Abstract
A lignin-related biphenyl compound, 5,5′-dehydrodivanillate (DDVA), is degraded to 5-carboxyvanillate (5CVA) by the enzyme reactions catalyzed by DDVA O-demethylase (LigX), meta-cleavage oxygenase (LigZ), and meta-cleavage compound hydrolase (LigY) in Sphingomonas paucimobilis SYK-6. 5CVA is then transformed to vanillate by a nonoxidative 5CVA decarboxylase and is further degraded through the protocatechuate 4,5-cleavage pathway. A 5CVA decarboxylase gene, ligW, was isolated from SYK-6 (X. Peng, E. Masai, H. Kitayama, K. Harada, Y, Katayama, and M. Fukuda, Appl. Environ. Microbiol. 68:4407-4415, 2002). However, disruption of ligW slightly affected the 5CVA decarboxylase activity and the growth rate on DDVA of the mutant, suggesting the presence of an alternative 5CVA decarboxylase gene. Here we isolated a second 5CVA decarboxylase gene, ligW2, which consists of a 1,050-bp open reading frame encoding a polypeptide with a molecular mass of 39,379 Da. The deduced amino acid sequence encoded by ligW2 exhibits 37% identity with the sequence encoded by ligW. Based on a gas chromatography-mass spectrometry analysis of the reaction product from 5CVA catalyzed by LigW2 in the presence of deuterium oxide, LigW2 was indicated to be a nonoxidative decarboxylase of 5CVA, like LigW. After disruption of ligW2, both the growth rate on DDVA and the 5CVA decarboxylase activity of the mutant were decreased to approximately 30% of the wild-type levels. The ligW ligW2 double mutant lost both the ability to grow on DDVA and the 5CVA decarboxylase activity. These results indicate that both ligW and ligW2 contribute to 5CVA degradation, although ligW2 plays the more important role in the growth of SYK-6 cells on DDVA.
Lignin is the most abundant natural aromatic compound that consists of phenylpropane units with various types of linkages. Lignin helps to strengthen and stiffen plant cell walls and is resistant to breakdown by microorganisms, but many microorganisms have evolved the ability to degrade lignin. The specific reactions of lignin metabolism by microorganisms are expected to enable the use of lignin for industrial purposes in order to supplement or replace traditional methods of chemical synthesis.
Lignin-derived compounds and their metabolic intermediates (e.g., hydroxylated benzoate derivatives) include carboxyl groups, and therefore the removal of carboxyl groups from the benzene nucleus by decarboxylases is an important reaction in lignin biodegradation. The aromatic acid decarboxylases have been classified into two groups, the nonoxidative and oxidative decarboxylases. Nonoxidative decarboxylation of aromatic acids involves the removal of the carboxyl group from the benzene nucleus via an enzymatic reaction that requires neither oxygen nor cofactors (26). The nonoxidative process results in complete removal of the carboxyl group, in contrast to the oxidative reaction, which substitutes a hydroxyl group at the relevant carbon atom (7). Nonoxidative decarboxylases are used not only in the degradation of lignin-related compounds, including 4-hydroxybenzoate (22, 25, 31), protocatechuate (PCA) (23), gallate (19, 58), vanillate (10, 24, 46), ferulate (57), and p-coumarate (8), but also in the degradation of such artificial compounds as phthalates (9, 15, 30, 38, 45, 47).
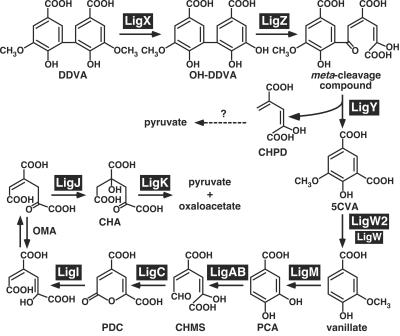
Sphingomonas paucimobilis SYK-6 was isolated with a lignin-related biphenyl compound, 5,5′-dehydrodivanillate (DDVA), as the sole carbon and energy source (28). This strain is able to mineralize various lignin-derived biaryls through the PCA 4,5-cleavage pathway using unique and specific enzymatic systems (21), which are thought to be good tools for conversion of lignin to valuable intermediate metabolites. The DDVA catabolic pathway of this strain has been determined, as shown in Fig. 1. DDVA is initially converted to a diol compound, 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA), by multicomponent DDVA O-demethylase, the oxygenase component of which is encoded by ligX (52). The ring fission of OH-DDVA to produce a meta-cleavage compound is catalyzed by OH-DDVA oxygenase encoded by ligZ (39). The resulting meta-cleavage compound is hydrolyzed to form 5-carboxyvanillate (5CVA) by a ligY-encoded hydrolase (40). 5CVA is further degraded to vanillate, which is finally converted to pyruvate and oxaloacetate via the PCA 4,5-cleavage pathway.
FIG. 1.
Proposed DDVA catabolic pathway of S. paucimobilis SYK-6. The following gene products are involved in this pathway: LigX, a component of DDVA O-demethylase; LigZ, OH-DDVA meta-cleavage dioxygenase; LigY, OH-DDVA meta-cleavage compound hydrolase; LigW and LigW2, 5CVA decarboxylase; LigM, vanillate/3-O-methylgallate O-demethylase; LigA and LigB, small and large subunits, respectively, of PCA 4,5-dioxygenase; LigC, 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase; LigI, 2-pyrone-4,6-dicarboxylate hydrolase; LigJ, 4-oxalomesaconate hydratase; LigK, 4-carboxy-4-hydroxy-2-oxoadipate aldolase. Abbreviations: CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; PDC, 2-pyrone-4,6-dicarboxylate; OMA, 4-oxalomesaconate; CHA, 4-carboxy-4-hydroxy-2-oxoadipate.
