environment of actinides in bacterial samples at dilute metal concentrations. XAS is a nondestructive method, and no sample reduction or digestion, which would alter the chemistry of the element of interest, is required.
The present work demonstrates that X-ray absorption spectroscopic methods (EXAFS and XANES) coupled with electron microscopy are particularly suited for differentiating among the large variety of uranium species that may be associated with S-layer sheets and vegetative cells of bacteria.
Interaction of B. sphaericus JG-A12 S-layer protein with U.
B. sphaericus JG-A12 cells possess an S-layer protein, which forms a tetragonally arranged structure on the surface of the cell wall with a lattice constant of 12.5 nm (47). The molecular mass of the protein monomers was estimated to be 126 kDa, and its primary structure differs significantly from that of all S-layer proteins previously known but shows the greatest similarity to that of the S-layer protein from the reference strain B. sphaericus NCTC 9602 (45, 46). The surface layer protein of B. sphaericus JG-A12 was studied in detail because this strain is able to selectively and reversibly bind metals from heavy-metal-contaminated U wastewaters.
Surprisingly, this S-layer (as the outermost cell wall component) possesses phosphate groups. To date, phosphorylation of the S-layer has been reported only in the pathogen Aeromonas hydrophila (55). In Lactobacillus helveticus, the exact location of demonstrable phosphorus in the cell envelope is uncertain (43). Compared to the S-layer protein of the uranium mining waste pile B. sphaericus JG-A12, the S-layer protein of its closest relative, the reference strain B. sphaericus NCTC 9602, contains about one-sixth as much phosphorus. This difference may reflect an adaptation of B. sphaericus JG-A12 to the extensive uranium contamination of its environment. The high affinity of phosphate groups on the surface of the cell may allow B. sphaericus JG-A12 to selectively bind large amounts of uranium before being damaged by this toxic radionuclide. The MIC of U for the growth of this bacterium is about 2 mM (unpublished data). EXAFS analysis of the uranium bound to this surface protein indicated that U is coordinated by phosphate groups in a monodentate mode and by carboxyl groups in a bidentate fashion. Kelly et al. (26) observed the same type of environment for U adsorbed by Bacillus subtilis cell walls between pH 1.67 and 4.80.
Previous studies (45) revealed that the amino acid composition of this surface protein included a high content of glutamic and aspartic acids and of other amino acids such as serine and threonine. The C-terminal part especially consists of stretches of glutamic acid and aspartic acid (both residues with carboxyl groups) and of serine and threonine (both residues with hydroxyl groups), the latter being potential phosphorylation sites. The carboxyl groups and phosphate groups of these amino acids are probably implicated in the complexation of uranium. The participation of the carboxyl groups of this S-layer in the binding of palladium has been demonstrated using EXAFS spectroscopy and attenuated total reflectant-FT-infrared spectroscopy (46, 51).
Detailed information about the functions of S-layers exists only for some bacteria. In the case of environmental isolates, the S-layer may function as a molecular sieve, ion trap, or protective shell (53). Because of the ability of the S-layer to self-assemble and replace the “older” S-layer sheets on the cell surface, one can speculate about the mechanism of its protective function against uranium and other toxic metals. The saturation with metals (in this case, uranium, as was demonstrated by TEM studies) may lead to denaturation of the S-layer lattice, which is then replaced by freshly synthesized protein monomers.
Interaction of B. sphaericus JG-A12 cells with U.
TEM analysis showed that U accumulated by the cells of B. sphaericus JG-A12 is located at the cell surface (areas of high contrast). Extracellular association of uranium with bacterial cell surfaces is primarily due to physical and chemical interactions involving adsorption, ion exchange, and complexation and does not directly depend on metabolism. Bacterial cell walls, exopolymers, proteins, and lipids contain functional groups such as carboxylate, hydroxyl, amino, and phosphate, which are capable of forming complexes with uranium (19). A similar accumulation profile was found using another strain of B. sphaericus, strain JG-7B, isolated from the same uranium mining waste (39). Krueger et al. (29) have shown that surface-bound uranium in Pseudomonas fluorescens was spread over the entire cell envelope in an outer membrane-peptidoglycan-plasma membrane complex as fine-grained, platy uranium minerals (10 nm to 1 μm). Other microorganisms such as Stenotrophomonas maltophilia JG-2 and Acidithiobacillus ferrooxidans (38-41) can accumulate U intracellularly via passive transport mechanisms, resulting in the formation of dense uranium deposits inside the cells.