In a previous study, we isolated and characterized the ligW gene, the product of which is a nonoxidative decarboxylase that catalyzes the conversion of 5CVA to vanillate (41). However, we found that ligW is not essential for the degradation of 5CVA by SYK-6. To identify the reaction step that converts 5CVA to vanillate, we isolated and characterized a second 5CVA decarboxylase gene, ligW2. The roles played by both ligW and ligW2 in DDVA catabolism were examined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and substrates.
The strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6 and its derivative strains were grown at 30°C in W minimal medium (39) containing 10 mM vanillate or 5 mM DDVA or in Luria-Bertani (LB) medium (Bacto Tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter). The methods used for preparation of DDVA (39) and 5CVA (41) have been described previously. Isophthalate, 4-hydroxyisophthalate, 3-methoxysalicylate, and deuterium oxide (D2O) (99.75%) were purchased from Wako Pure Chemical Industries (Osaka, Japan).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| S. paucimobilis strains | ||
| SYK-6 | Wild type; Nalr Smr | 29 |
| DW | SYK-6 derivative, ligW::kan Nalr Smr Kmr | This study |
| DW2 | SYK-6 derivative, ligW2::kan Nalr Smr Kmr | This study |
| DDW | SYK-6 derivative, ligW::kan ligW2::bla Nalr Smr Kmr Cbr | This study |
| Dorf1 | SYK-6 derivative, orf1::kan Nalr Smr Kmr | This study |
| Dorf3 | SYK-6 derivative, orf3::kan Nalr Smr Kmr | This study |
| P. putida PpY101 | Nalr Smr | 17 |
| E. coli strains | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 56 |
| S17-1 | recA; harbors the tra genes of plasmid RP4 in the chromosome, proA thi | 51 |
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 5 |
| Plasmids | ||
| pUC18 | Cloning vector; Apr | 56 |
| pBluescript II KS(+) | Cloning vector; Apr | 50 |
| pVK100 | Broad-host-range cosmid vector, Kmr Tetr | 14 |
| pRK2013 | Kmr Tra+ Mob+ | 16 |
| pDEC6 | pVK100 with an approximately 20-kb fragment carrying ligW2 | This study |
| pU351 | pUC18 with a 3.6-kb SalI fragment carrying ligW2 | This study |
| pU352 | pUC18 carrying the same fragment as pU351 in the opposite direction | This study |
| pA1, pA5, pA9, pA13, pA14 | Deletion derivatives of pU351 | This study |
| pB15, pB16, pB19, pB26, pB28, pB32 | Deletion derivatives of pU352 | This study |
| pW2 | KS(+) with a 1.1-kb SmaI-SacII fragment carrying ligW2 | This study |
| pVKW2 | pVK100 carrying the same fragment as pW2 | This study |
| pVK7E2 | pVK100 with a 7.0-kb EcoRI fragment carrying ligW | 41 |
| pUC4K | Apr/Kmr | 53 |
| pK19mobsacB | oriT sacB Kmr | 49 |
| pU351Km | pU351 with insertion of kan from pUC4K into an EcoRV site | This study |
| pDW2Km | pK19mobsacB with a 4.8-kb SalI fragment from pU351Km | This study |
| pK19U351 | pK19mobsacB with a 3.6-kb SalI fragment from pU351 | This study |
| pDW2Ap | pK19U351 with insertion of bla from pUC18 into an EcoRV site | This study |
| pKS7E2 | KS(+) with a 7.0-kb EcoRI fragment of pVK7E2 | 41 |
| pKV44 | Deletion derivative of pKS7E2 carrying ligW | 41 |
| pKV44Km | pKV44 with insertion of kan from pUC4K into a SalI site | This study |
| pDW | pK19mobsacB with a 3.6-kb SacI-XbaI fragment from pKV44Km | This study |
| pUC11 | pUC19 with a 2.8-kb BamHI fragment carrying orf1 | This study |
| pUC11Km | pUC11 with insertion of kan from pUC4K into a SalI site | This study |
| pDorf1 | pK19mobsacB with a 4.0-kb BamHI fragment from pUC11Km | This study |
| pSV322 | KS(+) with a 2.8-kb EcoRV-SalI fragment carrying orf3 | This study |
| pSV322Km | pSV322 with insertion of kan from pUC4K into a HindIII site | This study |
| pDorf3 | pK19mobsacB with a 4.0-kb EcoRI-SalI fragment from pSV322Km | This study |
| pHN139F | pUC18 with a 10.5-kb EcoRI fragment carrying ligJAB, part of ligC, ligK, and ligI | 35 |
| pDE20 | pVK100 with an approximately 20-kb EcoRI fragment carrying part of ligM, metF, and ligH | 37 |
Abbreviations: Nalr, Smr, Kmr, Cbr, Tetr and Apr, resistance to nalidixic acid, streptomycin, kanamycin, carbenicillin, tetracycline, and ampicillin, respectively.
Cloning procedure.
A gene library constructed with pVK100 and the partially SalI-digested DNA of SYK-6 (41) was introduced into Pseudomonas putida PpY101 by triparental mating. The transconjugants were grown in 3 ml of LB medium containing 25 mg of nalidixic acid/liter and 50 mg of kanamycin/liter at 30°C for 12 h. Cells were centrifuged at 4,500 × g for 10 min at 4°C, and they were washed twice with W medium. Then cells were suspended in 1 ml of the same medium containing 50 μM 5CVA. After incubation with shaking for 7 days at 30°C, each 200 μl of culture was acidified with 10 μl of 6 N hydrochloric acid and extracted with 60 μl of ethyl acetate. The disappearance of 5CVA from the resultant samples was analyzed by thin-layer chromatography (TLC) using Silica Gel 60 F254 (E. Merck, Darmstadt, Germany) with the solvent chloroform-ethyl acetate-formic acid (10:8:2). Compounds were visualized under UV light at 254 nm. 5CVA has an Rf of 0.52 in this system.
DNA manipulations and nucleotide sequencing.
DNA manipulations were carried out essentially as described previously (2, 48). Nucleotide sequences were determined by the dideoxy termination method with a Thermosequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and an ALF express DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.). Sequencing analysis and multiple alignment were carried out with the GeneWorks programs (IntelliGenetics, Inc., Mountain View, Calif.). Pairwise alignment was performed with the EMBOSS alignment tool at the homepage of the European Bioinformatics Institute (http://www.ebi.ac.uk/emboss/align).
Construction of insertion mutants of S. paucimobilis SYK-6.
To disrupt ligW, ligW2, orf1, and orf3 in SYK-6, the kanamycin resistance gene (kan) from pUC4K was inserted into each gene by the gene replacement technique. The ligW-, ligW2-, orf1-, and orf3-disrupted plasmids pDW, pDW2Km, pDorf1, and pDorf3, constructed with pK19mobsacB, were introduced into Escherichia coli S17-1 and then introduced into SYK-6 by conjugation. To disrupt both ligW and ligW2 in SYK-6, pDW2Ap, in which ligW2 was inactivated by the ampicillin resistance gene (bla) from pUC18, was introduced into a ligW mutant (DW). Selection of each gene-disrupted mutant was performed as described previously (35). To examine the disruption of each gene, a Southern hybridization analysis was carried out. Total DNA of the candidates for ligW, ligW2, ligW and ligW2, orf1, and orf3 insertion mutants were digested with SacI and XhoI, SalI, SalI, BamHI, and SalI, respectively. The 1.3-kb SalI fragment carrying kan, the 1.9-kb SacI-XhoI fragment carrying ligW, the 3.6-kb SalI fragment carrying ligW2 and orf3, the 1.0-kb BspHI fragment carrying bla, and the 2.8-kb BamHI fragment carrying orf1 were labeled with the DIG system (Roche Molecular Biochemicals, Mannheim, Germany) and used as probes.
Preparation of cell extracts.
E. coli JM109 cells harboring pW2 were grown in LB medium containing 100 mg of ampicillin/liter. Expression of ligW2 was induced for 4 h by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM when the turbidity of the culture at 600 nm reached 0.5. Cells were harvested by centrifugation at 3,000 × g for 10 min and were ruptured by passage through a French pressure cell in 50 mM Tris-HCl buffer (pH 7.5). The cell lysate was centrifuged at 15,000 × g for 15 min, and the supernatant was used as a crude enzyme. The expression of the enzymes was examined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined by the method of Bradford (6).
For preparation of cell extracts of S. paucimobilis SYK-6 and its mutants, cells were grown in W medium containing 10 mM sucrose, 10 mM glutamate, and 50 mg of methionine/liter. The expression of 5CVA decarboxylase genes was induced by addition of 5 mM DDVA when the turbidity of the culture at 600 nm reached 0.5, and the cells were incubated for 4 h. Cells were harvested by centrifugation at 5,000 × g for 10 min and washed with 50 mM Tris-HCl buffer (pH 7.5). Cells suspended in the same buffer were sonicated, and the cell lysate was centrifuged at 15,000 × g for 15 min. The resulting supernatant was used as the cell extract.
Enzyme assays.
The 5CVA decarboxylase activity was spectrophotometrically determined by measuring the decrease in the absorbance at 312 nm (ɛ312 = 3.5 × 103 M−1 cm−1; pH 7.5) with a DU-7500 spectrophotometer (Beckman, Fullerton, Calif.) at 30°C. Each 1-ml assay mixture contained 50 mM Tris-HCl buffer (pH 7.5), 100 μM 5CVA, and cell extract of E. coli JM109 harboring pW2 (100 μg of protein) or S. paucimobilis SYK-6 and its mutants (200 μg of protein). One unit of enzyme activity was defined as the amount that degraded 1 μmol of substrate per min at 30°C
To determine the activity of LigW2 toward 3-methoxysalicylate, 4-hydroxyisophthalate, and isophthalate, a high-pressure liquid chromatography system (Alliance 2690 separation module; Waters, Milford, Mass.) equipped with an octadecyl silica reverse-phase column (4.6 by 100 mm; Waters) was employed. The reaction was performed as described above, and filtrates of the reaction mixture were analyzed by high-performance liquid chromatography. The mobile phase consisted of 0.1% phosphoric acid in water (solvent A) and 0.1% phosphoric acid in acetonitrile (solvent B). The flow rate was 1 ml/min. After injection of a sample, solvent A was run through the column for 2 min, the solvent gradient was programmed to increase from 0 to 70% solvent B over 6 min, and then this mixture was maintained for 5 min. The absorbance between 230 and 280 nm of eluent was monitored with a photodiode array detector (Waters 2996) for the reaction mixtures containing 4-hydroxyphthalate and isophthalate. Absorbance at 276 nm and 317 nm was monitored for the reaction mixture containing 3-methoxysalicylate. The retention times of 4-hydroxyisophthalate, 4-hydroxybenzoate, isophthalate, benzoate, 3-methoxysalicylate, and guaiacol were 6.36, 5.67, 6.17, 6.94, 6.93, and 6.80 min, respectively.
Vanillin dehydrogenase activity was measured by the method of Gasson et al. (18). Each 1-ml reaction mixture contained 100 mM potassium phosphate buffer (pH 7.0), cell extract of E. coli JM109 harboring pUCI1 (200 μg of protein), 50 μM vanillin, 500 μM NAD+, 1.2 mM pyruvate, and 1.1 U lactate dehydrogenase. The reaction was carried out at 30°C, and the decrease in absorbance at 340 nm was monitored.
Conversion of 5CVA to vanillate by LigW2 in D2O.