Optimization of U(VI) bioremediation strategies requires an extensive understanding of the speciation of uranium associated with bacteria. If present in crystalline phases that are sufficiently concentrated, uranium speciation can be determined using XRD. However, when the concentration of uranium-containing crystalline phases is below that needed for identification by XRD (typically <1% by volume) or when uranium is present in a noncrystalline phase, other molecular-scale characterization methods are required, including ones not requiring long-range order. One of the most effective techniques for determining the molecular-level speciation of heavy metals and actinide ions at dilute concentrations in complex materials is XAS (XANES and EXAFS) spectroscopy.
In this study, XANES analysis allowed the identification of U(VI) as the main uranium species bound to the cell surface of B. sphaericus JG-A12 and to its purified and recrystallized S-layer protein, excluding the reduction of U(VI) to U(IV). The combination of the latter spectroscopic study with TEM analysis demonstrates that the biosorption at the cell surface is the main mechanism responsible for the interaction of uranium with this bacterium.
Our EXAFS data indicate that U in the whole-cell sample is coordinated by phosphate groups in monodentate bonding mode and by carboxyl groups in a bidentate fashion. These groups are associated with the biocomponents of the cell surface as was demonstrated by the TEM analysis. Typically, the cell walls of gram-positive bacteria are comprised of approximately 25 layers of cross-linked linear polymers of peptidoglycan which consist of up to 40 to 50 repeating dimers of N-acetylglucosamine and N-acetylmuramic acid rich in carboxylate groups. Teichoic acids are intermeshed in the peptidoglycan framework and contain phosphate groups as well as d-alanine (34). These groups can deprotonate to form negatively charged metal binding sites (57).
Numerous studies have examined the metal ion-cell wall interactions of gram-positive bacteria (particularly members of the genus Bacillus). The sites responsible for metal binding in these organisms are probably the carboxyl sites within the peptidoglycan as well as the phosphoryl groups of the teichoic acids and other secondary polymers (16).
In the case of gram-negative bacteria, Ferris and Beveridge (18) demonstrated that the phosphoryl residues of the polar head of phospholipids and LPS in the outer membrane were the most probable binding sites for metal cations in Escherichia coli K-12, whereas in Pseudomonas aeruginosa, the phosphoryl groups in the core-lipid A region of the LPS are mainly involved in metal binding (31). The main implication of phosphate groups in the complexation of uranium by representatives of gram-negative bacteria is confirmed by the use of different spectroscopic and microscopic techniques. In previous studies, the combination of EXAFS spectroscopy, TEM, and EDX analysis showed that phosphorus-containing residues of three ecotypes of A. ferrooxidans are involved in the complexation of uranium (40). In addition, the structural parameters of these uranium complexes appear similar to those arising from the complexation of uranium with organic phosphate compounds such as fructose 6-phosphate (28).
The pH of the uranium solution used in this study was 4.5. Under these conditions, it is expected that the cells of B. sphaericus JG-A12 are dominantly electronegative. Bacterial cell walls are negatively charged under acidic pH conditions, and the cell wall chemical functional groups (hydroxyl, carbonyl, carboxyl, thioether, amine, amide, phosphonate, phosphodiester, etc.) display a high affinity for metal ions in solution (13). It has been demonstrated that the bacterial cells of gram-positive bacteria (B. subtilis) display an isoelectric point of approximately pH 2.4 (57). At this pH condition, the concentration of negatively charged surface sites is equal to the concentration of positively charged amino acids, and the net surface charge is equal to zero. As the pH increases from the isoelectric point, the acidic cell wall functional groups progressively deprotonate, generating a net negative charge within the cell wall and an electronegative potential on the cell surface (57).