5CVA (400 μM) and cell extract of E. coli JM109 harboring pW2 (100 μg of protein) were incubated in 1-ml reaction mixtures containing 0 or 96% D2O. After incubation for 30 min at 30°C, the reaction product was acidified with 6 N hydrochloric acid and extracted with ethyl acetate. The organic phase was dried in vacuo and was treated with the trimethylsilyl reagent. The trimethylsilylated (TMS) derivatives of the reaction product were analyzed by gas chromatography-mass spectrometry (GC-MS) using a model 5971A instrument (Agilent Technologies, Palo Alto, Calif.) with an Ultra-2 capillary column (50 m by 0.2 mm; Agilent Technologies). The analytical conditions used have been described previously (41).
Mapping of ligW2.
Total DNA of SYK-6 prepared in an agarose block was digested with AseI and separated with a CHEF DRIII apparatus (Bio-Rad, Hercules, Calif.) as described in a previous study (41). For Southern hybridization analysis, the digoxigenin-labeled 3.6-kb SalI fragment carrying ligW2 was used as a probe. To determine the location of ligW2 and ligM-metF-ligH or the PCA 4,5-cleavage pathway genes, Southern hybridization of the SalI digests of pDEC6 was performed with digoxigenin-labeled pDE20 and pHN139F digested with SalI as probes.
Nucleotide sequence accession number.
The nucleotide sequences of ligW2, orf1, and orf3 have been deposited in the DDBJ, EMBL, and GenBank sequence databases under accession no. AB089690.
RESULTS AND DISCUSSION
Cloning of a second 5CVA decarboxylase gene.
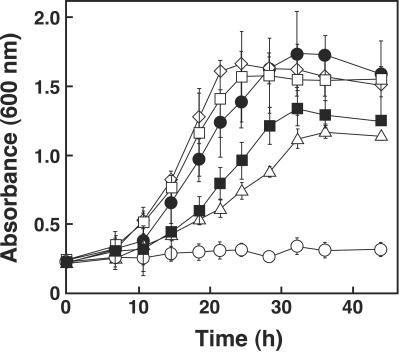
In order to examine the role played by ligW in the DDVA catabolism of S. paucimobilis SYK-6, ligW was disrupted by a gene replacement technique using pDW, in which ligW was inactivated by insertion of kan (Fig. 2). Disruption of ligW in the candidate mutants was confirmed by Southern hybridization analysis using a 1.9-kb SacI-XhoI fragment carrying ligW and a 1.3-kb SalI fragment carrying kan. The resulting ligW mutant (DW) and SYK-6 cells were grown in W medium containing 5 mM DDVA as the sole carbon and energy source. The growth rate of DW was slightly decreased (k = 0.079/h; 83% of the growth rate of the wild-type strain) (Fig. 3). Because the 5CVA decarboxylase activity of the SYK-6 cells incubated with DDVA (95 ± 15 mU/mg) was approximately three times higher than that of SYK-6 cells incubated without DDVA, the activity of DW was examined using cells incubated with 5 mM DDVA in order to induce the enzyme activity. It was found that the decarboxylase activity of the cell extract of DW was approximately 80% of the wild-type activity. These results indicated that ligW is not essential for the degradation of 5CVA, although it contributes to the total 5CVA decarboxylase activity in SYK-6 cells.
FIG. 2.
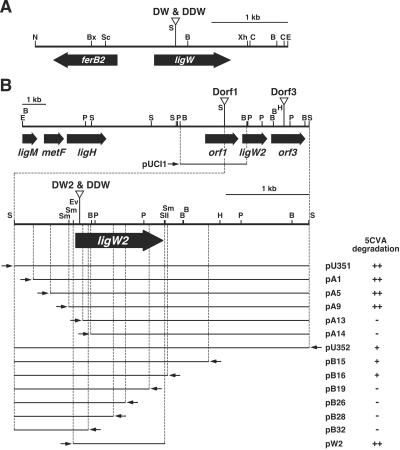
Restriction map of the 3.3-kb NotI-EcoRI fragment carrying ligW (A) and deletion analysis of the 3.6-kb SalI fragment carrying ligW2 (B). Triangles indicate the positions of the kan or bla gene insertion of the ligW mutant (DW), ligW2 mutant (DW2), ligW ligW2 double mutant (DDW), orf1 mutant (Dorf1), and orf3 mutant (Dorf3). Small arrows indicate the direction of transcription from the lac promoter. The 5CVA decarboxylase activities of E. coli containing each of the deletion plasmids are indicated on the right. ++, 5CVA was completely degraded; +, approximately one-half of 5CVA was degraded; −, 5CVA was not degraded. Abbreviations: B, BamHI; Bx, BstXI; C, ClaI; E, EcoRI; Ev, EcoRV; H, HindIII; N, NotI; P, PstI; S, SalI; Sc, SacI; Sm, SmaI; SII, SacII; Xh, XhoI.
FIG. 3.
Growth of the insertion mutants on DDVA. Cells of SYK-6 (open diamonds), DW (open squares), DW2 (open triangles), DDW (open circles), DDW harboring pVKW2 (solid circles), and DDW harboring pVK7E2 (solid squares) were grown in W medium containing 5 mM DDVA at 30°C. The values are averages ± standard deviations (error bars) of three independent experiments.
To isolate an alternative 5CVA decarboxylase gene, a cosmid gene library of SYK-6 constructed in E. coli was introduced into P. putida PpY101 by triparental mating. Approximately 2,000 transconjugants were screened for 5CVA transformation activity by TLC analysis. As a result, 13 positive clones that transformed 5CVA to vanillate were obtained. In a previous study, we obtained only one positive clone carrying ligW. This finding might have been due to the difference in the growth conditions used for the transconjugants in the assay. The transconjugants were grown in LB medium in this study, whereas vanillate-grown cells were used in the previous study. Because the growth of P. putida PpY101 on vanillate was somewhat poor, the activities of the positive clones might have been too weak for detection by TLC.
Southern hybridization analysis of the cosmid clones digested with SalI using a 3.3-kb NotI-EcoRI fragment carrying ligW as a probe suggested that 5 of the 13 clones contained ligW. The remaining eight clones were used for further experiments. These cosmid clones had a common 3.6-kb SalI fragment, and this fragment from pDEC6 was cloned into pUC18 in order to generate pU351 (Fig. 2). TLC analysis showed that the cell extract of E. coli JM109 harboring pU351 grown with IPTG was able to convert 5CVA to vanillate (Fig. 2), suggesting that the 3.6-kb SalI fragment contained a second 5CVA decarboxylase gene.
A series of subclones of the 3.6-kb SalI fragment was constructed by using restriction enzymes and E. coli exonuclease III, and these subclones were subjected to nucleotide sequencing. To determine the localization of the 5CVA decarboxylase gene in the 3.6-kb SalI fragment, the 5CVA decarboxylase activity of cell extracts of E. coli JM109 carrying each of the subclones was assayed by using TLC plates (Fig. 2). The 5CVA decarboxylase activity was detected in E. coli carrying pA9 and pB16 and yet was lost in E. coli carrying pA13 and pB19. This result suggested that the decarboxylase gene was limited to the DNA segment shared by pA9 and pB16. The presence of a promoter sequence upstream of ligW2 was considered, since the lac promoter of the vector was located downstream of ligW2 in pB16. Plasmid pW2 with the 1.1-kb SmaI-SacII fragment containing the DNA shared by pA9 and pB16 indeed conferred 5CVA decarboxylase activity on E. coli, and the specific activity for 5CVA decarboxylation was estimated to be 120 ± 32 mU/mg. The nucleotide sequence of the 3.6-kb SalI fragment revealed a 1,050-bp open reading frame (ORF) in the 1.1-kb SmaI-SacII segment, which was considered to be a second 5CVA decarboxylase gene and was designated ligW2. The molecular mass of the ligW2 gene product (LigW2) calculated from the deduced amino acid sequence was 39,379 Da.
Ishii et al. (27) reported that nonoxidative aromatic acid decarboxylases should be classified as members of the following enzyme families: the 3-octaprenyl-4-hydroxybenzoate carboxylyase (UbiD) family (59), which includes a 4-hydroxybenzoate decarboxylase, Ohb1 of Clostridium hydroxybenzoicum (25); the aldolase family, which includes 4,5-dihydroxyphthalate decarboxylases, including Pht5 of P. putida (38), PhtC of Arthrobacter keyseri (15), and PhtD of Comamonas testosteroni (30); the phenolic acid decarboxylase family, which includes the ferulic acid decarboxylase, Fdc, of Bacillus pumilus (57), and p-coumaric acid decarboxylase, PdcC, of Lactobacillus plantarum (8), as well as the phenolic acid decarboxylases of both Bacillus (3, 43) and Pediococcus (4); and a new family, which includes LigW and the 2,6-dihydroxybenzoic acid decarboxylase, Rdc, of Rhizobium radiobacter (27). LigW2 exhibited no homology with the enzymes in the UbiD, aldolase, and phenolic acid decarboxylase families, but it exhibited 37 and 28% identity with LigW and Rdc, respectively. In addition, LigW2 exhibited approximately 20% identity with the OH-DDVA meta-cleavage compound hydrolase (LigY) (40) and the 4-oxalomesaconate hydratase (LigJ) (15, 20, 32, 44, 55). Based on the finding that the decarboxylation activity of Rdc was inhibited by diethyl pyrocarbonate, a His residue was suggested to be one of the active site residues. The site-directed mutagenesis study suggested that His-164 and His-218 are essential for the decarboxylation activity catalyzed by Rdc. Interestingly, alignment of the amino acid sequences of Rdc with those of LigW2, LigW, LigY, and LigJ revealed that two corresponding His residues are completely conserved in these enzymes. Thus, these enzymes may have similar active sites.
Downstream of ligW2, 1,458-bp orf3 was found. The deduced amino acid sequence encoded by orf3 exhibited 51% and 30% identity with the sequences of the putative major facilitator superfamily transporters ZP_003051 and ZP_003018, respectively, of Novosphingobium aromaticivorans DSM 12444. However, orf3 exhibited only 15 to 19% identity with the benzoate transporter (BenK) (11, 12) and PCA/4-hydroxybenzoate transporter (PcaK) (13, 36) of P. putida and Acinetobacter, which are members of the major facilitator superfamily.
To obtain the sequence upstream of ligW2, the nucleotide sequence of the 2.8-kb BamHI fragment of pDEC6, which overlapped the 3.6-kb SalI fragment, was determined (Fig. 2); 1,428-bp orf1 was found in this region, and the deduced amino acid sequence encoded by orf1 exhibited ca. 30% identity with vanillin dehydrogenase (18, 42, 54). However, no vanillin dehydrogenase activity was detected in the cell extract of E. coli JM109 cells harboring pUCI1, which contains orf1 (data not shown).
Inactivation of ligW2, orf1, and orf3 in S. paucimobilis SYK-6.
In order to determine the involvement of ligW2 in 5CVA and DDVA degradation by SYK-6, the ligW2 gene in both SYK-6 and DW was disrupted by a gene replacement technique using ligW2-inactivated plasmids. The kan and bla genes were inserted into the ligW2 coding region in order to generate plasmids pDW2Km and pDW2Ap, respectively. These two plasmids were introduced into SYK-6 and DW, respectively. Disruption of ligW2 in the resulting ligW2 mutant (DW2) and the ligW ligW2 double mutant (DDW) was confirmed by Southern hybridization analysis using the 3.6-kb SalI fragment carrying ligW2 and the 1.3-kb SalI fragment carrying kan or the 1.0-kb BspHI fragment carrying bla as probes.