Environmental and application significance.
A key implication of our findings is that the elucidation of interaction mechanisms between uranium and B. sphaericus JG-A12 cells and S-layer protein, the determination of the uranium binding sites, and the structural parameters of the uranium complexes formed by these two biological structures may help in the optimization of bioremediation processes using this bacterium and its S-layer. Strategies for enhancing the uranium-binding capacities of the cells and S-layer proteins of this bacterium are needed. These include chemical or genetic insertion of amino acids possessing carboxyl groups such as aspartic and glutamic acids, as well as serine and threonine, into the S-layer protein sequence. The availability of genetic engineering technology provides the possibility of specially tailoring microbial biosorbents with the required high affinity for uranium. Searching for novel environmental bacterial isolates possessing S-layer proteins with high phosphate and carboxyl group content is also a promising strategy for the removal and recovery of uranium from contaminated water or soil, as is optimization of growth media by increasing the amounts of phosphates. Some of these studies are in progress in our laboratory.
Acknowledgments
This study was supported by grant DFG/SE 671/7-2 from DFG, Bonn, Germany, and by grant 7531.50-03.0370-01/5 from the SMWK, Dresden, Germany.
We are grateful to H. Funke for his assistance with the EXAFS measurements and U. Schafer for the ICP-MS measurements. We also acknowledge the assistance of Maria Teresa González-Muñoz, Jose Maria Arias Peñalver (Department of Microbiology), Maria del Mar Abad Ortega, and Concepcion Hernandez Castillo (Electron Microscopy Services), University of Granada, Granada, Spain.
REFERENCES
- 1.Allen, F. H., and O. Kennard. 1993. 3D search and research using the Cambridge structural database. Chem. Des. Autom. News 8:31-37. [Google Scholar]
- 2.Allen, P. G., D. K. Shuh, J. J. Bucher, N. M. Edelstein, T. Reich, M. A. Denecke, and H. Nitsche. 1996. EXAFS determinations of uranium structures: the uranyl ion complexed with tartaric, citric and malic acids. Inorg. Chem. 35:784-787. [Google Scholar]
- 3.Ankudinov, A. L., B. Ravel, J. J. Rehr, and S. D. Conradson. 1998. Real-space multiple-scattering calculation and interpretation of X-ray absorption near-edge spectra. Phys. Rev. B 58:7565-7575. [Google Scholar]
- 4.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. Academic Press, New York, N.Y.
- 5.Bargar, J. R., R. Reitmeyer, J. J. Lehnhart, and J. A. Davis. 2000. Characterization of U(VI)-carbonato ternary complexes on hematite: EXAFS and electrophoretic mobility measurements. Geochim. Cosmochim. Acta 64:2737-2749. [Google Scholar]
- 6.Beveridge, T. J. 1994. Bacterial S-layers. Curr. Opin. Struct. Biol. 4:204-212. [Google Scholar]
- 7.Beveridge, T. J., and R. J. Doyle. 1989. Metal ions and bacteria. John Wiley & Sons, Inc., New York, N.Y.