To test the ability of DW2 and DDW to grow on DDVA, these mutants were cultured in W medium containing 5 mM DDVA. DW2 retained the ability to grow on DDVA; however, the growth rate of DW2 on DDVA (k = 0.026/h) was markedly decreased (27% of the growth rate of the wild-type strain), and DDW completely lost the ability to grow on DDVA (Fig. 3). These results indicated that both ligW2 and ligW are involved in 5CVA degradation, but ligW2 plays a more important role in 5CVA degradation. The 5CVA decarboxylase activities of the mutants incubated with DDVA were also examined. Cell extract of DW2 exhibited 28% (27 ± 5.7 mU/mg) of the activity of the wild-type cells, whereas DDW almost completely lacked this activity (2.7 ± 1.5 mU/mg). The 5CVA decarboxylase activities of DW and DDW were in accord with the growth rates of these strains. The introduction of pVKW2 and pVK7E2, which carried ligW2 and ligW, respectively, into DDW restored the ability of DDW to grow on DDVA (Fig. 3), thus indicating that the deficiency of the 5CVA decarboxylase activity in DDW was truly caused by the disruption of ligW2 and ligW. Further investigations are needed to clarify the reason for the difference in the levels of participation of ligW2 and ligW in 5CVA degradation.
To gain a better understanding of the roles played by orf1 and orf3, these ORFs were disrupted by the strategy described above, using pDorf1 and pDorf3, respectively. No differences in either the growth rate on DDVA or 5CVA decarboxylase activity were observed between the insertion mutants of orf1 (Dorf1) and orf3 (Dorf3) and the wild-type cells, indicating that these ORFs are not involved in DDVA degradation. Due to the poor growth of SYK-6 on 5CVA, the growth of the mutants on 5CVA could not be tested. Therefore, we cannot exclude the possibility that orf3 encodes the transporter for 5CVA. On the other hand, it was suggested that orf1 does not encode vanillin dehydrogenase because Dorf1 grew normally on vanillin (data not shown).
Characterization of LigW2.
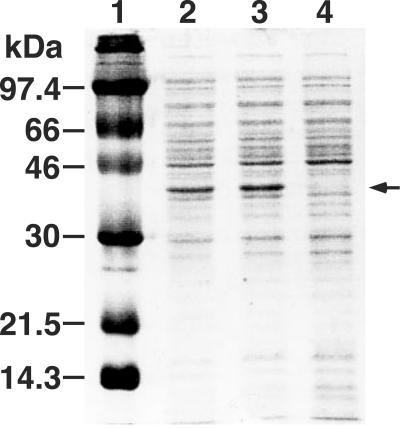
The ligW2 gene expression induced by IPTG in E. coli JM109 was examined with plasmids pU351 and pW2. Production of a 37-kDa protein in these transformants was observed by SDS-PAGE, and this size was in good agreement with the value calculated from the deduced amino acid sequence of ligW2 (Fig. 4).
FIG. 4.
Expression of ligW2 in E. coli demonstrated by SDS-PAGE. Lane 1, molecular size markers; lane 2, cell extract of E. coli JM109 harboring pU351; lane 3, cell extract of E. coli JM109 harboring pW2; lane 4, cell extract of E. coli JM109 harboring pBluescript II KS(+).
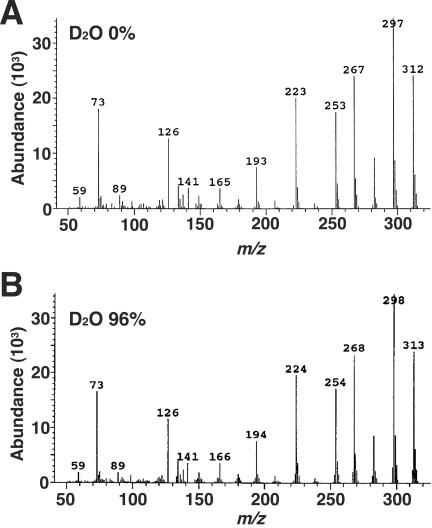
To confirm that LigW2 also acts as a nonoxidative decarboxylase, the cell extract of E. coli harboring pW2 was incubated with 400 μM 5CVA for 30 min in the presence of 0 or 96% D2O. The reaction product, vanillate, was extracted and subjected to GC-MS analysis. The mass spectrum of vanillate generated in the presence of 96% D2O was compared with that of vanillate produced in the absence of D2O (Fig. 5). Unlabeled TMS-vanillate showed an intense molecular ion at m/z 312, and the major ion fragments were observed at m/z 297 (M-CH3), 282 (M-2CH3), and 267 (M-3CH3). On the other hand, the abundance of these ion fragments of TMS-vanillate produced in the presence of D2O decreased, and that of the ion fragments at m/z 313, 298, 283, and 268 increased. This result indicated that the 5-carboxyl group in 5CVA was replaced by a deuterium atom that originated from D2O. LigW2 was concluded to be a nonoxidative decarboxylase, like LigW.
FIG. 5.
GC-MS analysis of the reaction product from 5CVA catalyzed by LigW2 in the presence of D2O. Mass spectra of TMS-vanillate generated in the presence of 0 and 96% D2O are shown in panels A and B, respectively.
No decarboxylation activity of LigW2 toward 5CVA analogs like 3-methoxysalicylate, 4-hydroxyisophthalate, and isophthalate was detected. The substrate specificity of LigW2 appeared to be restricted to 5CVA.
Localization of ligW2 on the 95-kb AseI fragment.