- 8.Beveridge, T. J., and S. F. Koval. 1981. Binding of metals to cell envelopes of Escherichia coli K12. Appl. Environ. Microbiol. 42:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beveridge, T. J., and R. G. E. Murray. 1980. Sites of metal deposition in the cell wall of Bacillus subtilis. J. Bacteriol. 141:876-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boswell, C. D., R. E. Dick, H. Eccles, and L. E. Macaskie. 2001. Phosphate uptake and release by Acinetobacter johnsonii in continuous culture and coupling of phosphate release to heavy metal accumulation. J. Ind. Microbiol. Biotechnol. 26:333-340. [DOI] [PubMed] [Google Scholar]
- 11.Burns, P. C. 1999. Uranium: mineralogy, geochemistry and the environment, p. 23-90. In P. C. Burns and R. Finch (ed.), Review in mineralogy, vol. 38. Mineralogical Society of America, Washington, D.C. [Google Scholar]
- 12.Clark, D. L., D. E. Hobart, and M. P. Neu. 1995. Actinide carbonate complexes and their importance in actinide environmental chemistry. Chem. Rev. 95:25-48. [Google Scholar]
- 13.Collins, Y. E., and G. Stotzky. 1992. Heavy metals alter the electrokinetic properties of bacteria, yeasts and clay minerals. Appl. Environ. Microbiol. 58:1592-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denecke, M. A., T. Reich, S. Pompe, M. Bubner, K. H. Heise, H. Nitsche, E. A. Hudson, P. G. Allen, L. J. Terminello, M. A. Denecke, and T. Reich. 1997. Differentiating between monodentate and bidentate carboxylate ligands coordinated to uranyl ions using EXAFS. J. Phys. IV France 7:637-638. [Google Scholar]
- 15.Douglas, S., and T. J. Beveridge. 1998. Mineral formation by bacteria in natural microbial communities. FEMS Microbiol. Rev. 26:79-88. [Google Scholar]
- 16.Doyle, R. J., T. H. Matthews, and U. N. Streips. 1980. Chemical basis for selectivity of metal ions by Bacillus subtilis cell wall. J. Bacteriol. 143:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekman, P., and O. Jäger. 1993. Quantification of subnanomolar amounts of phosphate bound to seryl and threonyl residues in phosphoproteins using alkaline hydrolysis and malachit green. Anal. Biochem. 214:138-141. [DOI] [PubMed] [Google Scholar]
- 18.Ferris, F. G., and T. J. Beveridge. 1986. Site specificity of metallic ion binding in Escherichia coli K-12 lipopolysaccharide. Can. J. Microbiol. 32:52-55. [DOI] [PubMed] [Google Scholar]
- 19.Francis, A. J., J. B. Gillow, C. J. Dodge, R. Harris, T. Beveridge, and H. W. Papenguth. 2004. Uranium association with halophilic and non-halophilic bacteria and archaea. Radiochim. Acta 92:481-488. [Google Scholar]
- 20.George, G. N., and I. J. Pickering. 1995. EXAFSPAK: a suite of computer programs for analysis of X-ray absorption spectra. Stanford Synchrotron Radiation Laboratory, Stanford, Calif.
- 21.Günther, A., G. Bernhard, G. Geipel, T. Reich, A. Rossberg, and H. Nitsche. 2003. Uranium speciation in plants. Radiochim. Acta 91:319-328. [Google Scholar]
- 22.Hennig, C., P. Panak, T. Reich, A. Roßberg, J. Raff, S. Selenska-Pobell, W. Matz, J. J. Bucher, G. Bernhard, and H. Nitsche. 2001. EXAFS investigation of uranium(VI) complexes formed at Bacillus cereus and Bacillus sphaericus surfaces. Radiochim. Acta 89:625-631. [Google Scholar]
- 23.Heptinstall, S., A. R. Archibald, and J. Baddiley. 1970. Teichoic acids and membrane function in bacteria. Nature 225:519-521. [DOI] [PubMed] [Google Scholar]
- 24.Hoare, D. G., and D. E. Koshland, Jr. 1967. A method for quantitative modification and estimation of carboxylic acid groups in proteins. J. Biol. Chem. 242:2447-2453. [PubMed] [Google Scholar]
- 25.Hudson, E. A., P. G. Allen, and L. J. Terminello. 1996. Polarized X-ray absorption spectroscopy of the uranyl ion: comparison of experiment and theory. Phys. Rev. B 54:156-165. [DOI] [PubMed] [Google Scholar]
- 26.Kelly, S. D., K. M. Kemner, J. B. Fein, D. A. Fowle, M. I. Boyanov, B. A. Bunker, and N. Yee. 2002. X-ray absorption fine structure determination of pH-dependent U-bacterial cell wall interactions. Geochim. Cosmochim. Acta 65:3855-3871. [Google Scholar]
- 27.Knopp, R., P. Panak, L. A. Wray, N. S. Renninger, J. D. Keasling, and H. Nitsche. 2003. Laser spectroscopic studies of interactions of U(VI) with bacterial phosphate species. Chem. Eur. J. 9:2812-2818. [DOI] [PubMed] [Google Scholar]
- 28.Koban, A., G. Geipel, A. Roßberg, and G. Bernhard. 2004. Uranyl(VI) complexes with sugar phosphates in aqueous solution. Radiochim. Acta 92:903-908. [Google Scholar]
- 29.Krueger, S., G. L. Olsen, D. Johnsonbaugh, and T. J. Beveridge. 1993. Characterization of the binding of gallium, platinum, and uranium to Pseudomonas fluorescens by small-angle X-ray scattering and transmission electron microscopy. Appl. Environ. Microbiol. 59:4056-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Langley, S., and T. J. Beveridge. 1999. Effect of O-side-chain lipopolysaccharide chemistry on metal binding. Appl. Environ. Microbiol. 65:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locock, A. J., and P. C. Burns. 2003. Crystal structure and synthesis of the copper-dominant members of the autunite and meta-autunite groups: torbernite, zeunerite, metatorbernite and metazeunerite. Can. Min. 41:489-502. [Google Scholar]
- 33.Lueng, W. C., H. Chua, and W. Lo. 2001. Biosorption of heavy metals by bacteria isolated from activated sludge. Appl. Biochem. Biotechnol. 91-93:171-184. [DOI] [PubMed] [Google Scholar]
- 34.Madigan, M., J. Martinko, and J. Parker. 2003. Brock biology of microorganisms, 10th ed., p. 74. Prentice Hall, Upper Saddle River, N.J.
- 35.Makarov, E. S., and V. I. Ivanov. 1960. The crystal structure of meta-autunite, Ca(UO2)2(PO4)2 · 6H2O. Doklady Akademi Nauk SSSR 132:573-577. [Google Scholar]
- 36.Matz, W., N. Schell, G. Bernhard, F. Prokert, T. Reich, J. Claußner, W. Oehme, R. Schlenk, S. Dienel, H. Funke, F. Eichhorn, M. Betzel, D. Pröhl, U. Strauch, G. Hüttig, H. Krug, W. Neumann, V. Brendler, P. Reichel, M. A. Denecke, and H. Nitsche. 1999. ROBL—a CRG beamline for radiochemistry and material research at the ESRF. J. Synchrotron. Rad. 6:1076-1085. [Google Scholar]
- 37.Merroun, M. L., G. Geipel, R. Nicolai, K. Heise, and S. Selenska-Pobell. 2003. Complexation of uranium(VI) by three eco-types of Acidithiobacillus ferrooxidans studied using time-resolved laser-induced fluorescence spectroscopy and infrared spectroscopy. Biometals 16:331-339. [DOI] [PubMed] [Google Scholar]
- 38.Merroun, M. L., C. Hennig, A. Rossberg, G. Geipel, T. Reich, and S. Selenska-Pobell. 2002. Molecular and atomic analysis of the uranium complexes formed by three eco-types of Acidithiobacillus ferrooxidans. Biochem. Soc. Trans. 30:669-672. [DOI] [PubMed] [Google Scholar]
- 39.Merroun, M. L., C. Hennig, A. Rossberg, T. Reich, G. Geipel, and S. Selenska-Pobell. 2003. Bacterial strains isolated from uranium mining waste piles and their interactions with uranium. In P. S. Hah (ed.), Proceedings of the 9th International Conference on Chemistry and Migration Behaviour of Actinides and Fission Products in the Geosphere. Korea Atomic Energy Research Institute, Gyeongju, Korea.