In our previous study (41), we demonstrated that the gene clusters for DDVA and β-aryl ether degradation lie in the 340-kb AseI fragment, and the PCA 4,5-cleavage pathway gene cluster and ligH, which is thought to encode the 10-formyltetrahydrofolate synthetase involved in one-carbon metabolism, are located in the 95-kb AseI fragment. Recently, we found that the vanillate/3-O-methylgallate O-demethylase gene (ligM) and the 5,10-methylenetetrahydrofolate reductase gene (metF) are located upstream of ligH (1). Southern hybridization analysis using the 3.6-kb SalI fragment carrying ligW2 as a probe indicated that ligW2 was located on the 95-kb AseI fragment (data not shown). To determine the localization of ligW2 and other genes in the 95-kb AseI fragment, Southern hybridization analysis of pDEC6 with the pHN139F and pDE20 probes carrying the PCA 4,5-cleavage pathway genes and part of ligM, metF, and ligH, respectively, was carried out. The pDE20 probe hybridized to the 1.0-, 2.2-, 2.3-, and 3.6-kb SalI fragments of pDEC6, thus demonstrating the overlap between pDEC6 and pDE20. Comparison of the restriction map of pDEC6 and that of pDE20 revealed that ligW2 was located approximately 6-kb downstream of ligH (Fig. 2). Further analysis is necessary in order to determine the distance between ligW2 and the PCA 4,5-cleavage pathway genes.
Implications for the DDVA catabolic pathway.
In this study, we found that two 5CVA decarboxylase genes are involved in 5CVA degradation, with ligW2 playing the more important role in the growth of SYK-6 cells on DDVA. It should be noted that in the course of this study, the question was raised regarding whether SYK-6 is able to utilize 4-carboxy-2-hydroxypent-2,4-dienoate (CHPD), which appeared to be generated from the meta-cleavage compound of OH-DDVA resulting from the reaction catalyzed by LigY (Fig. 1). Our preliminary experiment indicated that the ligI (35), ligJ (20), and ligK (21) mutants grew well on DDVA (data not shown). Because vanillate generated from 5CVA is degraded through the PCA 4,5-cleavage pathway, a pathway already known to be essential for vanillate degradation, the present findings suggested that CHPD supports the growth of SYK-6. On the other hand, the ligB and ligC mutants (33) exhibited poor growth on DDVA (data not shown). Recently, we found that generation of 5-methyltetrahydrofolate in the vanillate O demethylation catalyzed by the tetrahydrofolate-dependent O-demethylase, LigM, is essential for methionine biosynthesis in this strain. Therefore, SYK-6 is able to grow on PCA only when methionine is added to the medium (1, 34). Disruption of ligB and ligC seemed to cause the deficiency in ligM induction; however, DDW could not grow on DDVA in the presence of 50 mg/liter of methionine (data not shown). These findings may suggest that a metabolite of 5CVA, probably 2-pyrone-4,6-dicarboxylate, is required for induction of CHPD catabolic gene expression. The lack of induction of CHPD catabolic gene expression might explain why DDW lacked the ability to grow on DDVA. However, further study, including isolation of the CHPD catabolic genes and regulation of the all of the genes involved in DDVA catabolism, is necessary to critically assess this possibility.
Acknowledgments
This work was supported in part by Grant-in-Aid for the Encouragement of Young Scientists 09760077 from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Abe, T., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillin and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Barthelmebs, L., C. Divies, and J. F. Cavin. 2001. Expression in Escherichia coli of native and chimeric phenolic acid decarboxylases with modified enzymatic activities and method for screening recombinant E. coli strains expressing these enzymes. Appl. Environ. Microbiol. 67:1063-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthelmebs, L., B. Lecomte, C. Divies, and J. F. Cavin. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vector. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Buswell, J. A., K.-E. Eriksson, and B. Pettersson. 1981. Purification and partial characterization of vanillate hydroxylase (decarboxylase) from Sporotrichum pulverulentum. J. Chromatogr. 215:99-108. [Google Scholar]
- 8.Cavin, J. F., L. Barthelmebs, and C. Divies. 1997. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl. Environ. Microbiol. 63:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, H. K., and G. J. Zylstra. 1998. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J. Bacteriol. 180:6529-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, K. T., M. K. Pope, and J. Davies. 1999. Characterization of a vanillic acid non-oxidative decarboxylation gene cluster from Streptomyces sp. D7. Microbiology 145:2393-2403. [DOI] [PubMed] [Google Scholar]
- 11.Collier, L. S., N. N. Nichols, and E. L. Neidle. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179:5943-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowles, C. E., N. N. Nichols, and C. S. Harwood. 2000. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 182:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Argenio, D. A., A. Segura, W. M. Coco, P. V. Bunz, and L. N. Ornston. 1999. The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J. Bacteriol. 181:3505-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda, M., and K. Yano. 1985. Construction of broad host range cloning vectors for Gram-negative bacteria. Agric. Biol. Chem. 49:2719-2724. [Google Scholar]
- 18.Gasson, M. J., Y. Kitamura, W. R. McLauchlan, A. Narbad, A. J. Parr, E. L. Parsons, J. Payne, M. J. Rhodes, and N. J. Walton. 1998. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J. Biol. Chem. 273:4163-4170. [DOI] [PubMed] [Google Scholar]
- 19.Haddock, J. D., and J. G. Ferry. 1993. Initial steps in the anaerobic degradation of 3,4,5-trihydroxybenzoate by Eubacterium oxidoreducens: characterization of mutants and role of 1,2,3,5-tetrahydroxybenzene. J. Bacteriol. 175:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara, H., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2003. Characterization of the 4-carboxy-4-hydroxy-2-oxoadipate aldolase gene and operon structure of the protocatechuate 4,5-cleavage pathway genes in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 185:41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, Z., and J. Wiegel. 1995. Purification and characterization of an oxygen-sensitive reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. Eur. J. Biochem. 229:77-82. [DOI] [PubMed] [Google Scholar]