- 40.Merroun, M. L., C. Hennig, A. Rossberg, T. Reich, and S. Selenska-Pobell. 2003. Characterization of U(VI)-Acidithiobacillus ferrooxidans complexes by using EXAFS, transmission electron microscopy and energy-dispersive X-ray analysis. Radiochim. Acta 91:583-591. [Google Scholar]
- 41.Merroun, M. L., J. Raff, A. Rossberg, C. Hennig, T. Reich, and S. Selenska-Pobell. 2004. Interaction of U(VI) with bacterial strains isolated form uranium mining piles: spectroscopic and microscopic studies. Geochim. Cosmochim. Acta 68:A499. [Google Scholar]
- 42.Merroun, M. L., and S. Selenska-Pobell. 2001. Interactions of three eco-types of Acidithiobacillus ferrooxidans with U(VI). Biometals 14:171-179. [DOI] [PubMed] [Google Scholar]
- 43.Mozes, N., and S. Lortal. 1995. X-ray photoelectron spectroscopy and biochemical analysis of the surface of Lactobacillus helveticus ATCC 12046. Microbiology 141:11-19. [Google Scholar]
- 44.Panak, P., S. Selenska-Pobell, S. Kutschke, G. Geipel, G. Bernhard, and H. Nitsche. 1999. Complexation of U(VI) with cells of Thiobacillus ferrooxidans and Thiomonas cuprina of different geological origin. Radiochim. Acta 84:183-190. [Google Scholar]
- 45.Pollmann, K., J. Raff, M. Schnopfeil, G. Radeva, and S. Selenska-Pobell. Novel surface layer protein genes in Bacillus sphaericus associated with unusual insertion elements. Microbiology, in press. [DOI] [PubMed]
- 46.Pollmann, K., M. Schnorpfeil, G. Radeva, and S. Selenska-Pobell. 2004. Molecular analyses of the S-layer genes and proteins of Bacillus sphaericus JG-A12 and NCTC 9602. FZR Rep. 400:23. [Google Scholar]
- 47.Raff, J. 2002. Ph.D. thesis. University of Leipzig, Leipzig, Germany.
- 48.Redden, G., J. Bargar, and R. Bencheikh-Latmani. 2001. Citrate enhanced uranyl adsorption on geothite: an EXAFS analysis. J. Coll. Int. Sci. 244:211-219. [Google Scholar]
- 49.Renninger, N., K. D. McMahon, R. Knopp, H. Nitsche, D. S. Clark, and J. D. Keasling. 2001. Uranyl precipitation by biomass from an enhanced biological phosphorus removal reactor. Biodegradation 12:401-410. [DOI] [PubMed] [Google Scholar]
- 50.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savchuk, O., K. Pollmann, J. Raff, J. Philipp, S. Selenska-Pobell, and K. Fahmy. 2004. Structural stability and reactivity with heavy metals ions of S-layer protein from Bacillus sphaericus. FZR Rep. 401:47. [Google Scholar]
- 52.Selenska-Pobell, S., P. Panak, V. Miteva, I. Boudakov, G. Bernhard, and H. Nitsche. 1999. Selective accumulation of heavy metals by three indigeneous Bacillus strains, B. cereus, B. megaterium and B. sphaericus from drain waters of a uranium waste pile. FEMS Microbiol. Ecol. 29:59-67. [Google Scholar]
- 53.Sleytr, U. B. 1997. I. Basic and applied S-layer research: an overview. FEMS Microbiol. Rev. 20:5-12. [Google Scholar]
- 54.Teo, B. K. 1986. EXAFS: basic principles and data analysis. Springer-Verlag, New York, N.Y.
- 55.Thomas, S. R., and T. J. Trust. 1995. Tyrosine phosphorylation of the tetragonal paracrystalline array of Aeromonas hydrophila: molecular cloning and high-level expression of the S-layer protein gene. J. Mol. Biol. 245:568-581. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, H. L., G. E. Brown, Jr., and G. A. Parks. 1997. XAFS spectroscopic study of uranyl coordination in solids and aqueous solution. Am. Min. 82:483-496. [Google Scholar]
- 57.Yee, N., D. A. Fowle, and F. G. Ferris. 2004. A donnan potential model for metal sorption onto Bacillus subtilis. Geochim. Cosmochim. Acta 68:3657-3664. [Google Scholar]