- 23.He, Z., and J. Wiegel. 1996. Purification and characterization of an oxygen-sensitive, reversible 3,4-dihydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J. Bacteriol. 178:3539-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu, T. D., M. F. Lux, and H. L. Drake. 1990. Expression of an aromatic-dependent decarboxylase which provides growth-essential CO2 equivalents for the acetogenic (Wood) pathway of Clostridium thermoaceticum. J. Bacteriol. 172:5901-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, J., Z. He, and J. Wiegel. 1999. Cloning, characterization, and expression of a novel gene encoding a reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J. Bacteriol. 181:5119-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Z., L. Dostal, and J. P. Rosazza. 1993. Mechanisms of ferulic acid conversions to vanillic acid and guaiacol by Rhodotorula rubra. J. Biol. Chem. 268:23954-23958. [PubMed] [Google Scholar]
- 27.Ishii, Y., Y. Narimatsu, Y. Iwasaki, N. Arai, K. Kino, and K. Kirimura. 2004. Reversible and nonoxidative γ-resorcylic acid decarboxylase: characterization and gene cloning of a novel enzyme catalyzing carboxylation of resorcinol, 1,3-dihydroxybenzene, from Rhizobium radiobacter. Biochem. Biophys. Res. Commun. 324:611-620. [DOI] [PubMed] [Google Scholar]
- 28.Katayama, Y., S. Nishikawa, A. Murayama, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1988. The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 233:129-133. [DOI] [PubMed] [Google Scholar]
- 29.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 30.Lee, J.-H., T. Omori, and T. Kodama. 1994. Identification of the metabolic intermediates of phthalate by Tn5 mutants of Pseudomonas testosteroni and analysis of the 4,5-dihydroxyphthalate decarboxylase gene. J. Ferment. Bioeng. 77:591-597. [Google Scholar]
- 31.Li, T., P. Juteau, R. Beaudet, F. Lepine, R. Villemur, and J. G. Bisaillon. 2000. Purification and characterization of a 4-hydroxybenzoate decarboxylase from an anaerobic coculture. Can. J. Microbiol. 46:856-859. [PubMed] [Google Scholar]
- 32.Maruyama, K., M. Miwa, N. Tsujii, T. Nagai, N. Tomita, T. Harada, H. Sobajima, and H. Sugisaki. 2001. Cloning, sequencing, and expression of the gene encoding 4-hydroxy-4-methyl-2-oxoglutarate aldolase from Pseudomonas ochraceae NGJ1. Biosci. Biotechnol. Biochem. 65:2701-2709. [DOI] [PubMed] [Google Scholar]
- 33.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masai, E., M. Sasaki, Y. Minakawa, T. Abe, T. Sonoki, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J. Bacteriol. 186:2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4, 6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols, N. N., and C. S. Harwood. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishikawa, S., T. Sonoki, T. Kasahara, T. Obi, S. Kubota, S. Kawai, N. Morohoshi, and Y. Katayama. 1998. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl. Environ. Microbiol. 64:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura, Y., M. Nakagawa, N. Ogawa, S. Harashima, and Y. Oshima. 1992. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J. Ferment. Bioeng. 74:333-344. [Google Scholar]
- 39.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, X., E. Masai, Y. Katayama, and M. Fukuda. 1999. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 65:2789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng, X., E. Masai, H. Kitayama, K. Harada, Y. Katayama, and M. Fukuda. 2002. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priefert, H., J. Rabenhorst, and A. Steinbüchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prim, N., F. I. Pastor, and P. Diaz. 2003. Biochemical studies on cloned Bacillus sp. BP-7 phenolic acid decarboxylase PadA. Appl. Microbiol. Biotechnol. 63:51-56. [DOI] [PubMed] [Google Scholar]
- 44.Providenti, M. A., J. Mampel, S. MacSween, A. M. Cook, and R. C. Wyndham. 2001. Comamonas testosteroni BR6020 possesses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology 147:2157-2167. [DOI] [PubMed] [Google Scholar]
- 45.Pujar, B. G., and D. W. Ribbons. 1985. Phthalate metabolism in Pseudomonas fluorescens PHK: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl. Environ. Microbiol. 49:374-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahouti, M., F. Seigle-Murandi, R. Steiman, and K.-E. Eriksson. 1989. Metabolism of ferulic acid by Paecilomyces variotii and Pestalotia palmarum. Appl. Environ. Microbiol. 55:2391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rani, M., D. Prakash, R. C. Sobti, and R. K. Jain. 1996. Plasmid-mediated degradation of o-phthalate and salicylate by a Moraxella sp. Biochem. Biophys. Res. Commun. 220:377-381. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 50.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vivo constructed Tn-5 mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 52.Sonoki, T., T. Obi, S. Kubota, M. Higashi, E. Masai, and Y. Katayama. 2000. Coexistence of two different O demethylation systems in lignin metabolism by Sphingomonas paucimobilis SYK-6: cloning and sequencing of the lignin biphenyl-specific O-demethylase (LigX) gene. Appl. Environ. Microbiol. 66:2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venturi, V., F. Zennaro, G. Degrassi, B. C. Okeke, and C. V. Bruschi. 1998. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 144:965-973. [DOI] [PubMed] [Google Scholar]
- 55.Wattiau, P., L. Bastiaens, R. van Herwijnen, L. Daal, J. R. Parsons, M. E. Renard, D. Springael, and G. R. Cornelis. 2001. Fluorene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152:861-872. [DOI] [PubMed] [Google Scholar]
- 56.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 57.Zago, A., G. Degrassi, and C. V. Bruschi. 1995. Cloning, sequencing, and expression in Escherichia coli of the Bacillus pumilus gene for ferulic acid decarboxylase. Appl. Environ. Microbiol. 61:4484-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeida, M., M. Wieser, T. Yoshida, T. Sugio, and T. Nagasawa. 1998. Purification and characterization of gallic acid decarboxylase from Pantoea agglomerans T71. Appl. Environ. Microbiol. 64:4743-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, H., and G. T. Javor. 2000. Identification of the ubiD gene on the Escherichia coli chromosome. J. Bacteriol. 182:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]